ABSTRACT
The World Health Organization estimates that nearly 500 million malaria tests are performed annually. While microscopy and rapid diagnostic tests (RDTs) are the main diagnostic approaches, no single method is inexpensive, rapid, and highly accurate. Two recent studies from our group have demonstrated a prototype computer vision platform that meets those needs. Here we present the results from two clinical studies on the commercially available version of this technology, the Sight Diagnostics Parasight platform, which provides malaria diagnosis, species identification, and parasite quantification. We conducted a multisite trial in Chennai, India (Apollo Hospital [n = 205]), and Nairobi, Kenya (Aga Khan University Hospital [n = 263]), in which we compared the device to microscopy, RDTs, and PCR. For identification of malaria, the device performed similarly well in both contexts (sensitivity of 99% and specificity of 100% at the Indian site and sensitivity of 99.3% and specificity of 98.9% at the Kenyan site, compared to PCR). For species identification, the device correctly identified 100% of samples with Plasmodium vivax and 100% of samples with Plasmodium falciparum in India and 100% of samples with P. vivax and 96.1% of samples with P. falciparum in Kenya, compared to PCR. Lastly, comparisons of the device parasite counts with those of trained microscopists produced average Pearson correlation coefficients of 0.84 at the Indian site and 0.85 at the Kenyan site.
KEYWORDS: malaria, diagnosis, computer vision, machine learning
INTRODUCTION
Accurate malaria diagnosis prior to treatment is imperative to reduce mortality rates, to prevent antimalarial resistance, and to limit unnecessary side effects from improperly prescribed drugs (1). As a result, several governmental health organizations now require malaria testing prior to the use of antimalarials, prompting a projected increase in demand from 500 million tests in 2012 to 1 billion tests by 2020 (1, 2).
Recent reports show that a large number of malaria carriers maintain low levels of parasitemia, increasing the need for highly sensitive diagnostic modalities (3, 4). Microscopy remains the most widely used malaria test worldwide (5, 6). Under controlled laboratory conditions, it is possible to reach a limit of detection of 250 parasites/μl for thin smears. In most settings, however, microscopy can be very inaccurate, with typical limits of detection ranging from 300 parasites/μl to 1,000 parasites/μl, and requires highly trained staff (7, 8). Several studies have demonstrated poor sensitivity of microscopy resulting from improperly trained technicians and fatigue due to high test volumes (9, 10). Recently, rapid diagnostic tests (RDTs) have increased in popularity since they require minimal training and are considered more consistent than microscopy (11). However, RDTs have poor sensitivity at low levels of parasitemia, detect residual malaria antigens, and are very inconsistent among brands and batches (12, 13). The arrival of more sensitive PCR and loop-mediated isothermal amplification (LAMP) assays has shown that many infected individuals are not detected by microscopy and RDTs. However, these tools are impractical for the vast majority of areas in which malaria is endemic, due to their high costs and long turnaround times (14, 15).
An automated microscopist could significantly improve malaria diagnosis by identifying live intracellular parasites at low levels of parasitemia with improved speed, cost, and consistency. Previous attempts at designing an automated microscopist did not progress past the development stage (16, 17). Recently, our group published two studies showing that a prototype of such a system could be successful. Srivastava et al. presented the prototype system (P1), which showed performance results comparable to those of many marketed RDTs (18). A follow-up article introduced the improved system (P2), which achieved sensitivity, specificity, and species identification results comparable to those of expert human microscopists (19).
Here we describe the performance of the commercial Parasight device. We provide an overview of the technology, as well as the results of clinical studies performed at Apollo Hospital (Chennai, India) and Aga Khan University Hospital (AKUH) (Nairobi, Kenya) to evaluate the sensitivity, specificity, species identification, and parasite count results, compared to standard diagnostic procedures.
RESULTS
Analytical process.
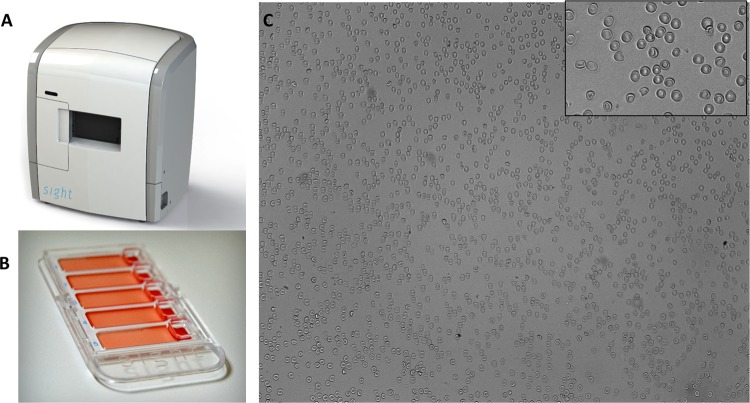
The Sight Diagnostics Parasight device is a desktop system for computerized malaria diagnosis (Fig. 1A). Blood samples are stained and then loaded into a cartridge that holds five patient samples (Fig. 1B). After sample loading, a monolayer forms in the cartridge, with minimal overlap between the blood cells (Fig. 1C). During scanning, the device analyzes stained white blood cells, platelets, red blood cells (RBCs), and malaria ring-stage parasites, trophozoites, schizonts, and gametocytes (Fig. 2). In total, the platform records images of ∼1.5 million red blood cells (0.3 μl of blood). The results are processed by a machine learning algorithm that performs feature extraction by a computer vision support vector machine (SVM) classifier. The algorithm examines unique morphological features to reach a final diagnosis that detects, enumerates, and identifies the malaria species (Fig. 3).
FIG 1.
Sight Diagnostics malaria platform. (A) Desktop scanning device. (B) Loading cartridge, which holds five patient samples. (C) Image of the monolayer at ×20, which evenly disperses the blood cells with minimal overlap. Inset, enlarged image of the monolayer.
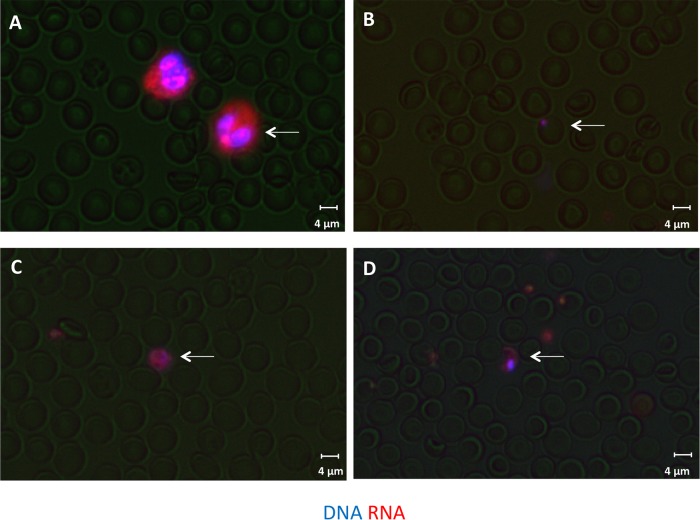
FIG 2.
Images of stained blood monolayers. Blood is stained with DNA and RNA dyes and formed into a monolayer. The device searches for stained white blood cells (A), ring-stage P. falciparum malaria (B), P. vivax gametocytes (C), and P. falciparum gametocytes (D), as shown by the white arrows.
FIG 3.
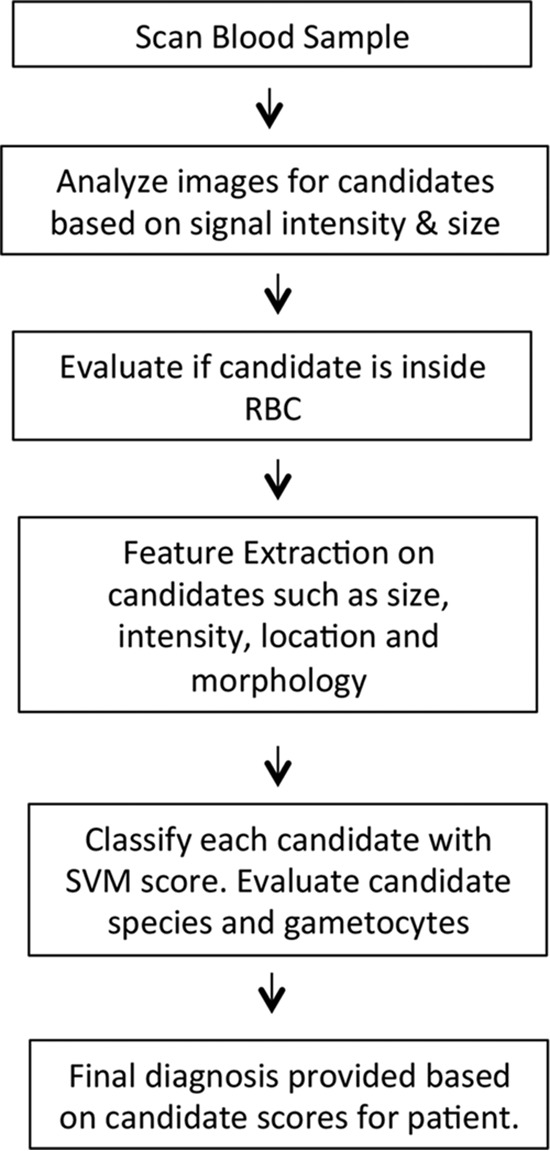
Algorithm flow diagram. The algorithm begins by identifying candidates, after which it extracts specific features and provides a SVM score for each possible parasite. This information is then used to provide a final diagnosis.
Sensitivity and specificity.
At Apollo Hospital and at AKUH, microscopy and PCR were used as two independent standards for comparison (Tables 1 and 2). At Apollo Hospital, 205 samples were collected and 5 were discarded due to blood hemolysis; at AKUH, 263 samples were collected and 6 were discarded due to blood hemolysis. At Apollo Hospital, the sensitivity of the RDT was 100% (95% confidence interval [CI], 92.8 to 100%) and the sensitivity of the Parasight system was 99% (95% CI, 94.6 to 99.9%), in comparison with both microscopy and PCR. The specificity of the RDT was 97.9% (95% CI, 92.8 to 99.7%) and the specificity of the Parasight system was 100% (95% CI, 96.3 to 100%), in comparison with both microscopy and PCR.
TABLE 1.
Device sensitivities versus microscopy and PCR
| Trial site and method | No. positive/total no. tested (sensitivity [95% CI] [%]) |
|
|---|---|---|
| Versus microscopy | Versus PCR | |
| Apollo Hospital | ||
| RDT | 101/101 (100 [92.8–100]) | 101/101 (100 [92.8–100]) |
| Microscopy | NAa | 101/101 (100 [92.8–100]) |
| Parasight | 100/101 (99 [94.6–99.9]) | 100/101 (99 [94.6–99.9]) |
| AKUH | ||
| RDT | 155/160 (96.8 [92.8–98.9]) | 155/160 (96.8 [92.8–98.9]) |
| Microscopy | NA | 160/160 (100 [97.7–100]) |
| Parasight | 159/160 (99.3 [95.6–99.8]) | 159/160 (99.3 [95.6–99.8]) |
NA, not applicable.
TABLE 2.
Device specificities versus microscopy and PCR
| Trial site and method | No. positive/total no. tested (specificity [95% CI] [%]) |
|
|---|---|---|
| Versus microscopy | Versus PCR | |
| Apollo Hospital | ||
| RDT | 97/99 (97.9 [92.8–99.7]) | 97/99 (97.9 [92.8–99.7]) |
| Microscopy | NAa | 99/99 (100 [96.3–100]) |
| Parasight | 99/99 (100 [96.3–100]) | 99/99 (100 [96.3–100]) |
| AKUH | ||
| RDT | 92/97 (94.8 [88.3–98.3]) | 92/97 (94.8 [88.3–98.3]) |
| Microscopy | NA | 97/97 (100 [96.1–100]) |
| Parasight | 96/97 (98.9 [94.4–99.7]) | 96/97 (98.9 [94.3–99.9]) |
NA, not applicable.
At AKUH, the sensitivity of the RDT was 96.8% (95% CI, 92.8 to 98.9%) and the sensitivity of the Parasight system was 99.3% (95% CI, 97.7 to 100%), in comparison with microscopy and PCR. The specificity of the RDT was 94.8% (95% CI, 88.3 to 98.3%) and the specificity of the Parasight system was 98.9% (95% CI, 94.4 to 99.7%), in comparison with microscopy and PCR.
Species identification.
Species identification studies were conducted with samples provided at Apollo Hospital and AKUH, and the results were compared with PCR findings (Table 3). At Apollo Hospital, the RDT showed species identification of 98.8% (95% CI, 93.9 to 99.9%) for Plasmodium vivax and 91.6% (95% CI, 61.5 to 99.7%) for Plasmodium falciparum, while the Parasight system showed species identification of 100% (95% CI, 95.9 to 100%) for P. vivax and 100% (95% CI, 73.5 to 100%) for P. falciparum. At AKUH, the RDT showed species identification of 100% (95% CI, 97.5 to 100%) for P. falciparum and 0% (95% CI, 0 to 26.4%) for P. vivax, while the Parasight system showed species identification of 100% (95% CI, 73.5 to 100%) for P. vivax and 96.1% (95% CI, 91.7 to 98.6%) for P. falciparum.
TABLE 3.
Species identification accuracy
| Trial site and method | No. identified/total no. tested (identification [95% CI] [%]) |
|
|---|---|---|
| P. falciparum | P. vivax | |
| Apollo | ||
| RDT | 11/12 (91.6 [61.52–99.7]) | 88/89 (98.8 [93.9–99.9]) |
| Microscopy | 12/12 (100 [73.5–100]) | 89/89 (100 [95.9–100]) |
| Parasight | 12/12 (100 [73.5–100]) | 89/89 (100 [95.9–100]) |
| AKUH | ||
| RDT | 148/148 (100 [97.5–100]) | 0/7 (0 [0–26.4]) |
| Microscopy | 147/153 (96.1 [91.7–98.6]) | 7/7 (100 [73.5–100]) |
| Parasight | 147/153 (96.1 [91.7–98.6]) | 7/7 (100 [73.5–100]) |
Parasitemia.
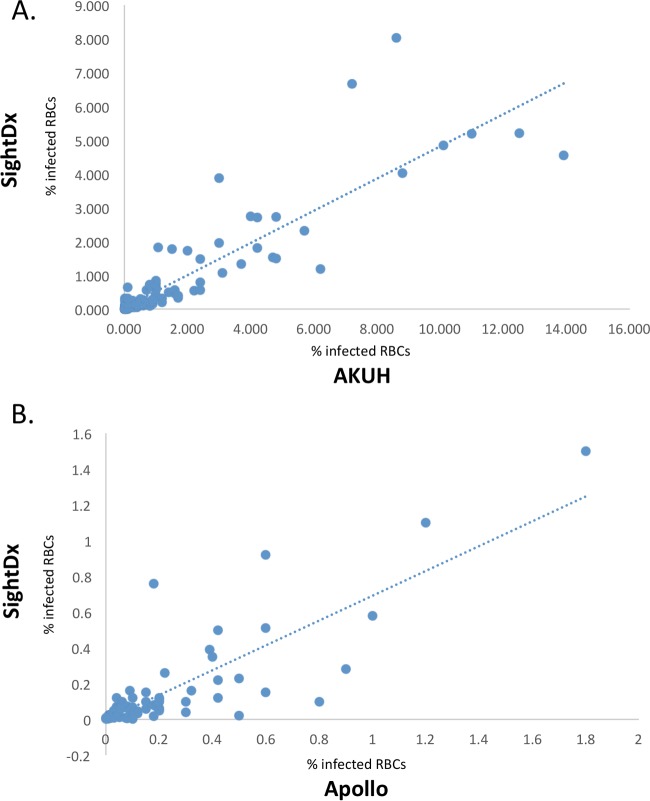
At both locations, thin-smear microscopy was performed and parasitemia levels were compared to values obtained from the device (Fig. 4A and B). At Apollo Hospital, the device showed a Pearson correlation coefficient of 0.84, compared to microscopy (correlation coefficients of 0.98 for P. falciparum and 0.83 for P. vivax); at AKUH, the device showed a Pearson correlation coefficient of 0.85, compared to microscopy (correlation coefficients of 0.86 for P. falciparum and 0.55 for P. vivax).
FIG 4.
Microscopy results versus device results. (A) At AKUH, microscopy parasitemia levels correlated with Sight Diagnostics (SightDx) parasitemia levels with a Pearson correlation coefficient of 0.85. (B) At Apollo Hospital, microscopy parasitemia levels correlated with Sight Diagnostics parasitemia levels with a Pearson correlation coefficient of 0.84.
DISCUSSION
The increasing demand for highly accurate and rapid malaria testing has created an unmet need in the malaria diagnostic market. Numerous studies have shown poor accuracy for both RDTs and microscopists, necessitating the development of new technologies.
The Parasight platform is a novel malaria test system that is able to provide highly sensitive malaria evaluations faster than current malaria tests. The system uses a combination of DNA and RNA fluorescent dyes to stain various blood components. By using rapidly staining dyes, the device is able to provide rapid diagnoses, as opposed to antibody-based detection, which requires extensive incubation to detect specific antigens. As several cellular entities are stained by the dye combination, the algorithms use a feature library to distinguish the malaria inside RBCs and to establish the infected species. The features are based on a database of over 2,500 positive and negative patient samples of several different species that were collected from institutions in Europe, Africa, and India.
Since the publication of the studies by Srivastava et al. and Houri-Yafin et al. (18, 19), we have significantly enlarged the algorithm database of positive and negative samples, leading to vastly improved performances in all categories. Additionally, we expanded the features involved in the candidate assessment (as shown in Fig. 3) to increase the accuracy of the classifier malaria detection and to allow us to lower the limit of detection to 20 parasites/μl. These modifications have improved the accuracy of the device from equivalent to a skilled microscopist to equivalent to PCR.
The performance of the device was consistent across different species and use conditions. Eighty-nine percent of positive samples at Apollo Hospital were P. vivax and 11% were P. falciparum, while 4.3% of positive samples at AKUH were P. vivax and 95.7% were P. falciparum, reflecting the different species distributions found in India and Africa. Additionally, samples from AKUH showed significantly higher levels of parasitemia, with 15% of samples (24/160 samples) having RBC parasite burdens of over 2%, whereas none of the samples from Apollo Hospital had parasitemia over this level. This discrepancy is likely a function of the higher levels of parasitemia found intrinsically in P. falciparum infections, compared with other varieties of malaria.
While the device performed very accurately, there were several notable cases of discrepancies for which we found explanations. One patient sample at Apollo Hospital and one at AKUH were incorrectly diagnosed as negative by the Parasight system, due to levels of parasitemia below 20 parasites/μl. At AKUH, several P. falciparum cases were erroneously identified as P. vivax both by the local microscopists and by the device. Careful review of those samples found that the errors were caused by large trophozoites that resembled P. vivax instead of P. falciparum. One sample at AKUH was assessed as a false-positive sample when the Parasight system result was compared to the PCR result, due to an extremely high level of Howell-Jolly bodies. Finally, while most of the correlation coefficients for the parasitemia levels were high (>0.8), the correlation coefficient for P. vivax in the AKUH experiment was only 0.55; this stemmed from the very small number of P. vivax samples included in the study (8) and the lack of familiarity of the microscopists with this strain, due to its rarity in the region. It is important to note also that these discrepancies might have emerged due to changes in cell morphology that occurred in >24-h-old samples. Additionally, future adaptions will be made in the algorithm to flag samples with high Howell-Jolly body levels for additional testing.
It is important to note that the high sensitivity and specificity values for microscopy at these sites do not reflect standard practices and results. Three expert microscopists reviewed all slides, with a minimum of 10 views per slide, while at most locations a single technician briefly reviews each slide. Therefore, the true sensitivity of microscopy in normal laboratory settings is closer to 500 parasites/μl, as cited elsewhere in the literature (20). The performance of the RDT at AKUH was in line with previously reported findings, while the RDT at Apollo Hospital showed superior accuracy, compared to prior results; this was also likely due to the highly controlled environment in which the test was performed, which is not reflective of standard field results. The RDT species identification at AKUH was very poor for P. vivax, but the RDT at Apollo Hospital performed well in species identification of both P. vivax and P. falciparum. This again shows great interbrand and interspecies variability among RDTs, which makes them difficult for clinicians to interpret.
Several updates are currently under development to improve the diagnostic performance of the Parasight system, due to the limitations of the current study. The current device has a limit of detection of 20 parasites/μl. Future versions of the algorithm will provide detection capabilities that could be as low as 5 parasites/μl, by better differentiating between malaria parasites and objects with similar morphological features (such as Howell-Jolly bodies). Additionally, the device is currently unable to distinguish between P. vivax and Plasmodium ovale, as few P. ovale samples have been collected to train the algorithm. While this is less problematic clinically, as the two forms of malaria infections require the same medication regimens, distinguishing between the two is important for epidemiological considerations. With a significant number of operational devices, we will be able to build a large enough library of P. ovale samples to enable the classifier to distinguish the species.
Taken together, these findings indicate that the Parasight platform demonstrates improved accuracy, compared with our previous prototype devices, and provides significant value for the malaria diagnostic community. The distribution of the device at strategic locations will lead to improved disease treatment and screening, ultimately resulting in an expedited roadmap to eradication.
MATERIALS AND METHODS
Study design.
The study was a two-center, prospective, blinded trial conducted with blood samples from patients with clinically suspected malaria, at Apollo Hospital (Chennai, India) (n = 205) and at Aga Khan University Hospital (Nairobi, Kenya) (n = 263). Ethics committee approval was obtained for the use of all samples.
Sample collection.
Determination of eligibility for malaria treatment was based solely on the standard diagnostic protocols of the clinics and the patient treatment courses and was not altered due to the study or results from the Sight Diagnostics diagnostic device. At all locations, venous blood samples were collected in EDTA-containing Vacutainer tubes and were analyzed by RDTs, microscopy, and the Parasight device within 48 h after collection.
RDTs.
For the purpose of comparison, RDTs were performed with all samples. At Apollo Hospital, the Alere Trueline test was used for RDT analysis, according to the manufacturer's directions. At AKUH, the SD Bioline test was used for RDT analysis, according to the manufacturer's directions.
Microscopy.
At both locations, thin-smear microscopy was performed independently by three expert microscopists, who agreed upon a final diagnosis. Parasite counts were performed via light microscopy for all samples in a standardized manner, representing ideal (rather than typical) conditions. The microscopists analyzed at least 10 fields at ×100 magnification, with approximately 100 RBCs being counted per field. Parasitemia levels were calculated as the ratio of infected RBCs to total RBCs. Gametocytes were not included in the final parasite counts.
PCR.
At Apollo Hospital and at AKUH, all samples were reviewed by real-time PCR according to a previously described protocol (19). Briefly, real-time PCR was performed with Fast Sybr Green master mix in a volume of 10 μl (Applied Biosystems), to identify P. falciparum, P. vivax, and Plasmodium in general. All reactions were performed in 384-well quantitative PCR plates (Bio-Rad) in a CFX384 real-time PCR system (Bio-Rad).
Sight Diagnostics device analysis.
At both locations, digital imaging was carried out onsite with the commercially available Parasight platform. Five microliters of each blood sample was mixed with 500 μl of a fluorescent dye solution, and the sample was loaded into a plastic cartridge. The cartridge was then incubated at room temperature for 10 min, during which time the cells formed a monolayer. The cartridge was inserted into the device and scanned with three different light-emitting diode (LED) light sources (370 nm, 475 nm, and 530 nm). The total scan time per sample was 4 min and the device held up to 30 samples, which could be loaded in a batch or individually. Approximately 800 images were scanned per sample, and the algorithm processed individual images in real time. If a sample was incorrectly prepared, an error resulted and the process was repeated. Errors could result from too little or too much blood used in sample preparation, dirt on the cartridge, or improper preparation of the working stain solution. Computer vision and statistical models were used to determine infection status, parasitemia levels, and species.
Analysis.
Both microscopy and PCR were used as a basis for comparison for the results. In many settings, microscopy is considered more accurate than PCR, as it identifies live intracellular parasites and is not subject to errors caused by circulating DNA from ablated parasites. Therefore, sensitivity and specificity were calculated in comparison with microscopy and PCR independently. In all calculations for which the comparison standard is not noted, PCR is used as the standard for comparison.
ACKNOWLEDGMENTS
Y.E., A.H.-Y., H.B., N.L., S.L.-S., C.L.C., J.J.P., D.G., and S.J.S. developed and designed the system. M.S., M.C., J.S., H.S., and P.S. performed experiments in India. Z.P., C.M., Z.N., S.O., and D.M. performed experiments in Africa.
Y.E., A.H.-Y., H.B., N.L., S.L.-S., C.L.C., J.J.P., D.G., and S.J.S. declare competing financial interests as employees of Sight Diagnostics.
REFERENCES
- 1.UNITAID. 2014. Malaria diagnostics technology and market landscape, 2nd ed UNITAID, Geneva, Switzerland. [Google Scholar]
- 2.Kiszewski A, Johns B, Schapira A, Delacollette C, Crowell V, Tan-Torres T, Ameneshewa B, Teklehaimanot A, Nafo-Traore F. 2007. Estimated global resources needed to attain international malaria control goals. Bull World Health Organ 85:623–630. doi: 10.2471/BLT.06.039529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dal-Bianco MP, Köster KB, Kombila UD, Kun JF, Grobusch MP, Ngoma GM, Matsiegui PB, Supan C, Salazar CL, Missinou MA, Issifou S, Lell B, Kremsner P. 2007. High prevalence of asymptomatic Plasmodium falciparum infection in Gabonese adults. Am J Trop Med Hyg 77:939–942. [PubMed] [Google Scholar]
- 4.Klein EY, Smith DL, Boni MF, Laxminarayan R. 2008. Clinically immune hosts as a refuge for drug-sensitive malaria parasites. Malar J 7:67. doi: 10.1186/1475-2875-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan J, Li N, Wei X, Li P, Zhao Z, Wang L. 2013. Performance of two rapid diagnostic tests for malaria diagnosis at the China-Myanmar border area. Malar J 12:73. doi: 10.1186/1475-2875-12-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arya SC, Agarwal N. 2013. Laboratory tests for malaria: a diagnostic conundrum? S Afr Med J 103:701. doi: 10.7196/SAMJ.7433. [DOI] [PubMed] [Google Scholar]
- 7.Harchut K, Standley C, Dobson A, Klaassen B, Rambaud-Althaus C, Althaus F, Nowak K. 2013. Over-diagnosis of malaria by microscopy in the Kilombero Valley, Southern Tanzania: an evaluation of the utility and cost-effectiveness of rapid diagnostic tests. Malar J 12:159. doi: 10.1186/1475-2875-12-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fançony C, Sebastião YV, Pires JE, Gamboa D, Nery SV. 2013. Performance of microscopy and RDTs in the context of a malaria prevalence survey in Angola: a comparison using PCR as the gold standard. Malar J 12:284. doi: 10.1186/1475-2875-12-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reyburn H, Mbakilwa H, Mwangi R, Mwerinde O, Olomi R, Drakeley C, Whitty CJ. 2007. Rapid diagnostic tests compared with malaria microscopy for guiding outpatient treatment of febrile illness in Tanzania: randomised trial. BMJ 334:403. doi: 10.1136/bmj.39073.496829.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ansah EK, Narh-Bana S, Epokor M, Akanpigbiam S, Quartey AA, Gyapong J, Whitty CJ. 2010. Rapid testing for malaria in settings where microscopy is available and peripheral clinics where only presumptive treatment is available: a randomised controlled trial in Ghana. BMJ 340:c930. doi: 10.1136/bmj.c930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar A, Chery L, Biswas C, Dubhashi N, Dutta P, Dua VK, Kacchap M, Kakati S, Khandepakar A, Kour D, Mahajan SN, Maji A, Majumder P, Mohanta J, Mohaptra PK, Narayanasamy K, Roy K, Shastri J, Valecha N, Vikash R, Wani R, White J, Rathod PK. 2012. Malaria in South Asia: prevalence and control. Acta Trop 121:246–255. doi: 10.1016/j.actatropica.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mouatcho JC, Goldring JP. 2013. Malaria rapid diagnostic tests: challenges and prospects. J Med Microbiol 62:1491–1505. doi: 10.1099/jmm.0.052506-0. [DOI] [PubMed] [Google Scholar]
- 13.Alonso PL, Tanner M. 2013. Public health challenges and prospects for malaria control and elimination. Nat Med 19:150–155. doi: 10.1038/nm.3077. [DOI] [PubMed] [Google Scholar]
- 14.Hopkins H, González IJ, Polley SD, Angutoko P, Ategeka J, Asiimwe C, Agaba B, Kyabayinze DJ, Sutherland CJ, Perkins MD, Bell D. 2013. Highly sensitive detection of malaria parasitemia in a malaria-endemic setting: performance of a new loop-mediated isothermal amplification kit in a remote clinic in Uganda. J Infect Dis 208:645–652. doi: 10.1093/infdis/jit184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coleman RE, Sattabongkot J, Promstaporm S, Maneechai N, Tippayachai B, Kengluecha A, Pachapaew N, Zollner G, Miller RS, Vaughan JA, Thimasarn K, Khuntirat B. 2006. Comparison of PCR and microscopy for the detection of asymptomatic malaria in a Plasmodium falciparum/vivax endemic area in Thailand. Malar J 5:121. doi: 10.1186/1475-2875-5-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaewkamnerd S, Uthaipibull C, Intarapanich A, Pannarut M, Chaotheing S, Tongsima S. 2012. An automatic device for detection and classification of malaria parasite species in thick blood film. BMC Bioinformatics 13(Suppl 17):S18. doi: 10.1186/1471-2105-13-S17-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vink JP, Laubscher M, Vlutters R, Silamut K, Maude RJ, Hasan MU, De Haan G. 2013. An automatic vision-based malaria diagnosis system. J Microsc 250:166–178. doi: 10.1111/jmi.12032. [DOI] [PubMed] [Google Scholar]
- 18.Srivastava B, Anvikar AR, Ghosh SK, Mishra N, Kumar N, Houri-Yafin A, Pollak JJ, Salpeter SJ, Valecha N. 2015. Computer-vision-based technology for fast, accurate and cost effective diagnosis of malaria. Malar J 14:526. doi: 10.1186/s12936-015-1060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houri-Yafin A, Eshel Y, Lezmy N, Larbi B, Wypkema E, Dayanand V, Levy-Schreier S, Cohen CL, Pollak JJ, Salpeter SJ. 2016. An enhanced computer vision platform for clinical diagnosis of malaria. Malar Control Elimin 5:138. [Google Scholar]
- 20.Milne LM, Chiodini PL, Warhurst DC. 1994. Accuracy of routine laboratory diagnosis of malaria in the United Kingdom. J Clin Pathol 47:740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]