ABSTRACT
Advances in laboratory and information technologies are transforming public health microbiology. High-throughput genome sequencing and bioinformatics are enhancing our ability to investigate and control outbreaks, detect emerging infectious diseases, develop vaccines, and combat antimicrobial resistance, all with increased accuracy, timeliness, and efficiency. The Advanced Molecular Detection (AMD) initiative has allowed the Centers for Disease Control and Prevention (CDC) to provide leadership and coordination in integrating new technologies into routine practice throughout the U.S. public health laboratory system. Collaboration and partnerships are the key to navigating this transition and to leveraging the next generation of methods and tools most effectively for public health.
KEYWORDS: microbiology, communicable diseases, public health, laboratory science, molecular, sequencing, bioinformatics, molecular epidemiology
INTRODUCTION
Rapid advances in both laboratory and information technologies are transforming public health microbiology. Ten years ago, shotgun sequencing a single bacterial genome containing 3 to 5 million nucleotides was a process that took several months and thousands of dollars to complete; today, next-generation sequencing (NGS) instruments can produce dozens of bacterial genomes per day at a fraction of the cost (1). By using increasingly sophisticated bioinformatics tools and curated databases of microbial sequences, public health microbiologists can examine and compare the entire genomes of pathogens and microbial populations within days or even hours of specimen receipt. Increasingly affordable, high-throughput laboratory methods and bioinformatics are being rapidly translated from research techniques into practical clinical and public health applications.
State-of-the-art molecular technologies promise to revolutionize our ability to identify pathogens and diagnose infectious diseases, investigate and control outbreaks, understand transmission patterns and dynamics, predict antimicrobial susceptibility and virulence characteristics, and develop and target vaccines, all with increased accuracy, timeliness, and efficiency (2–4). Integrating advanced molecular methods into routine public health practice faces a major practical challenge: the need to make changes in large interconnected systems without interrupting system functions. Laboratories will have to retool longstanding work practices and revise the flow of biological samples and information. New information technology infrastructure will be needed to handle the scale and complexity of genomic data, including enhanced systems for data management, analysis, sharing, and reporting.
NEW TECHNOLOGIES: OPPORTUNITIES AND CHALLENGES
Public health laboratory-based surveillance provides the essential data for monitoring trends, detecting outbreaks, and initiating the public health response to control many infectious diseases. Most current surveillance systems rely on clinical laboratories to culture, isolate, and identify pathogens from ill patients; they then report the results or send the isolates to state health department laboratories for further characterization using specialized assays, e.g., for strain typing, virulence, antimicrobial resistance, or antigenic determinants. In addition to supporting surveillance and outbreak investigations, state laboratories provide critical reference functions to support the diagnosis and treatment of unusual or reportable pathogens.
Until recently, public health laboratories identified pathogens by using growth, biochemical, phenotypic, or molecular assays that entailed hours to days of work, even for skilled laboratorians. Additional phenotypic or molecular assays for further subtyping and characterization were often subjective, required specialized expertise, and could take days, weeks, or even months to obtain and confirm critical results. Today, NGS and other high-throughput laboratory methods are replacing many traditional microbiological techniques for identifying, typing, and characterizing pathogens. Because they are often faster, more comprehensive, and more accurate, these methods can help public health authorities recognize and stop outbreaks earlier, preventing illness and saving lives (5). Whole-genome sequencing (WGS) has higher resolving power than older molecular methods for distinguishing clusters of related cases and can greatly improve our ability to match clinical and environmental isolates during epidemiologic investigations. WGS also offers crucial insights into rapidly evolving pathogen characteristics, detecting the emergence of antimicrobial resistance or highly virulent strains. Another important advantage is in the generalizability of sequencing and bioinformatics workflows: laboratories using NGS can often consolidate workflows for multiple pathogens, or pathogens that are emerging or difficult to identify and characterize, improving overall efficiency in terms of throughput and cost.
New technology is also transforming clinical laboratories, where next-generation diagnostic tests based on the detection of nucleic acid sequences or other molecular markers are rapidly replacing traditional culture-based methods for many routine diagnostic assays. These culture-independent diagnostic tests (CIDTs) are based on molecular features of pathogens and can deliver fast and accurate diagnoses, either in clinical laboratories or potentially at the point of care, bypassing the public health laboratory entirely. The U.S. Food and Drug Administration (FDA) has already approved many syndromic CIDT panels for enteric, respiratory, and invasive pathogens (http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/ucm330711.htm). In theory, CIDTs could become a source of more complete data on infectious disease incidence than are presently available because they are generally more sensitive and specific than culture-based diagnostic tests; furthermore, they are faster, more comprehensive, and more convenient for clinicians, who might now choose to test more patients than before (6).
Together, NGS and CIDT clearly offer unprecedented opportunities to improve infectious disease diagnosis and expedite pathogen identification and characterization. On the other hand, both present major challenges to public health laboratories and programs. Incorporating NGS into routine public health activities will require fundamental changes in laboratory practice at multiple levels. For example, as consolidated workflows reduce the need for many labor-intensive laboratory processes, laboratories will need to reorganize, retrain, and recruit a workforce with new expertise. As molecular methods produce vast and rapidly growing amounts of complex data, sophisticated new computing infrastructure and bioinformatics capacity will be needed to manipulate and analyze them. Because even a single bacterial genome may represent several gigabytes of raw sequence data, “big data” approaches are needed for storing, sharing, and analyzing WGS data and integrating them with other laboratory and epidemiologic data. Systems for quality assurance, data standards, and data security are also essential elements of the infrastructure needed to communicate molecular data within the public health system reliably and efficiently (7).
The growing use of CIDTs also presents immediate and important challenges to many well-established public health surveillance activities. The number and types of settings where such tests can be performed are growing to include doctors' offices, pharmacies, and even people's homes. Decentralized testing will complicate the systematic data collection needed to maintain a consistent baseline for surveillance. As CIDT technologies continue to evolve, the changing array of available tests is likely to affect case definitions and trends in ways that are not entirely predictable (6). More importantly, widespread use of CIDTs requires clinical laboratories to perform fewer primary cultures for many organisms, reducing the supply of isolates that can be sent to state and local public health laboratories for detailed characterization and reporting. Because CIDTs are designed to provide results for clinical decision-making, they do not produce all the data, such as strain type, virulence profile, and antimicrobial susceptibility, that are needed for public health surveillance, outbreak investigation, and treatment guidelines. Furthermore, because CIDT panels are highly targeted to specific molecular features, they may produce false-negative results for pathogens that have evolved sufficiently to evade detection. Laboratory methods, such as direct metagenomic sequencing of clinical specimens, may offer a future strategy for obtaining some of these missing data; however, many technical and logistical obstacles must be overcome before these techniques are practical and economical for routine testing (8).
Longstanding cooperation between clinical and public health laboratories forms the basis for infectious disease surveillance and control, and this cooperation is now more important than ever. During the transition from traditional microbiology to molecular detection, all parties can benefit from active engagement and constructive dialog (9). For example, the American Society for Microbiology, the Association of Public Health Laboratories, and the CDC have all worked together to produce interim guidelines to help clinical laboratories performing CIDTs continue their participation in public health surveillance for enteric pathogens (10). These guidelines will be revised as additional data and experience become available.
The CDC's Emerging Infections Program (EIP [http://www.cdc.gov/ncezid/dpei/eip/]) offers another means to engage a wide array of partners in evaluating the public health impact of new technologies and practices. The EIP is a network of state health department epidemiologists and laboratorians and their collaborators in local public health departments, academic institutions, other federal agencies, public health and clinical laboratories, and health care settings. The EIP can help assess the potential value of new diagnostic tests for surveillance of infections with emerging pathogens or for those for which reliable detection methods were previously unavailable. The EIP is well positioned to evaluate how changes in factors, such as test performance and case definitions, impact surveillance and population-based estimates of disease burden (6). For example, EIPs in many states conduct regular systematic surveys of clinical, commercial, and public health laboratories to evaluate the use of CIDTs and changing diagnostic practices at the population level.
CDC'S ADVANCED MOLECULAR DETECTION INITIATIVE
The Centers for Disease Control and Prevention (CDC) is responding to these new opportunities and challenges by leading the transformation of public health systems in a way that will maximize the timely availability, utility, and quality of the information needed for action at every level. Beginning in fiscal year 2014, Congress appropriated funds for the Advanced Molecular Detection and Response to Infectious Disease Outbreaks (AMD) initiative to allow the CDC to coordinate the integration of NGS and advanced bioinformatics with traditional epidemiologic methods to enhance infectious disease prevention and control (http://www.cdc.gov/amd/pdf/amd-overview-2016-508.pdf). The overarching goals for the AMD initiative include improving pathogen identification and detection; developing new diagnostics to meet evolving public health needs; supporting states to coordinate current and future reference testing needs; implementing enhanced, sustainable, and integrated laboratory information systems; and developing tools for the prediction, modeling, and early recognition of emerging infectious diseases. A range of activities is under way in CDC laboratories and programs and in state and local health departments to incorporate NGS and other new laboratory technologies into routine public health practice. Recognizing that no single approach will serve every purpose, the overall strategy in AMD has been to balance innovation and flexibility with the need for standardization and sustainability.
The AMD initiative supports cross-cutting efforts that focus on (i) building shared computing and laboratory capacity for genomics, bioinformatics, and next-generation diagnostic testing, including standardization of workflows, data systems, and quality management processes; (ii) developing training in bioinformatics and other essential skills, including fellowships and career paths for public health laboratory staff; and (iii) supporting continued innovation and coordinated application of genomics and other emerging technologies to meet public health challenges.
NGS technology is already transforming the workflow and output of many CDC laboratories and is expected to expand rapidly in state and local public health laboratories. Now in its fourth year, the AMD initiative is accelerating the rollout of NGS technologies to state public health laboratories. Much of this effort is being led by individual CDC programs, such as PulseNet and the tuberculosis and influenza programs, with the support of AMD initiative funding. The rationale for this approach is that it is more likely to deliver the technology at a time when it meets an existing demand, along with the training needed to get started and program resources to sustain it. These program-led efforts are strategically coordinated to identify common resource needs, opportunities for shared information technology and laboratory infrastructure, protocol and data analysis methods, and common requirements for training and workforce development. The AMD initiative has enabled the CDC to build vital core capacity in NGS, bioinformatics, and scientific computing and has provided the public health workforce at the CDC and in state, county, and local laboratories with training and support to help navigate this fundamental technological shift in public health microbiology. Health departments in all 50 states, the District of Columbia, and Puerto Rico are already sending samples to the CDC for one or more AMD projects; of these, more than 20 are also performing next-generation sequencing on site.
The AMD program promotes coordination and standardization of next-generation sequencing protocols and bioinformatic methods across public health programs. For example, multiple CDC programs that are implementing bacterial whole-genome sequencing have agreed to use standardized procedures for extracting and preparing DNA and to establish consistent quality standards and metrics for library preparation and quality control in public health laboratories. In addition, AMD funding has been extended to state partners to support the development of training networks and courses for next-generation sequencing, bioinformatics, and other skills that will advance the adoption of AMD technologies nationwide. State and local public health departments are also being encouraged to form local or regional training networks, with one laboratory taking the lead. These networks will be able to partner with local universities to build local capacity, foster long-term collaboration, and promote innovation.
In the following sections, we present some examples of how we are implementing AMD to improve the detection and control of infectious diseases. In addition to highlighting some exciting progress, we use these examples to point out a few of the challenges encountered so far.
DETECTING, INVESTIGATING, AND CONTROLLING FOODBORNE OUTBREAKS
Approximately 48 million foodborne illnesses occur in the United States each year, resulting in about 128,000 hospital admissions and 3,000 deaths. Most cases are not associated with recognized outbreaks, even though state and local health departments investigate more than 1,000 outbreaks annually (http://www.cdc.gov/foodsafety/foodborne-germs.html). The National Molecular Subtyping Network for Foodborne Disease Surveillance, also known as PulseNet, was established by the CDC and the Association of Public Health Laboratories in 1996 (11). PulseNet introduced the use of then-new technology, pulsed-field gel electrophoresis (PFGE), a type of “DNA fingerprinting,” as a standardized approach to subtype and categorize foodborne bacterial pathogens. PulseNet USA currently includes 83 participating laboratories, including state and local health departments, federal agencies, and international partners. Each laboratory follows a set of standardized laboratory and analysis protocols to generate PFGE patterns from foodborne bacterial isolates and upload them to electronic databases at the CDC; there, they can be compared to patterns from other bacteria that have been isolated from humans, animals, and foods throughout the country. By looking for pattern matches, investigators can identify clusters of related illnesses and potential sources, even those resulting from a widely distributed food or other vehicle. Once a potential source has been identified, public health authorities work with regulatory agencies, such as the FDA or the U.S. Department of Agriculture (USDA), to implement control measures that can limit the number of additional cases and improve outcomes among affected persons. Furthermore, by identifying unsafe food production, processing, and preparation practices and vehicles of contamination that would otherwise not be recognized, foodborne disease outbreak investigations give the food industry and regulatory authorities the information they need to prevent future outbreaks and improve the safety of the food supply. An economic analysis recently published by authors from academia, state public health, and the CDC estimated that PulseNet activities prevent at least 260,000 cases of foodborne disease each year in the United States, saving the U.S. economy one-half billion dollars (12).
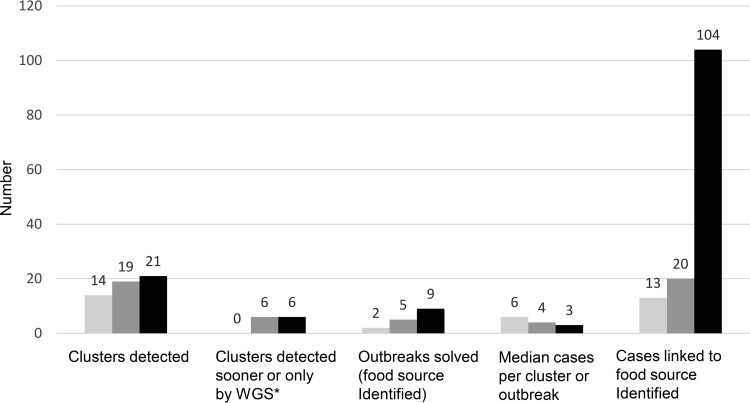
In 2013, the CDC helped to establish the nationwide Listeria Whole Genome Sequencing Project, a collaboration with the FDA, USDA, the National Institutes of Health (NIH) National Center for Biotechnology Information (NCBI), and state and local health departments. The goal was to evaluate the feasibility and usefulness of real-time WGS to enhance surveillance and outbreak investigation for listeriosis, one of the most deadly foodborne infections (http://www.cdc.gov/listeria/). The project built on earlier efforts by PulseNet and the FDA GenomeTrakr network to sequence strains from highly characterized outbreaks, foods, and production environments (13), along with the Listeria Initiative, an enhanced epidemiology approach for surveillance of listeriosis pioneered by the Institut de Veille Sanitaire en France (14). The Listeria Whole Genome Sequencing Project aimed to conduct WGS in parallel with PFGE on every clinical isolate of Listeria monocytogenes in the United States, along with related food and environmental isolates from USDA and the FDA GenomeTrakr program, and to deposit them in near-real time into a shared sequence data repository at NCBI. The goal was to detect outbreaks earlier, to distinguish clusters of related cases more precisely, and to link illnesses to a potential contaminated food source more quickly. During the first 2 years of the project, the number of outbreaks detected increased, as did the number solved, while the median cluster size decreased (Fig. 1) (5). These results indicate that more clusters were being identified and that WGS was effective in attributing these infections to their sources; most importantly, they suggest that outbreak clusters were being caught much earlier, preventing many severe illnesses and deaths. During the fall of 2014, for example, WGS was used to help investigate an outbreak of listeriosis that resulted in 34 hospitalizations and 7 deaths across 12 states, ultimately tracing two distinct Listeria strains to caramel apples made from fruit from a single supplier. Caramel apples and other produce had not historically been associated with Listeria infections, and using conventional laboratory and epidemiologic methods, the source of this outbreak might not have been conclusively identified.
FIG 1.
Listeriosis clusters detected and outbreaks solved before and after implementation of real-time whole-genome sequencing (WGS) of Listeria monocytogenes isolates from patients, food, and the environment, United States, September 2012 to August 2015. *, Cluster detection sooner or only by WGS, compared with pulsed-field gel electrophoresis. Based on figure published in reference 5).
Experience with the Listeria Whole Genome Sequencing Project has laid the groundwork for the next generation of PulseNet as a generalizable platform for genome-scale epidemiologic surveillance of foodborne illness. The CDC is currently helping to build WGS capacities in state health departments and develop whole-genome multilocus sequence typing (wgMLST) databases for standardized WGS analysis of Campylobacter, Vibrio, Shigella spp., Salmonella, Shiga toxin-producing Escherichia coli, and other E. coli pathotypes. Replacing PFGE with WGS promises to provide not only higher-resolution data for comparison in epidemiologic investigations but additional information on pathogen characteristics, like serotype, virulence, and antimicrobial resistance, without further testing. Although the per-isolate reagent cost for WGS is approximately 10- to 20-fold higher than for PFGE, it continues to fall; furthermore, even greater savings can be achieved through laboratory automation and workflow consolidation, particularly if WGS replaces reference methods for identifying and characterizing enteric bacteria. Making the transition to these methods presents significant organizational challenges, however, including the need to build appropriate informatics systems, software, and network infrastructure for managing, analyzing, and transmitting data and harmonizing them with laboratory and program operations. While providing a richer substrate for analysis, WGS data also raise scientific and policy questions. For example, is it possible to develop standard criteria for defining a cluster with WGS data? How should WGS data be integrated and analyzed with epidemiologic data? What information can be shared with other public health agencies, researchers, and the public?
DEVELOPING SEASONAL INFLUENZA VIRUS VACCINE
The CDC's Influenza Division serves as the U.S. National Influenza Center and as a WHO Collaborating Center for Surveillance, Epidemiology and Control of Influenza (http://www.cdc.gov/flu/weekly/who-collaboration.htm), where laboratory scientists and epidemiologists monitor the evolution of influenza viruses year-round as they spread through human and other animal populations. The division's laboratory analyzes 8,000 to 12,000 influenza virus samples annually to support surveillance and the selection of candidate vaccine strains. Using current technologies, seasonal influenza virus vaccine is produced in a “just in time” fashion every year, with 150 million doses per year needed in the United States alone. Because the influenza virus evolves very rapidly through antigenic drift and reassortment, it is critical to identify prevalent variants quickly and to monitor for emerging strains and changes in the cocirculating viral population dynamics. Preparedness for pandemic influenza virus also depends on the rapid identification of emerging antigenic variants while they are still rare, which sometimes signals the arrival of immune escape variants that have increased fitness in the population.
Current laboratory surveillance procedures entail unavoidable delays, requiring approximately 1 month from specimen collection to complete antigenic analysis for most domestic isolates and up to 2 months or more for those that are collected by international partners. The CDC's Influenza Division and other public health global partners are implementing a sequencing first approach for genetic surveillance of cocirculating virus variants. This fundamental change in the laboratory surveillance model for seasonal influenza produces significantly more data for candidate vaccine selection and public health decision-making. By using advanced bioinformatics to infer antigenicity, they can help expedite and improve candidate vaccine strain selection. Changing the sequence of laboratory procedures, that is, performing genetic analysis first and using the results to select a subset of samples for isolation, propagation, and phenotyping, changes the surveillance paradigm, allowing the laboratory to acquire more higher-quality data in less time and at lower cost.
This approach was particularly helpful in 2014, when new H3N2 influenza virus subtypes were found to have drifted; notable changes in the hemagglutinin structures of these viruses made them difficult to characterize antigenically. The CDC's Influenza Division performed deep sequencing of these viruses using a new NGS pipeline that employs multisegment reverse transcription-PCR and an iterative refinement meta-assembler (IRMA) (15). Analysis of the sequence data confirmed the expansion of cocirculating variant H3N2 viruses in clades 3C.2a and 3C.3a; the data also allowed inference of antigenicity, improving the knowledge base for upcoming vaccine recommendations for the Southern Hemisphere. At the Southern Hemisphere Vaccine Consultation Meeting in September 2014, the recommended H3N2 vaccine component was changed to include one of the major variant viruses (http://www.who.int/influenza/vaccines/virus/recommendations/2015_south/en/).
GUIDING HIV PREVENTION
The human immunodeficiency virus (HIV) epidemic emerged 35 years ago and remains a major public health problem. Currently, more than 1.2 million people in the United States are living with HIV infection, and about 40,000 people become newly infected each year (http://www.cdc.gov/hiv/statistics/overview/ataglance.html). In 2015, an outbreak of HIV infections occurred in a rural county of southeastern Indiana, where historically fewer than 5 cases of HIV infection had been reported per year (16). Most of the people newly infected with HIV resided in the same community and were linked by sharing syringes and paraphernalia used to inject the prescription narcotic oxymorphone. The CDC integrated HIV genomic sequences with epidemiologic data in phylodynamic analyses to infer transmission dynamics and characterize the outbreak in near real-time (Fig. 2). Network analysis showed that both sexual and injection drug use (IDU) contact were correlated with HIV infection; however, many more potential transmission events corresponded with a reported IDU contact (82%) than with reported sexual contacts (11%) (17). The public health response led by the Indiana State Health Department included a public education campaign and community outreach, short-term authorization of syringe exchange, and support for comprehensive medical care and substance abuse counseling and treatment. The potential for high-resolution genome sequence data to improve outbreak response now and in the future depends on its timely integration with epidemiologic data to help focus response efforts where they will have the greatest impact, for example, by recognizing unreported contacts and foci of ongoing transmission.
FIG 2.
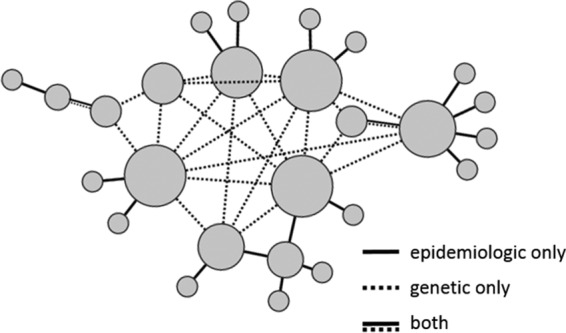
Schematic of inferred relationships among cases of HIV infection based on epidemiologic information, genetic information, or both. Each circle represents one case, with circle size proportional to the number of epidemiologic or genetic connections.
DETECTING EMERGING INFECTIONS
The causative agent of Middle East respiratory syndrome (MERS), first reported from the Kingdom of Saudi Arabia in 2012, was found to be a previously unknown coronavirus (18), now called Middle East respiratory syndrome coronavirus (MERS-CoV). Following the first case reports, CDC laboratory scientists used NGS data to develop a specific real-time PCR assay for MERS-CoV, which the FDA authorized for emergency use in June 2013. The CDC subsequently developed and validated additional diagnostic tests for MERS-CoV and distributed them to public health laboratories throughout the United States and in 48 other countries to date (19). By developing methods that can use small-volume samples without the need for cultured virus, the CDC has also been able to generate complete and accurate MERS-CoV genome sequences both rapidly and cost-effectively. Sequence data have been deposited in GenBank (http://www.ncbi.nlm.nih.gov/GenBank/) for 54 complete or nearly complete genomes obtained from human cases or from camels, the suspected primary reservoir of MERS-CoV. Full-genome sequence data are useful for epidemiologic investigations to describe chains of transmission and serve as an important resource for identifying mutations with the potential to increase the virulence or transmissibility of MERS-CoV.
Genomic and metagenomics sequencing has been vital in the identification and characterization of other novel pathogens, such as the tick-borne Heartland virus, a Bunyavirus that was first identified in two Missouri farmers in 2012 (20). In 2014, a Kansas man presented with a presumed tick-borne illness that was negative for molecular and serologic test results for known tick-borne pathogens, including Heartland virus. CDC scientists used NGS to identify a previously unknown Thogotovirus, the first of its kind known to have caused human disease in North America (21). It was named Bourbon virus after the county where it was collected. MERS-CoV and Heartland and Bourbon viruses are RNA viruses, the most commonly implicated class of emerging human pathogens. Improved surveillance, including the use of NGS and broad-based molecular approaches, are likely to identify more such viruses (22).
COMBATTING ANTIMICROBIAL RESISTANCE
Widespread use of antimicrobial drugs has led multiresistant strains of bacteria to emerge and spread, threatening to undermine control of infectious diseases. In the United States alone, antimicrobial-resistant (AR) bacteria cause at least 2 million infections and 23,000 deaths (23). A comprehensive federal plan has been developed to combat this threat (24), accompanied by an action plan released in March 2015 (25). The goals of this plan are to slow the emergence and spread of AR bacteria, improve surveillance, develop advanced diagnostic tests, accelerate research on antibiotics and vaccines, and improve international capacity and collaboration. NGS and other advanced molecular technologies are poised to play an essential role in supporting these efforts by detecting and characterizing resistance faster and more accurately, and shedding light on mechanisms and transmission patterns.
Investigation and control of health care-associated infections are crucial to stemming the spread of antimicrobial resistance. By clarifying transmission links with more confidence and precision, WGS is serving as a force multiplier for epidemiologic investigation of health care-associated outbreaks. For example, in 2012, multiple cases of infection with New Delhi metallo-beta-lactamase-1 (NDM-1) Klebsiella pneumoniae strains occurred in a Colorado health care facility (26). Epidemiologic investigation found many potential links among cases, which also appeared to be highly related (>90%) by PFGE analysis. WGS made it possible to compare the genomes of every isolate from this outbreak with databases of antimicrobial resistance genes and marker sequences, simultaneously screening for more than 1,000 different acquired resistance genes. Ultimately, the cases could be sorted into three transmission clusters in separate parts of the facility.
ADDRESSING THE CHALLENGES OF NEW TECHNOLOGIES
Partnerships and collaboration are critically important in the CDC's efforts to integrate advanced molecular and bioinformatics technologies into public health practice. Our partners include other federal agencies, state and local public health departments, major professional organizations, academic centers, and industry groups. These partnerships lend specialized expertise to public health programs and expand the range of work that is possible, for example, in bioinformatics, which is a relatively new discipline for public health. The CDC has engaged with several academic institutions and expert groups to develop and deliver bioinformatics training to the CDC laboratory workforce through seminars, coursework, and hands-on technical workshops. In partnership with the Association of Public Health Laboratories (APHL), the CDC has also introduced a new postdoctoral and postmasters fellowship program that gives bioinformaticians a chance to apply their skills to challenging public health problems and to explore potential careers in public health (https://www.aphl.org/fellowships/Pages/Bioinformatics.aspx). To date, 17 bioinformatics fellows have been placed in CDC programs and, beginning in 2016, in several state public health laboratories. Collaborations that pair state health departments with local academic experts (such as the Integrated Food Safety Centers of Excellence) are also developing training in AMD concepts and methods for public health epidemiologists.
In addition to leveraging partnerships through existing programs and cooperative agreements, the CDC's AMD program has used special funding mechanisms to foster innovation. For example, the “No Petri Dish” test challenge grant focused on developing methods to rapidly identify and characterize pathogens directly from complex clinical samples (http://www.cdc.gov/amd/achievements/index.html). A CDC broad agency announcement (http://go.usa.gov/x94fN) was used to seek proposals on integration of epidemiologic and genomics data; as a result, the CDC is now working with the developers of Microreact, a tool for genomic epidemiology, to develop new applications for public health data integration and visualization (27).
The CDC's partnerships with other federal agencies are crucial in ensuring that federal resources are used efficiently to support public health efforts at state and local levels. Major efforts currently focus on harmonizing methods for laboratory proficiency testing and bioinformatic analysis of clinical and environmental samples for surveillance and investigation of foodborne disease. Other priorities include sharing resources and capabilities and promoting interagency collaboration among communications groups to ensure that public information uses consistent terminology and is cross-cleared. The CDC participates in work led by other federal agencies, such as the effort to establish standards for pathogen detection via next-generation sequencing led by the National Institute of Standards and Technology and the FDA (28). Partnerships with the NIH on scientific and technical issues help the CDC transfer cutting-edge knowledge and expertise from the research domain to the front lines of public health. The CDC's most important collaboration in this area has been with NCBI, which has developed new data structures, a repository of pathogen sequence information, and useful tools for the public health community to use for analyzing pathogen sequences (https://www.ncbi.nlm.nih.gov/pathogens). In addition, the CDC has consulted with the National Cancer Institute (NCI) on the development and optimization of computational methods and with the National Human Genome Research Institute (NHGRI) on the analysis of antimicrobial resistance. Several CDC projects have also engaged the genome sequencing centers and bioinformatics resource centers funded by the National Institute of Allergy and Infectious Diseases (NIAID).
Rather than a one-time overhaul, the integration of advanced molecular methods and bioinformatics into public health practice will require ongoing adaptation to fast-paced technological innovation and significant changes to public health informatics and workforce capacity. This adaptation must be further propagated and coordinated among diverse public health systems in state and local health departments, federal agencies, and public health organizations around the world. Laboratory and bioinformatics methods for WGS, proteomics, and other high-throughput technologies are undergoing active development for application to microbiology (29). Multiple platforms are currently in use, sometimes side-by-side within the same laboratories; all of them generate vast quantities of complex data. To assimilate, analyze, and integrate these data with epidemiologic information, public health programs need enhanced and new capacities in bioinformatics and data science, supported by specialized data systems and software. As laboratory costs for generating comprehensive data on microbial pathogens continue to decrease, the costs of storing, managing, exchanging, and interpreting the data continue to increase; as a result, information technology and the management and governance of large-scale data sets are increasingly critical considerations for public health.
The diffusion of molecular technology throughout the network of U.S. public health laboratories means that data will eventually replace biological specimens as the currency in many routine transactions. Reference laboratories will need to add substantial “dry lab” capabilities, including databases and analytic tools, to complement their sample repositories. Although still at an early stage, the development of data standards, analytical standards, and quality assurance measures is crucial in their implications for the uses and usefulness of data beyond their laboratory of origin (7, 30, 31). Efficient and reliable communication channels are needed to transmit data from point to point. Feedback on data quality must also be built into reporting systems to preserve the integrity of data that will be used to make public health decisions. The success of this process ultimately depends on harmonizing both technical and policy efforts.
The utility of WGS data for public health depends on the availability of comprehensive and well-curated reference databases for a comparison to understand pathogen diversity and characteristics. NCBI maintains databases, such as the Sequence Read Archive (SRA [http://www.ncbi.nlm.nih.gov/sra]), which store raw genetic sequence data and alignment information for many pathogens, as well as for other organisms. Curated databases for particular pathogens (e.g., Los Alamos National Laboratory Pathogen Research Databases [http://www.lanl.gov/collaboration/pathogen-database/]), groups of pathogens (e.g., PATRIC, the Bacterial Bioinformatics Resource Center [https://www.patricbrc.org]), or pathogens and their associated vectors or hosts (e.g., PHI-base [http://www.phi-base.org/]) are maintained by academic research groups with government funding to support basic and clinical research. Most genetic sequence data, however, are either not actively archived, not curated, or inaccessible, leading some to propose community-based curation models (32).
The collaborative Listeria Whole Genome Sequencing Project demonstrated the feasibility of a population-based approach to collecting, curating, and sharing Listeria monocytogenes genome sequences from clinical and environmental isolates. In this project, the participating agencies submitted raw sequence data immediately following initial quality control, along with laboratory and epidemiologic metadata (e.g., date, location, submitting laboratory, sequencing parameters, and source of isolate), to publicly accessible NCBI data archives, where they are associated with a shared BioProject identifier (http://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA211456). NCBI continuously updates and shares phylogenies constructed using these and other publicly available Listeria monocytogenes genome sequences. This data submission model, which is the basis for the next generation of PulseNet, marked an important departure from traditional public health surveillance, where primary laboratory and epidemiologic data are collected and analyzed in secure systems without public access.
Data sharing and collaboration are fundamental to the success of public health surveillance and interventions, as well as applied research to develop antibiotics and vaccines. An open data model for real-time sharing of pathogen genome sequences enables rapid access for basic science research and diagnostic development, even as an outbreak unfolds. Developing an effective model requires careful consideration of appropriate metadata standards and patient privacy concerns. Public health agencies can play an important role in facilitating data sharing and encouraging research transparency while maintaining necessary controls over private health care information. A recent review of barriers to data sharing in public health, conducted by an independent policy institute based in London (United Kingdom), concluded that only a complex process based on trust, advocacy, and capacity building could overcome existing barriers; these included legal, political, economic, technical, motivational, and ethical barriers, with the first three presenting the biggest challenges (33). The reviewers concluded that a single global system was probably not feasible; instead, the model most likely to succeed might be “a global data governance or ethical framework, supplemented by local memoranda of understanding that take into account the local context.” Collaborations, like the Global Microbial Identifier (http://www.globalmicrobialidentifier.org/), help maintain ongoing international dialog and consensus on increasingly complicated technical and policy issues, and help develop a shared vision for aggregating, sharing, and using microbial genomic data to improve global public health.
CONCLUSION
High-throughput technologies and bioinformatics have demonstrated potential to improve public health control of infectious diseases by speeding outbreak detection and response, improving preventive interventions, detecting emerging infectious diseases, and combatting antimicrobial resistance. The Advanced Molecular Detection (AMD) initiative is helping the CDC accelerate and coordinate the integration of new technologies into practice. Collaboration and partnerships are the key to navigating this transition and leveraging the next generation of methods and tools for public health.
ACKNOWLEDGMENTS
We thank Gregory Armstrong, John Besser, Lia Haynes, William Switzer, and David Wentworth for their input, including reviewing selected sections of the manuscript.
This article was prepared by the authors in their personal capacity. The opinions expressed in this article are the authors' own and do not reflect the view of the Centers for Disease Control and Prevention, the Department of Health and Human Services, or the U.S. Government. The use of trade names is for identification only and does not imply endorsement by the U.S. Department of Health and Human Services.
REFERENCES
- 1.Loman NJ, Pallen MJ. 2015. Twenty years of bacterial genome sequencing. Nat Rev Microbiol 13:787–794. doi: 10.1038/nrmicro3565. [DOI] [PubMed] [Google Scholar]
- 2.Luheshi L, Raza S, Moorthie S, Hall A, Blackburn L, Rands C, Sagoo G, Chowdhury S, Kroese M, Burton H. 2015. Pathogen genomics into practice. PHG Foundation, Cambridge, United Kingdom: http://www.phgfoundation.org/file/16848/. [Google Scholar]
- 3.Köser CU, Ellington MJ, Cartwright EJ, Gillespie SH, Brown NM, Farrington M, Holden MT, Dougan G, Bentley SD, Parkhill J, Peacock SJ. 2012. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog 8:e1002824. doi: 10.1371/journal.ppat.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palm D, Johansson K, Ozin A, Friedrich A, Grundmann H, Larsson J, Struelens M. 2012. Molecular epidemiology of human pathogens: how to translate breakthroughs into public health practice, Stockholm, November 2011. Euro Surveill 17:pii=20054 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20054. [PubMed] [Google Scholar]
- 5.Jackson BR, Tarr C, Strain E, Jackson KA, Conrad A, Carleton H, Katz LS, Stroika S, Gould LH, Mody RK, Silk BJ, Beal J, Chen Y, Timme R, Doyle M, Fields A, Wise M, Tillman G, Defibaugh-Chavez S, Kucerova Z, Sabol A, Roache K, Trees E, Simmons M, Wasilenko J, Kubota K, Pouseele H, Klimke W, Besser J, Brown E, Allard M, Gerner-Smidt P. 2016. Implementation of nationwide real-time whole-genome sequencing to enhance listeriosis outbreak detection and investigation. Clin Infect Dis 63:380–386. doi: 10.1093/cid/ciw242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langley G, Besser J, Iwamoto M, Lessa FC, Cronquist A, Skoff TH, Chaves S, Boxrud D, Pinner RW, Harrison LH. 2015. Effect of culture-independent diagnostic tests on future Emerging Infections Program surveillance. Emerg Infect Dis 21:1582–1588. doi: 10.3201/eid2109.150570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gargis AS, Kalman L, Bick DP, da Silva C, Dimmock DP, Funke BH, Gowrisankar S, Hegde MR, Kulkarni S, Mason CE, Nagarajan R, Voelkerding KV, Worthey EA, Aziz N, Barnes J, Bennett SF, Bisht H, Church DM, Dimitrova Z, Gargis SR, Hafez N, Hambuch T, Hyland FC, Luna RA, MacCannell D, Mann T, McCluskey MR, McDaniel TK, Ganova-Raeva LM, Rehm HL, Reid J, Campo DS, Resnick RB, Ridge PG, Salit ML, Skums P, Wong LJ, Zehnbauer BA, Zook JM, Lubin IM. 2015. Good laboratory practice for clinical next-generation sequencing informatics pipelines. Nat Biotechnol 33:689–693. doi: 10.1038/nbt.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldberg B, Sichtig H, Geyer C, Ledeboer N, Weinstock GM. 2015. Making the leap from research laboratory to clinic: challenges and opportunities for next-generation sequencing in infectious disease diagnostics. mBio 6(6):e01888-15. doi: 10.1128/mBio.01888-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shea S, Kubota KA, Maguire H, Gladbach S, Woron A, Atkinson-Dunn R, Couturier MR, Miller MB. 2017. Clinical microbiology laboratories' adoption of culture-independent diagnostic tests is a threat to foodborne-disease surveillance in the United States. J Clin Microbiol 55:10–19. doi: 10.1128/JCM.01624-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Association of Public Health Laboratories. 2016. Submission of enteric pathogens from positive culture-independent diagnostic test specimens to public health: interim guidelines. Association of Public Health Laboratories, Silver Spring, MD: https://www.aphl.org/AboutAPHL/publications/Documents/FS-Enteric_Pathogens_Guidelines_0216.pdf. [Google Scholar]
- 11.Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV, CDC PulseNet Task Force. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg Infect Dis 7:382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scharff RL, Besser J, Sharp DJ, Jones TF, Peter GS, Hedberg CW. 2016. An economic evaluation of PulseNet: a network for foodborne disease surveillance. Am J Prev Med 50(5 Suppl 1):S66–S73. [DOI] [PubMed] [Google Scholar]
- 13.Allard MW, Strain E, Melka D, Bunning K, Musser SM, Brown EW, Timme R. 2016. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J Clin Microbiol 54:1975–1983. doi: 10.1128/JCM.00081-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goulet V, Jacquet C, Laurent E, Rocourt J, Vaillant V, de Valk H. 2001. Surveillance of human listeriosis in France in 1999. Bul Epid Heb 34:161–165. (In French.) [Google Scholar]
- 15.Shepard SS, Meno S, Bahl J, Wilson MM, Barnes J, Neuhaus E. 2016. Viral deep sequencing needs an adaptive approach: IRMA, the iterative refinement meta-assembler. BMC Genomics 17:708. doi: 10.1186/s12864-016-3030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conrad C, Bradley HM, Broz D, Buddha S, Chapman EL, Galang RR, Hillman D, Hon J, Hoover KW, Patel MR, Perez A, Peters PJ, Pontones P, Roseberry JC, Sandoval M, Shields J, Walthall J, Waterhouse D, Weidle PJ, Wu H, Duwve JM, Centers for Disease Control and Prevention (CDC). 2015. Community outbreak of HIV infection linked to injection drug use of oxymorphone–Indiana, 2015. MMWR Morb Mortal Wkly Rep 64:443–444. [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell EM, Galang RR, Heneine W, Switzer W, Peters P, Spiller MT, Jia H, Masciotra S, HIV Outbreak Investigation Team. 2015. Infer and characterize a transmission network in an opioid-driven HIV-1 outbreak, abstr 215 Conference on Retroviruses and Opportunistic Infections (CROI), 22 to 25 February 2016, Seattle, WA http://www.croiconference.org/sessions/infer-and-characterize-transmission-network-opioid-driven-hiv-1-outbreak. [Google Scholar]
- 18.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 19.Bialek SR, Allen D, Alvarado-Ramy F, Arthur R, Balajee A, Bell D, Best S, Blackmore C, Breakwell L, Cannons A, Brown C, Cetron M, Chea N, Chommanard C, Cohen N, Conover C, Crespo A, Creviston J, Curns AT, Dahl R, Dearth S, DeMaria A, Echols F, Erdman DD, Feikin D, Frias M, Gerber SI, Gulati R, Hale C, Haynes LM, Heberlein-Larson L, Holton K, Ijaz K, Kapoor M, Kohl K, Kuhar DT, Kumar AM, Kundich M, Lippold S, Liu L, Lovchik JC, Madoff L, Martell S, Matthews S, Moore J, Murray LR, Onofrey S, Pallansch MA, Pesik N, Pham H, et al. . 2014. First confirmed cases of Middle East respiratory syndrome coronavirus (MERS-CoV) infection in the United States, updated information on the epidemiology of MERS-CoV infection, and guidance for the public, clinicians, and public health authorities–May 2014. MMWR Morb Mortal Wkly Rep 63:431–436. (Erratum, 63:554. [PMC free article] [PubMed] [Google Scholar]
- 20.Savage HM, Godsey MS Jr, Lambert A, Panella NA, Burkhalter KL, Harmon JR, Lash RR, Ashley DC, Nicholson WL. 2013. First detection of heartland virus (Bunyaviridae: Phlebovirus) from field collected arthropods. Am J Trop Med Hyg 89:445–452. doi: 10.4269/ajtmh.13-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambert AJ, Velez JO, Brault AC, Calvert AE, Bell-Sakyi L, Bosco-Lauth AM, Staples JE, Kosoy OI. 2015. Molecular, serological and in vitro culture-based characterization of Bourbon virus, a newly described human pathogen of the genus Thogotovirus. J Clin Virol 73:127–132. doi: 10.1016/j.jcv.2015.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg R. 2014. Detecting the emergence of novel, zoonotic viruses pathogenic to humans. Cell Mol Life Sci 72:1115–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/drugresistance/threat-report-2013/. [Google Scholar]
- 24.The White House. 2014. National strategy for combating antibiotic resistant bacteria. The White House, Washington, DC: https://www.whitehouse.gov/sites/default/files/docs/carb_national_strategy.pdf. [Google Scholar]
- 25.The White House. 2015. National action plan for combating antibiotic-resistant bacteria. The White House, Washington, DC: https://www.whitehouse.gov/sites/default/files/docs/national_action_plan_for_combating_antibotic-resistant_bacteria.pdf. [Google Scholar]
- 26.Epson EE, Pisney LM, Wendt JM, MacCannell DR, Janelle SJ, Kitchel B, Rasheed JK, Limbago BM, Gould CV, Kallen AJ, Barron MA, Bamberg WM. 2014. Carbapenem-resistant Klebsiella pneumoniae producing New Delhi metallo-β-lactamase at an acute care hospital, Colorado, 2012. Infect Control Hosp Epidemiol 35:390–397. doi: 10.1086/675607. [DOI] [PubMed] [Google Scholar]
- 27.Argimón S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, Glasner C, Feil EJ, Holden MTG, Yeats CA, Grundmann H, Spratt BG, Aanensen DM. 2016. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom 2:11. doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Institute of Standards and Technology. 2016. NIST-FDA Workshop report: standards for pathogen detection via next generation sequencing. National Institute of Standards and Technology, Gaithersburg, MD: https://www.nist.gov/mml/bbd/microbial-metrology/nistfda-workshop-standards-pathogen-detection-nextgeneration-sequencing. [Google Scholar]
- 29.American Academy of Microbiology. 2016. Applications of clinical microbial next-generation sequencing, February 2016. American Academy of Microbiology, Washington, DC: http://academy.asm.org/index.php/genetics-genomics-molecular-microbiology/5416-applications-of-clinical-microbial-next-generation-sequencing-2. [PubMed] [Google Scholar]
- 30.Dugan VG, Emrich SJ, Giraldo-Calderón GI, Harb OS, Newman RM, Pickett BE, Schriml LM, Stockwell TB, Stoeckert CJ Jr, Sullivan DE, Singh I, Ward DV, Yao A, Zheng J, Barrett T, Birren B, Brinkac L, Bruno VM, Caler E, Chapman S, Collins FH, Cuomo CA, Di Francesco V, Durkin S, Eppinger M, Feldgarden M, Fraser C, Fricke WF, Giovanni M, Henn MR, Hine E, Hotopp JD, Karsch-Mizrachi I, Kissinger JC, Lee EM, Mathur P, Mongodin EF, Murphy CI, Myers G, Neafsey DE, Nelson KE, Nierman WC, Puzak J, Rasko D, Roos DS, Sadzewicz L, Silva JC, Sobral B, Squires RB, Stevens RL, et al. . 2014. Standardized metadata for human pathogen/vector genomic sequences. PLoS One 9:e99979. doi: 10.1371/journal.pone.0099979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Food and Drug Administration. 2016. Infectious disease next generation sequencing based diagnostic devices: microbial identification and detection of antimicrobial resistance and virulence markers: draft guidance for industry and Food and Drug Administration staff. U.S. Food and Drug Administration, Silver Spring, MD: http://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/UCM500441.pdf. [Google Scholar]
- 32.Putman TE, Burgstaller-Muehlbacher S, Waagmeester A, Wu C, Su AI, Good BM. 2016. Centralizing content and distributing labor: a community model for curating the very long tail of microbial genomes. Database (Oxford) 2016:baw028. doi: 10.1093/database/baw028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edelstein M, Sane J. 2015. Overcoming barriers to data sharing in public health: a global perspective. Chatham House: The Royal Institute of International Affairs, London, UK: https://www.chathamhouse.org/sites/files/chathamhouse/field/field_document/20150417OvercomingBarriersDataSharingPublicHealthSaneEdelstein.pdf. [Google Scholar]