ABSTRACT
New rapid molecular diagnostic technologies for infectious diseases enable expedited accurate microbiological diagnoses. However, diagnostic stewardship and antimicrobial stewardship are necessary to ensure that these technologies conserve, rather than consume, additional health care resources and optimally affect patient care. Diagnostic stewardship is needed to implement appropriate tests for the clinical setting and to direct testing toward appropriate patients. Antimicrobial stewardship is needed to ensure prompt appropriate clinical action to translate faster diagnostic test results in the laboratory into improved outcomes at the bedside. This minireview outlines the roles of diagnostic stewardship and antimicrobial stewardship in the implementation of rapid molecular infectious disease diagnostics.
KEYWORDS: rapid diagnostics, diagnostic stewardship, antimicrobial stewardship, implementation, utilization, appropriate use criteria
INTRODUCTION
The clinical microbiology laboratory is in the midst of a diagnostic revolution. New molecular diagnostic technologies have the potential to transform the modern microbiology laboratory and the care of patients with suspected infections by providing more rapid and robust microbiological diagnoses. However, these technological advances come with practical challenges for the laboratory and for clinicians.
Many of the challenges we face today with molecular diagnostics were first noted by Dr. Raymond C. Bartlett, a visionary clinical pathologist, in the era of conventional culture-based microbiology (1). In 1974, Dr. Bartlett observed that “our technical capabilities are exceeding our ability to apply them effectively and economically to human problems” (2). Similar to Dr. Bartlett's observation, the microbiology laboratory today is exceedingly “faced with a superabundance of academic information and pressure to perform exhaustive, expensive, clinically irrelevant [testing],” which, when misguided, “misleads physicians into erroneous diagnosis and inappropriate therapy” (2). These observations have become more relevant than ever in the emerging era of rapid molecular diagnostics. The time has come to heed Dr. Bartlett's call for a “more practical, economical, clinically meaningful approach” (2). This minireview introduces the role of diagnostic and antimicrobial stewardship in the implementation of rapid molecular infectious disease diagnostics, echoing the timeless principles of Dr. Bartlett.
OVERVIEW OF RAPID MOLECULAR INFECTIOUS DISEASE DIAGNOSTICS
Clinicians seek three basic truths from the clinical microbiology laboratory, i.e., (i) whether the patient is infected, (ii) if so, with what, and (iii) what will treat it (3). In the window between specimen collection, organism identification, and antimicrobial susceptibility determination, broad empirical antimicrobials are given to many patients to avoid the grave consequences of untreated infections in a few. Properly implemented and applied, molecular diagnostic technologies enable the microbiology laboratory to provide these three basic truths faster and more accurately than ever before. The ability of rapid diagnostics to shrink the window creates the potential to provide earlier effective, targeted antimicrobials and to decrease the use of unnecessary empirical therapies.
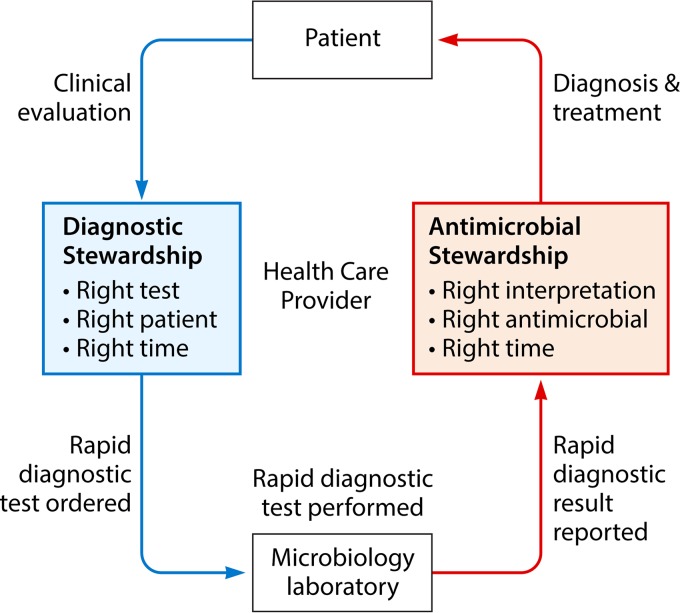
Bacteria, viruses, fungi, and parasites can now be rapidly identified using molecular methods, to diagnose causative agents in infections of the bloodstream, respiratory tract, urinary tract, gastrointestinal tract, and central nervous system. Similarly, new gene-based resistance detection platforms are rapidly emerging to guide antimicrobial use. New techniques used in these rapid diagnostic technologies include nucleic acid-based diagnostics (such as monoplex PCR testing and multiplex PCR panels) (4–8), microarray panels (9), peptide nucleic acid fluorescent in situ hybridization (FISH) technologies (10), magnetic resonance-based testing (11), matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (12), and next-generation sequencing (13). The scope of these technologies ranges from single-target pathogen-specific or resistance gene-specific testing to syndromic panels containing many common pathogens causing a disease process to unbiased sequencing with the ability to detect unsuspected or novel pathogens, as topically reviewed elsewhere. This minireview aims to introduce concepts in diagnostic and antimicrobial stewardship that can be applied to implement these technologies in the clinical setting to optimally affect patient care (Fig. 1).
FIG 1.
Roles of diagnostic and antimicrobial stewardship in the implementation of rapid molecular infectious disease diagnostics in the clinical setting.
DIAGNOSTIC STEWARDSHIP
The goal of diagnostic stewardship is to select the right test for the right patient, generating accurate, clinically relevant results at the right time to optimally influence clinical care and to conserve health care resources. The process of diagnostic stewardship begins with evaluation, selection, and implementation of appropriate diagnostic tests for the clinical setting, incorporates guidance for health care providers regarding judicious use of testing for appropriate patients, and ensures timely sample collection, transport, and processing and timely reporting of results. Key considerations and strategies for each step of the diagnostic stewardship process are outlined below and summarized in Table 1.
TABLE 1.
Key diagnostic stewardship considerations for implementation of rapid infectious disease diagnostics
| Goal | Key question | Key considerations and potential strategies |
|---|---|---|
| Right test | Is the test appropriate for the clinical setting? | Sensitivity and specificity |
| Predictive values | ||
| Testing volumes | ||
| Diagnostic yield | ||
| Laboratory feasibility | ||
| Cost | ||
| Clinical impact | ||
| Right patient | Will the clinical care of the patient be affected by the test result? | Laboratory test utilization committee |
| Automatic laboratory reflex | ||
| CPOE decision support | ||
| Appropriate use criteria | ||
| Indication selection | ||
| Prior authorization | ||
| Benchmarking | ||
| Specimen rejection | ||
| Right time | Will the result be available in time to optimally affect care? | Time to specimen receipt |
| Centralized vs point-of-care testing | ||
| On-demand vs batched testing | ||
| Specimen preparation time | ||
| Run time | ||
| Result reporting time |
Selecting the right test for the clinical setting involves the evaluation of test performance, laboratory feasibility, and cost versus value. Evaluation of basic intrinsic test characteristics, such as sensitivity and specificity, is essential, but findings can be challenging to interpret when there is no clear “gold standard.” This is a particular challenge for molecular methods when conventional culture-based or serological techniques are unable to confirm the presence of infection and reliance on clinical findings may be necessary. For instance, a multiplex PCR assay detecting pneumococcal DNA in a spinal fluid sample from a patient with suspected meningitis who was pretreated with antibiotics and has negative culture results could indicate a true-positive result or a false-positive result, depending on the clinical scenario. Even more important than sensitivity and specificity in predicting clinical utility are the positive and negative predictive values of a test, specific to the local incidence of disease. Clinically, these statistics assess whether a clinician can trust a positive test result and whether the test can be used to rule out an infection. Multiplex panels, however, challenge traditional notions of positive and negative predictive values, as values can be calculated for individual pathogens, for all pathogens in the panel, or for a clinical syndrome. While a rapid multiplex PCR assay may have a high negative predictive value for an individual pathogen or for all pathogens included in the panel, it may have a low overall negative predictive value for the clinical condition of meningitis if bacteria that commonly cause meningitis are not included in the panel, limiting the clinical utility of the assay for ruling out bacterial meningitis.
To ensure that the diagnostic technology selected is appropriate for the clinical setting, it is important to consider testing volumes, diagnostic yield, and the feasibility of performing the test in the laboratory setting. Clinician guidance can be useful to determine the potential utility of newly emerging diagnostic tests with regard to medical decision-making in a particular setting. Retrospective data can be collected to predict testing volumes and diagnostic yield in that setting. For instance, a single-pathogen, point-of-care, influenza PCR test may be appropriate to triage and to treat patients with respiratory symptoms in an outpatient setting, whereas a more extensive rapid multiplex PCR assay for respiratory pathogens may be appropriate to diagnose ventilator-associated pneumonia in the critical care unit of a tertiary care referral center. Factors determining the laboratory feasibility of implementing a new rapid diagnostic test include the technologist training required, the hands-on time for performing the test, and the way in which testing would fit into the laboratory workflow. It is important to consider whether the testing would supplant or be conducted in addition to existing diagnostic tests. For example, with multiplex PCR platforms for cerebrospinal fluid testing, culture is still needed for detection of bacterial pathogens not detected by the panel, confirmation of positive results, and susceptibility testing. However, multiplex PCR assays for stool testing may supplant monoplex PCR assays for viral and parasitic pathogens and save the time and expense of conducting multiple monoplex tests with a single sample.
Cost-effectiveness and potential clinical impact are important factors for deciding whether to adopt an emerging diagnostic technology in the clinical microbiology laboratory. Cost analysis is often a priority of hospital administrators but can be challenging, given the paucity and lag in timing of published cost-effectiveness data (14). While a simple analysis can be used to compare costs and charges for existing diagnostic tests versus new rapid diagnostic tests, a true cost-value analysis should include “back-end” cost savings of decreases in resource utilization (antimicrobials, unnecessary admissions, and lengths of stays), as well as effects on morbidity and mortality rates (14). Assuming reported turnaround times for the rapid diagnostic technology, the potential clinical impact can be estimated by using minimal chart reviews to determine what changes in medical decision-making could occur if results were available at that time point. For instance, a rapid multiplex PCR test of spinal fluid for which results were available in 2 h was predicted to decrease acyclovir exposure by 2 to 3 days for nearly one-half of children undergoing lumbar punctures at our institution (15).
Once the appropriate rapid diagnostic test has been selected for implementation, the next step in diagnostic stewardship is directing testing toward the right patients, for whom the test results will affect clinical care. Overuse of rapid diagnostic tests can add to health care costs without having a significant impact on patient care, whereas underuse may lead to suboptimal clinical outcomes. Inappropriate use of these tests in cases with low pretest probability can lead to misinformation from false-positive results potentially misleading clinicians. Laboratory test utilization committees, modeled after pharmacy and therapeutics committees, with clinical content experts and key stakeholders have been successful in implementing and overseeing many diagnostic stewardship strategies (16).
Provider education efforts are essential for introducing end users to new diagnostic tests being offered and can be used to suggest appropriate testing strategies. Targeted educational interventions can be used to reduce overutilization among providers and trainees who are high test users, as has been demonstrated for Clostridium difficile PCR testing in pediatric populations (17, 18). Provider education is most effective when conducted as one component of a multifaceted approach to diagnostic stewardship.
Automatic laboratory reflex testing without requiring clinician ordering is appropriate for organism identification technologies that are used when culture or initial Gram stain results are positive. For instance, laboratories may choose to automatically perform multiplex PCR, microarray, or FISH testing for rapid identification of organisms in blood cultures that are flagged positive with positive Gram stain results. Similarly, MALDI-TOF MS may be used as a tool for rapid identification of pure organism colony growth on solid media, from a variety of specimen sources.
Diagnostic stewardship is more challenging for direct-from-specimen rapid diagnostic tests, for which the responsibility for appropriate ordering traditionally rests on the clinician. Development of diagnostic algorithms and inclusion of computerized order entry (CPOE) decision support can be used to direct clinicians toward the appropriate test for the clinical situation and curb unnecessary duplication of diagnostic testing (19–21). For example, a diagnostic algorithm can require negative culture results and multiplex PCR panel testing of spinal fluid before next-generation sequencing. CPOE decision support can recommend more cost-effective test ordering, such as a multiplex PCR panel in place of multiple monoplex PCR tests, or highlight redundant testing, such as an order for a monoplex PCR and a multiplex panel that includes the monoplex PCR.
Criteria for appropriate use can be developed as a mechanism to identify clinical situations in which a diagnostic test has significant clinical impact, for inclusion in clinical practice guidelines (22). Indication selection can be used in CPOE decision support, requiring clinicians to input an approved indication from a list of appropriate use criteria before ordering the diagnostic test, to prevent overutilization of tests by clinicians in low-impact situations. Prior authorization by infectious disease or antimicrobial stewardship providers can be required for testing in cases that do not meet appropriate use criteria. For instance, to order a rapid multiplex gastrointestinal diagnostic panel, a provider could be required to select an indication from the approved use criteria of bloody diarrhea, concern for bacterial enteritis, or travel outside the United States. If requesting this test for a patient who does not meet any of those criteria, the provider must call an infectious disease physician for prior authorization. Benchmarking with provider-specific feedback on diagnostic test ordering not meeting appropriate use criteria, compared to that of peers, can be used to curb overutilization by individual providers. For example, a monthly email with the percentage of children with bronchiolitis for whom the provider ordered a respiratory pathogen multiplex PCR panel, compared to the provider's peers, could be used to decrease test overutilization and encourage compliance with national guidelines. Adding measures of test order appropriateness to the existing systems of laboratory benchmarking of quality measures would allow institution-level comparisons of diagnostic stewardship (23). Lastly, laboratory rejection of samples can be used to decrease testing of low-yield samples, based on objective cutoff values. For instance, in one study, implementing a cutoff value of 10 white blood cells (WBCs) in cerebrospinal fluid led to a 46% reduction in herpes simplex virus, varicella-zoster virus, enterovirus, and cytomegalovirus testing without affecting the diagnostic yield (24).
The selection of the right test for the right patient does not affect clinical care unless the result is reported at the right time to influence medical decision-making. Turnaround time is critical for the clinical impact of rapid diagnostic tests. The first factor to consider is the time to specimen receipt in the clinical microbiology laboratory. Sending samples to reference laboratories for testing often negates the potential impact of rapid diagnostic technologies due to transport time alone. While centralization of microbiological testing allows more rapid diagnostic technologies to be offered, this must be balanced against logistical delays in receiving samples that could be mitigated by point-of-care testing, if available, at the bedside or in the clinic. The successful execution of on-demand testing, in contrast to batched testing, in the microbiology laboratory requires appropriate staffing models with trained personnel who are able to perform expedient testing regardless of the time of day. Considerations of specimen preparation time, run-time on the rapid diagnostic platform, and result reporting time are also important considerations in turnaround time. The timely communication of test results, once testing is complete, is a responsibility that requires collaboration between the laboratory and antimicrobial stewardship groups, as outlined below.
ANTIMICROBIAL STEWARDSHIP
The goal of antimicrobial stewardship for rapid diagnostic implementation is to ensure that the right interpretation of the result occurs at the right time, leading to the right antimicrobial therapy to improve clinical outcomes and to decrease unnecessary antimicrobial use. A functioning collaborative partnership between diagnostic stewardship in the laboratory and antimicrobial stewardship on the clinical side is essential for successful implementation of rapid diagnostic technologies to achieve this goal. Antimicrobial stewardship strategies to ensure rapid communication, correct interpretation, and appropriate adjustment of antimicrobial regimens to translate rapid diagnostic test results into improved patient care are outlined below and summarized in Table 2.
TABLE 2.
Key antimicrobial stewardship considerations for implementation of rapid infectious disease diagnostics
| Goal | Key question | Key considerations and potential strategiesa |
|---|---|---|
| Right interpretation | Will the clinician understand the test result? | Result report language |
| Selective reporting of relevant results | ||
| AS prospective audit and feedback | ||
| AS real-time decision support | ||
| Right antimicrobial | Will the clinician appropriately modify antimicrobials based on the test result? | Clinical practice guidelines |
| EMR-based decision support with result reporting | ||
| AS prospective audit and feedback | ||
| AS real-time decision support | ||
| Right time | Will the clinician act upon the test result promptly? | EMR reporting |
| Results called with readback reporting | ||
| AS prospective audit and feedback | ||
| AS real-time decision support |
AS, antimicrobial stewardship.
For a rapid diagnostic test to have rapid impact, it is essential that providers be alerted to results in real time. Active readback reporting ensures that results have been received in a timely manner and is superior to passive reporting that requires the clinician to continually check the medical record for results. A rapid diagnostic test result that remains unchecked in the medical record is the tree that falls in the forest with no one around to hear it, wasting expensive health care resources and potentially leading to delayed optimization of care.
As the complexity of molecular diagnostic testing for infectious diseases increases, the communication of clinically relevant results becomes more important in ensuring proper interpretation by clinicians. A recent survey of infectious disease physicians demonstrated that 67.5% think that new diagnostic testing is becoming too complex for non-infectious disease physicians (25). The reporting of increasingly specific, species-level identification of pathogens not widely familiar to general clinicians can create confusion. This confusion may be avoided by grouping species and using more familiar, clinically relevant nomenclature or providing interpretive decision support. For example, a report of Streptococcus sobrinus from MALDI-TOF MS analysis may be better communicated as the more familiar viridans Streptococcus group. A report of parechovirus detection in cerebrospinal fluid may be better interpreted if the results are accompanied by a description of parechoviruses in the comments or are communicated by an antimicrobial steward with contextual interpretation and decision support.
Clinicians may also require decision support to navigate some of the pitfalls in the interpretation of rapid molecular infectious disease diagnostics. The detection of nucleic acids by molecular diagnostics does not always equate to detection of a viable organism that is the cause of the patient's disease process. Pathogens detected may represent colonization, asymptomatic infection, reactivation in the setting of acute infection, chromosomal integration, or prolonged shedding after an unrelated prior infection. For instance, detection of a rhinovirus/enterovirus in a respiratory pathogen panel for a febrile infant without respiratory symptoms may reflect shedding from a prior respiratory infection, an asymptomatic infection, or a true source of fever. With more broadly based syndromic testing panels and next-generation sequencing, pathogens that are unlikely to be the cause of disease may be detected, creating clinical confusion. For instance, gastrointestinal panels may detect Escherichia coli species that are asymptomatically carried in a large portion of the population or Clostridium difficile in children less than 1 year of age, for whom this result more likely represents carriage than infection requiring treatment. Selective reporting of relevant pathogens is one approach to deal with these issues, but it requires full disclosure of the pathogens being reported versus withheld from reporting for the panel. The detection of multiple pathogens within a single specimen, some of which may be causing disease and others of which may be contaminants or innocent bystanders, can also be challenging for clinicians to interpret. The decision support strategies outlined below can assist clinicians in dealing with these new challenges posed by rapid molecular diagnostics.
Once a test result is rapidly communicated and properly interpreted, prompt and appropriate clinical action must be taken to optimally affect care. The first step in this process is creating evidence-based clinical practice guideline recommendations based on test results. National guidelines and local antibiogram data can be useful in creating consensus empirical treatment recommendations to standardize care. Providing antimicrobial stewardship decision support tools with rapid diagnostic results is more effective than traditional methods of reporting results alone in the electronic medical record (EMR) or having microbiology laboratory technicians call providers with results (26). Templated comments, with evidence-based clinical practice guideline recommendations to guide antimicrobial prescribing, accompanying results have been used successfully in settings with few antimicrobial stewardship resources (27). However, direct contact with real-time antimicrobial stewardship decision support appears to be a superior approach (26).
Rapid diagnostic technologies present unique opportunities and unique challenges for antimicrobial stewardship programs. Current guideline-recommended antimicrobial stewardship strategies include formulary restriction or prior authorization requirements targeting specific antimicrobials and prospective auditing and feedback, i.e., reviewing antimicrobial prescriptions at specified time points (typically 48 and 72 h after treatment initiation) and intervening when antimicrobial modifications are warranted (28). Interventions based on these strategies are triggered by antimicrobial use rather than rapid diagnostic test results. Alternatively, a proactive strategy of real-time antimicrobial stewardship decision support is optimized when used with rapid diagnostic technologies, providing intervention at the time of rapid diagnostic test result reporting, which often is the time of medical decision-making. Incorporating real-time decision support at the time of result reporting ensures that the right interpretation of the test result leads to the right antimicrobial being prescribed from the start, rather than correcting misinterpretations and rectifying suboptimal antimicrobial use after treatment has reached the patient. This approach has been shown to be acceptable to providers and has not led to decreases in infectious disease consultations (29). Although this approach requires a significant investment of antimicrobial stewardship resources, it capitalizes on the investment made in expensive rapid diagnostic technologies, ensuring that these laboratory tests are optimally influencing patient care.
One advantage of providing personalized antimicrobial stewardship decision support is the adaptability of these standardized recommendations to specific patient situations. For instance, although cefazolin may be first-line therapy for methicillin-susceptible Staphylococcus aureus bacteremia in a guideline, this would not be an ideal therapy for central nervous system penetration in a patient with brain abscesses and bacteremia, which may be caught only through direct personalized antimicrobial stewardship intervention. Drug allergies, drug-drug interactions, and dosing modifications are other examples of antimicrobial stewardship support factors that can be individualized by antimicrobial stewardship intervention but may not be easily communicated in clinical practice guidelines. The “handshake stewardship” approach of engaging in a two-way conversation, with an infectious disease-trained physician or pharmacist learning about the specific patient scenario from the provider directly caring for the patient and providing specialized antimicrobial recommendations, allows for the optimal exchange of information (30).
As new molecular diagnostic tests for infectious diseases are implemented, real-time feedback from end users and troubleshooting mechanisms are needed to optimize practices. Stakeholder involvement in the implementation planning process assists in designing protocols and algorithms that take into account the diverse perspectives of the microbiology laboratory, antimicrobial stewardship, infectious diseases, infection control, and, most importantly, end user clinicians. These stakeholder liaisons can be helpful in gathering ongoing feedback during the implementation process. Provider surveys are another method to obtain ongoing feedback regarding the accuracy, timeliness, impact, and acceptability of the newly implemented test (29). During the rollout period, telephone and email hotlines can be helpful for troubleshooting unforeseen issues and providing education and support for end users who may not be familiar with the new technology.
FUTURE DIRECTIONS
Although rapid molecular infectious disease diagnostics are revolutionizing the ability of the microbiology laboratory to more rapidly provide the three basic truths required by clinicians, unmet challenges remain. Current technologies are excellent at affirmatively answering the questions of whether the patient is infected and, if so, with what, by identifying organisms directly from specimens using syndromic panels, MALDI-TOF MS, and next-generation sequencing. However, technology currently cannot, and perhaps will never be able to, provide a definitive negative result to rule out infection. There will always be a need for clinicians, infectious disease specialists, and antimicrobial stewards to take into account the clinical history, examination, biomarker, and imaging findings to determine the patient's risk of an infection not detected by microbiological testing of the biological specimens obtained. In the future, instead of looking for an organism causing infection, there may be a role for analyzing the host response to infection. Promising preliminary studies of RNA biosignature profiles have been successful in differentiating bacterial infections, viral infections, and other noninfectious processes. More attainable, but lagging significantly behind the advances made in organism identification, is the ability of rapid diagnostic technologies to answer the question of what will treat the infection. Infectious disease physicians ranked resistant Gram-negative infections as the most important unmet need in pathogen diagnosis (25). With the increasing incidence and complexity of antimicrobial resistance, more rapid susceptibility data have great potential to have large impacts on antimicrobial stewardship and patient outcomes.
CONCLUSIONS
Dr. Raymond Bartlett had great foresight in 1974 when he adapted the classic adage of “just because you can does not necessarily mean you should” to the microbiology laboratory by stressing the importance of “careful integration of what is technically feasible with what is clinically important” (2). As increasing numbers of diagnostic technologies emerge from research laboratories into the clinical realm, it is essential to conduct clinical impact and cost-effectiveness analyses prior to adoption in the clinical setting, to avoid Dr. Bartlett's fear of “production of a substantial amount of useless information at considerable cost” (31). Rapid molecular diagnostics have the potential to revolutionize the clinical care of patients with suspected infections. However, careful diagnostic and antimicrobial stewardship is essential for successful implementation in the clinical setting. As Dr. Bartlett noted, “It has long been recognized, that frequent contact between the microbiologist and the clinician results in a free exchange of information which facilitates both laboratory and clinical decisions” (2). The concepts of diagnostic and antimicrobial stewardship outlined in this minireview will help to ensure that clinical relevance remains the driving force during this diagnostic revolution in the microbiology laboratory and that we never forget that “good [microbiology] is clinically relevant [microbiology].”
REFERENCES
- 1.Onderdonk AB. 2015. Biographical feature: Raymond C. Bartlett, MD. J Clin Microbiol 53:1464–1466. doi: 10.1128/JCM.00167-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartlett RC. 1974. Medical microbiology: quality cost and clinical relevance. Wiley, New York, NY. [Google Scholar]
- 3.Baron EJ, Miller JM, Weinstein MP, Richter SS, Gilligan PH, Thomson RB Jr, Bourbeau P, Carroll KC, Kehl SC, Dunne WM, Robinson-Dunn B, Schwartzman JD, Chapin KC, Snyder JW, Forbes BA, Patel R, Rosenblatt JE, Pritt BS. 2013. A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2013 recommendations by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM). Clin Infect Dis 57:e22–e121. doi: 10.1093/cid/cit278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poritz MA, Blaschke AJ, Byington CL, Meyers L, Nilsson K, Jones DE, Thatcher SA, Robbins T, Lingenfelter B, Amiott E, Herbener A, Daly J, Dobrowolski SF, Teng DH, Ririe KM. 2011. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One 6:e26047. doi: 10.1371/journal.pone.0026047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaschke AJ, Heyrend C, Byington CL, Fisher MA, Barker E, Garrone NF, Thatcher SA, Pavia AT, Barney T, Alger GD, Daly JA, Ririe KM, Ota I, Poritz MA. 2012. Rapid identification of pathogens from positive blood cultures by multiplex polymerase chain reaction using the FilmArray system. Diagn Microbiol Infect Dis 74:349–355. doi: 10.1016/j.diagmicrobio.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buss SN, Leber A, Chapin K, Fey PD, Bankowski MJ, Jones MK, Rogatcheva M, Kanack KJ, Bourzac KM. 2015. Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol 53:915–925. doi: 10.1128/JCM.02674-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leber AL, Everhart K, Balada-Llasat JM, Cullison J, Daly J, Holt S, Lephart P, Salimnia H, Schreckenberger PC, DesJarlais S, Reed SL, Chapin KC, LeBlanc L, Johnson JK, Soliven NL, Carroll K, Miller JA, Dien Bard J, Mestas J, Bankowski M, Enomoto T, Hemmert AC, Bourzac K. 2016. Multicenter evaluation of BioFire FilmArray meningitis/encephalitis panel for detection of bacteria, viruses and yeast in cerebrospinal fluid specimens. J Clin Microbiol 54:2251–2261. doi: 10.1128/JCM.00730-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claas EC, Burnham CA, Mazzulli T, Templeton K, Topin F. 2013. Performance of the xTAG(R) gastrointestinal pathogen panel, a multiplex molecular assay for simultaneous detection of bacterial, viral, and parasitic causes of infectious gastroenteritis. J Microbiol Biotechnol 23:1041–1045. doi: 10.4014/jmb.1212.12042. [DOI] [PubMed] [Google Scholar]
- 9.Buchan BW, Ginocchio CC, Manii R, Cavagnolo R, Pancholi P, Swyers L, Thomson RB Jr, Anderson C, Kaul K, Ledeboer NA. 2013. Multiplex identification of Gram-positive bacteria and resistance determinants directly from positive blood culture broths: evaluation of an automated microarray-based nucleic acid test. PLoS Med 10:e1001478. doi: 10.1371/journal.pmed.1001478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stender H. 2003. PNA FISH: an intelligent stain for rapid diagnosis of infectious diseases. Expert Rev Mol Diagn 3:649–655. doi: 10.1586/14737159.3.5.649. [DOI] [PubMed] [Google Scholar]
- 11.Mylonakis E, Clancy CJ, Ostrosky-Zeichner L, Garey KW, Alangaden GJ, Vazquez JA, Groeger JS, Judson MA, Vinagre YM, Heard SO, Zervou FN, Zacharioudakis IM, Kontoyiannis DP, Pappas PG. 2015. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: a clinical trial. Clin Infect Dis 60:892–899. doi: 10.1093/cid/ciu959. [DOI] [PubMed] [Google Scholar]
- 12.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 13.Naccache SN, Federman S, Veeraraghavan N, Zaharia M, Lee D, Samayoa E, Bouquet J, Greninger AL, Luk KC, Enge B, Wadford DA, Messenger SL, Genrich GL, Pellegrino K, Grard G, Leroy E, Schneider BS, Fair JN, Martinez MA, Isa P, Crump JA, DeRisi JL, Sittler T, Hackett J Jr, Miller S, Chiu CY. 2014. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res 24:1180–1192. doi: 10.1101/gr.171934.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caliendo AM, Gilbert DN, Ginocchio CC, Hanson KE, May L, Quinn TC, Tenover FC, Alland D, Blaschke AJ, Bonomo RA, Carroll KC, Ferraro MJ, Hirschhorn LR, Joseph WP, Karchmer T, MacIntyre AT, Reller LB, Jackson AF. 2013. Better tests, better care: improved diagnostics for infectious diseases. Clin Infect Dis 57(Suppl 3):S139–S170. doi: 10.1093/cid/cit578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Messacar K, Breazeale G, Robinson CC, Dominguez SR. 2016. Potential clinical impact of the film array meningitis encephalitis panel in children with suspected central nervous system infections. Diagn Microbiol Infect Dis 86:118–120. doi: 10.1016/j.diagmicrobio.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warren JS. 2013. Laboratory test utilization program: structure and impact in a large academic medical center. Am J Clin Pathol 139:289–297. doi: 10.1309/AJCP4G6UAUXCFTQF. [DOI] [PubMed] [Google Scholar]
- 17.Kociolek LK, Bovee M, Carter D, Ciolino JD, Patel R, O'Donnell A, Rupp AH, Zheng X, Shulman ST, Patel SJ. 2016. Impact of a healthcare provider educational intervention on frequency of Clostridium difficile polymerase chain reaction testing in children: a segmented regression analysis. J Pediatr Infect Dis Soc doi: 10.1093/jpids/piw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klatte JM, Selvarangan R, Jackson MA, Myers AL. 2016. Reducing overutilization of testing for Clostridium difficile infection in a pediatric hospital system: a quality improvement initiative. Hosp Pediatr 6:9–14. doi: 10.1542/hpeds.2015-0116. [DOI] [PubMed] [Google Scholar]
- 19.Krasowski MD, Chudzik D, Dolezal A, Steussy B, Gailey MP, Koch B, Kilborn SB, Darbro BW, Rysgaard CD, Klesney-Tait JA. 2015. Promoting improved utilization of laboratory testing through changes in an electronic medical record: experience at an academic medical center. BMC Med Inform Decis Mak 15:11. doi: 10.1186/s12911-015-0137-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bates DW, Kuperman GJ, Rittenberg E, Teich JM, Fiskio J, Ma'luf N, Onderdonk A, Wybenga D, Winkelman J, Brennan TA, Komaroff AL, Tanasijevic M. 1999. A randomized trial of a computer-based intervention to reduce utilization of redundant laboratory tests. Am J Med 106:144–150. doi: 10.1016/S0002-9343(98)00410-0. [DOI] [PubMed] [Google Scholar]
- 21.Wilson ML. 2015. Decreasing inappropriate laboratory test utilization: controlling costs and improving quality of care. Am J Clin Pathol 143:614–616. doi: 10.1309/AJCPHQODM9XYWLZ9. [DOI] [PubMed] [Google Scholar]
- 22.Bhatia RS, Alabousi M, Dudzinski DM, Weiner RB. 2016. Appropriate use criteria: a review of need, development and applications. Expert Rev Cardiovasc Ther 14:281–290. doi: 10.1586/14779072.2016.1131125. [DOI] [PubMed] [Google Scholar]
- 23.Shahangian S, Snyder SR. 2009. Laboratory medicine quality indicators: a review of the literature. Am J Clin Pathol 131:418–431. doi: 10.1309/AJCPJF8JI4ZLDQUE. [DOI] [PubMed] [Google Scholar]
- 24.Wilen CB, Monaco CL, Hoppe-Bauer J, Jackups R Jr, Bucelli RC, Burnham CA. 2015. Criteria for reducing unnecessary testing for herpes simplex virus, varicella-zoster virus, cytomegalovirus, and enterovirus in cerebrospinal fluid samples from adults. J Clin Microbiol 53:887–895. doi: 10.1128/JCM.03161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blaschke AJ, Hersh AL, Beekmann SE, Ince D, Polgreen PM, Hanson KE. 2015. Unmet diagnostic needs in infectious disease. Diagn Microbiol Infect Dis 81:57–59. doi: 10.1016/j.diagmicrobio.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerjee R, Teng CB, Cunningham SA, Ihde SM, Steckelberg JM, Moriarty JP, Shah ND, Mandrekar JN, Patel R. 2015. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. Clin Infect Dis 61:1071–1080. doi: 10.1093/cid/civ447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dekmezian M, Beal SG, Damashek MJ, Benavides R, Dhiman N. 2015. The SUCCESS model for laboratory performance and execution of rapid molecular diagnostics in patients with sepsis. Proc (Bayl Univ Med Cent) 28:144–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barlam TF, Cosgrove SE, Abbo LM, MacDougall C, Schuetz AN, Septimus EJ, Srinivasan A, Dellit TH, Falck-Ytter YT, Fishman NO, Hamilton CW, Jenkins TC, Lipsett PA, Malani PN, May LS, Moran GJ, Neuhauser MM, Newland JG, Ohl CA, Samore MH, Seo SK, Trivedi KK. 2016. Implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis 62:e51–e77. doi: 10.1093/cid/ciw118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messacar K, Hurst AL, Child J, Campbell K, Palmer C, Hamilton S, Dowell E, Robinson CC, Parker SK, Dominguez SR. 2016. Clinical impact and provider acceptability of real-time antimicrobial stewardship decision support for rapid diagnostics in children with positive blood culture results. J Pediatric Infect Dis Soc doi: 10.1093/jpids/piw047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurst AL, Child J, Pearce K, Palmer C, Todd JK, Parker SK. 2016. Handshake stewardship: a highly effective rounding-based antimicrobial optimization service. Pediatr Infect Dis J 35:1104–1110. doi: 10.1097/INF.0000000000001245. [DOI] [PubMed] [Google Scholar]
- 31.Bartlett RC. 1974. A plea for clinical relevance in medical microbiology. Am J Clin Pathol 61:867–872. doi: 10.1093/ajcp/61.6.867. [DOI] [PubMed] [Google Scholar]