ABSTRACT
As one of the first lines of host defense, monocytes play important roles in clearing infected microbes. The defensive response is triggered by recognition of diverse microbial moieties, including released factors, which modulate host immune responses to establish a harsh environment for clinically important bacterial pathogens. In this study, we found that the expression of PTX3, a soluble form of pattern recognition receptor, was induced by infection with live Pseudomonas aeruginosa or treatment of cells with its supernatant. P. aeruginosa GroEL, a homolog of heat shock protein 60, was identified as one of the factors responsible for inducing the expression of PTX3 in host cells. GroEL induced PTX3 expression by activating the Toll-like receptor 4 (TLR4)-dependent pathway via nuclear factor-kappa B (NF-κB), while simultaneously inhibiting expression of microRNA-9, which targets the PTX3 transcript. Finally, by acting as an opsonin, GroEL-induced PTX3 promoted the association and phagocytosis of Staphylococcus aureus into macrophages. These data suggest that the host defensive environment is supported by the production of PTX3 in response to GroEL, which thus has therapeutic potential for clearance of bacterial infections.
KEYWORDS: GroEL, microRNA-9, NF-κB, Pseudomonas aeruginosa, PTX3
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic bacterial pathogen that causes acute and chronic respiratory infections in patients with underlying conditions, such as immunodeficiency, cystic fibrosis, or ventilator-associated pneumonia (1). The morbidity and mortality of P. aeruginosa infection are influenced by the presence of coinfecting microbes such as Staphylococcus aureus, a well-known human pathogen that has acquired resistance to the majority of clinically used antibiotics (2). To clear such clinically important bacterial pathogens from infection sites, host inflammatory responses sense diverse pathogen-associated molecular patterns (PAMPs) via pattern recognition receptors (PRRs), including Toll-like receptors (TLRs) (3). Macrophages expressing TLRs play important roles in the induction of innate immune responses, as well as in subsequent stimulation of adaptive immune responses (4). Host immune responses are also modulated by recognition of released microbial moieties, which promote survival or development of intricate competition between pathogens in the same infection sites. Together, these observations imply that the microbial moieties responsible for immune modulation could have therapeutic potential for the control of bacterial infections.
Heat shock proteins (HSPs) are chaperonin proteins that facilitate proper assembly of unfolded polypeptides (5). HSP60, a cytosolic protein that is released into extracellular spaces as a damage-associated molecular pattern, promotes inflammatory responses (6). The Escherichia coli homolog of HSP60, GroEL, is essential for cell viability at all temperatures (7) and stimulates the production of proinflammatory cytokines interleukin-1β (IL-1β) and IL-6 in human monocytes (8, 9). Given that HSP60 is involved in promoting the immune responses, bacterial GroEL has the potential to stimulate the production of inflammatory cytokines and adhesion molecules (8, 9), likely by acting as a PAMP to modulate immune responses.
Pentraxins, which are humoral soluble PRRs of the innate immune responses, are classified into two categories, short and long (10). PTX3, a long pentraxin, is produced and released by multiple cell types, including monocytes, dendritic cells, and endothelial cells, in response to diverse stimuli, such as inflammatory cytokines and microbial moieties; this process is mediated by TLRs (11). PTX3 promotes immune responses by producing inflammatory cytokines in monocytes and macrophages (11). In addition, based on its capacity for opsonization and phagocytosis, PTX3 is widely recognized as an important molecule for the clearance of damaged tissue, certain fungal pathogens such as Aspergillus fumigatus, and bacterial pathogens such as P. aeruginosa (12, 13). PTX3 has a therapeutic effect against P. aeruginosa-mediated lung infection (13), but it is unclear whether P. aeruginosa can promote production of PTX3 (and, if so, what the underlying molecular mechanism is).
In this study, we investigated the role of P. aeruginosa in upregulating expression of PTX3 in human monocytes. GroEL released into the culture supernatant of P. aeruginosa stimulated the expression of PTX3 via the TLR4 and NF-κB signaling pathways. Interestingly, GroEL also suppressed the expression of a microRNA, miR-9, which is associated with the stability of the PTX3 transcript. PTX3 induced by P. aeruginosa-derived GroEL facilitated phagocytosis of S. aureus. Overall, our results suggest that GroEL has the potential to promote innate immune responses against clinically important bacterial pathogens by modulating the production of PTX3.
RESULTS
Supernatants of P. aeruginosa induce the expression of PTX3.
Previous studies demonstrated the therapeutic potential of PTX3 in P. aeruginosa lung infection (13). However, it remains unclear whether P. aeruginosa is capable of inducing PTX3 expression. To determine whether expression of PTX3 is induced by P. aeruginosa infection, we treated THP-1 human monocyte cells with P. aeruginosa strain PAO1 at a multiplicity of infection (MOI) of 5 or 10. As shown in Fig. 1A, PTX3 expression was significantly elevated in response to the treatment, indicating that P. aeruginosa is a potent inducer of this gene. We next investigated whether expression would be stimulated by treatment with supernatants obtained from stationary-phase cultures of PAO1. PTX3 expression gradually increased in a time- and a dose-dependent manner in response to the supernatants, reaching a maximum at 4 h (Fig. 1B). To determine whether the ability to induce PTX3 is a general feature of P. aeruginosa strains, we compared supernatants obtained from cultures of P. aeruginosa strains PAK, PA103, and PA14. As shown in Fig. 1C, all supernatants tested were able to induce PTX3 expression, suggesting that the bacterial factors contributing to expression of PTX3 are produced and released from all P. aeruginosa strains.
FIG 1.
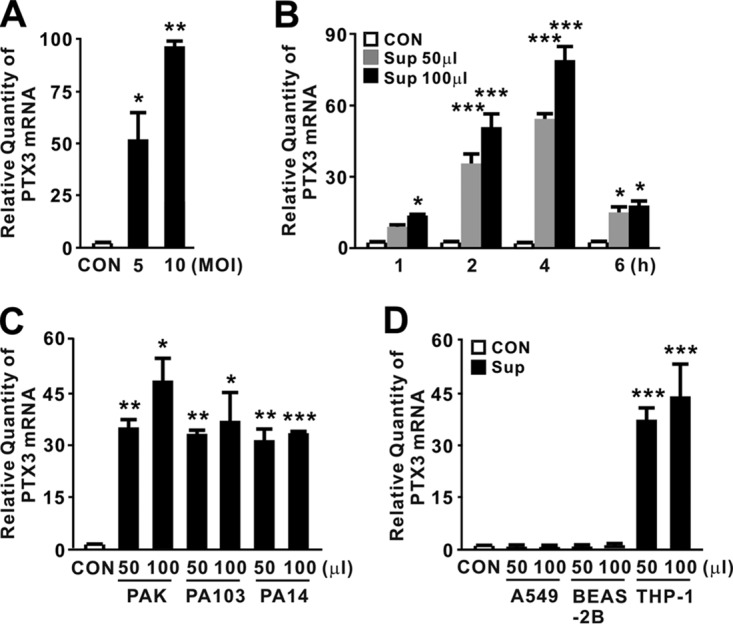
P. aeruginosa supernatant-mediated induction of PTX3. (A) THP-1 cells were treated with P. aeruginosa strain PAO1 at an MOI of 5 or 10 for 4 h. (B) THP-1 cells were treated for the indicated times with 50 or 100 μl of culture supernatants (Sup) obtained from PAO1 cultivated in MMA. (C) THP-1 cells were treated for 4 h with 50 or 100 μl of Sup obtained from P. aeruginosa strains PAK, PA103 and PA14 cultivated in MMA. (D) A549, BEAS-2B, and THP-1 cells were treated for 4 h with 50 or 100 μl of Sup obtained from PAO1 cultivated in MMA. After treatment, PTX3 mRNA levels were quantitated by qRT-PCR. Data in panels A to D are expressed as means ± the standard deviations (SD; n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus CON. MOI, multiplicity of infection. PBS (A) or MMA (B to D) was used as a control (CON).
During a respiratory tract infection, P. aeruginosa encounters epithelial cells and monocytes, which represent the first lines of host defense (1). Therefore, we investigated whether the expression of PTX3 in response to bacterial secreted factors would occur in both cell types. As shown in Fig. 1D, elevated expression of the PTX3 was clearly observed in THP-1 cells but not in A549 and BEAS-2B human lung epithelial cells. Taken together, these observations demonstrate that P. aeruginosa potently induces expression of PTX3 in monocytes and that the responsible molecules are released into the culture supernatant.
P. aeruginosa GroEL is involved in the induction of PTX3.
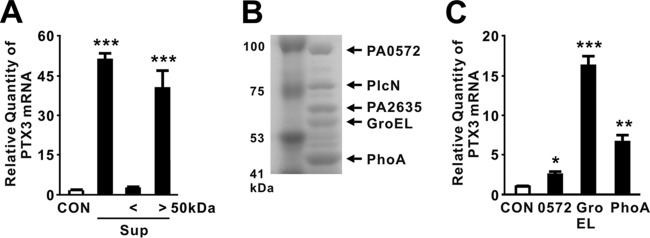
To identify the PTX3-inducing molecules in the culture supernatant of P. aeruginosa, we fractionated the bacterial supernatant by filtration through a centrifugal membrane with a 50-kDa cutoff. The resultant fractions were used to treat THP-1 cells, and their ability to induce PTX3 was compared to that of the original culture supernatant. As shown in Fig. 2A, fractions containing molecules larger than 50 kDa induced the expression of PTX3. Separation of molecules larger than 50 kDa by SDS-PAGE identified five major protein bands (Fig. 2B), which we further analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry. The proteins were identified as PlcN (nonhemolytic phospholipase C precursor), GroEL, PhoA (alkaline phosphatase), and two hypothetical proteins (PA0572 and PA2635). To evaluate their potential to induce expression of PTX3, these proteins (but not PlcN) were cloned, expressed, purified, and used individually to treat THP-1 cells. We were unable to purify PA2635 due to the formation of inclusion bodies. In these assays, GroEL was identified as the most active inducer because it increased expression to a greater extent than the other candidates (Fig. 2C).
FIG 2.
P. aeruginosa-derived GroEL is involved in induction of PTX3. (A) THP-1 cells were treated for 4 h with 100 μl of culture supernatants (Sup) obtained from PAO1 cultivated in MMA or size fractionated on a membrane filter with a 50-kDa cutoff. (B) Concentrated filtrates of P. aeruginosa cultures grown in MMA were loaded onto SDS-PAGE gels. (C) THP-1 cells were treated with expressed and purified protein candidates for 4 h. After treatment, PTX3 mRNA levels were measured by qRT-PCR (A and C). Data in panels A and C are expressed as means ± the SD (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus CON (A and C). MMA (A) or a control extract (C) was used as a control (CON).
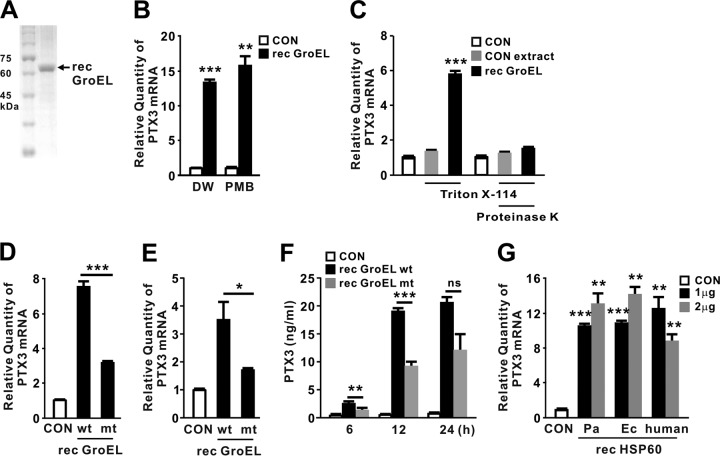
Because the recombinant protein was produced in E. coli, lipopolysaccharide (LPS) and nonspecific E. coli-derived proteins might have been introduced during the purification procedure, thereby possibly contributing to induction of PTX3 (although the GroEL protein preparations did not contain many nonspecific proteins) (Fig. 3A). To eliminate this possibility, recombinant GroEL protein was pretreated with the LPS inhibitor polymyxin B (PMB) and Triton X-114. In addition, we used E. coli harboring an empty vector to obtain a control extract, which was generated using the same purification procedure and used as a control alongside proteinase K treatment. As shown in Fig. 3B and C, treatment with PMB and Triton X-114 did not affect the induction ability of recombinant GroEL. In addition, Triton X-114-pretreated control extract did not induce expression of PTX3, suggesting that LPS and nonspecific proteins play no role in induction. This was further supported by application of proteinase K-treated recombinant GroEL, which resulted in a clear reduction in expression. Therefore, we concluded that GroEL plays a central role in induction and used the control extract and PMB treatment to examine the effect of recombinant GroEL throughout the study.
FIG 3.
Purified recombinant GroEL is sufficient in induction of PTX3. (A) Recombinant (rec) GroEL was purified from E. coli and loaded onto SDS-PAGE gels. (B) THP-1 cells were treated for 4 h with recombinant P. aeruginosa GroEL (1 μg/ml) that was itself pretreated with 20 μg of PMB/ml. DW, distilled water. (C) THP-1 cells were treated with either a control extract or recombinant GroEL (both pretreated with Triton X-114). (D and E) THP-1 (D) or U937 (E) cells were treated with 1 μg/ml of either wild-type recombinant P. aeruginosa GroEL (wt) or mutant (mt; D398A) protein for 4 h. (F) THP-1 cells were treated with 1 μg of either recombinant P. aeruginosa GroEL wt or mt protein/ml for the indicated periods of time. (G) THP-1 cells were treated for 4 h with 1 or 2 μg of recombinant HSP60 derived from P. aeruginosa (Pa), E. coli (Ec), or human. After treatment, the PTX3 mRNA levels were quantitated by qRT-PCR (B to E and G) and the level of PTX3 protein released from THP-1 cells was measured by ELISA analysis (F). Data in panels B to G are expressed as means ± the SD (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus CON (B, C, and G) and versus rec GroEL wt (D to F). PBS (C) or a control extract (B, D to G) was used as a control (CON).
ATP is required in the folding cycle of chaperonin (14), and the ATP hydrolysis-defective mutant GroEL (D398A) protein was less able to induce expression of PTX3 (Fig. 3D), implying that a particular folded structure of GroEL plays a role in induction of PTX3 expression. A similar result was also obtained in human monocyte U937 cells (Fig. 3E). Furthermore, the fold induction of PTX3 production was smaller in response to mutant GroEL (D398A) than intact GroEL protein (Fig. 3F). Because GroEL is a highly conserved protein that plays an important role as an intercellular signal in innate immune responses (15), we investigated whether recombinant HSP60 proteins derived from E. coli and human could also induce PTX3 as efficiently as the P. aeruginosa homolog. As shown in Fig. 3G, all recombinant proteins tested induced PTX3 to similar extents. These findings suggest that GroEL induces expression of PTX3 following its release into the culture supernatant.
GroEL-induced expression of PTX3 is mediated by the TLR4 and NF-κB signaling pathways.
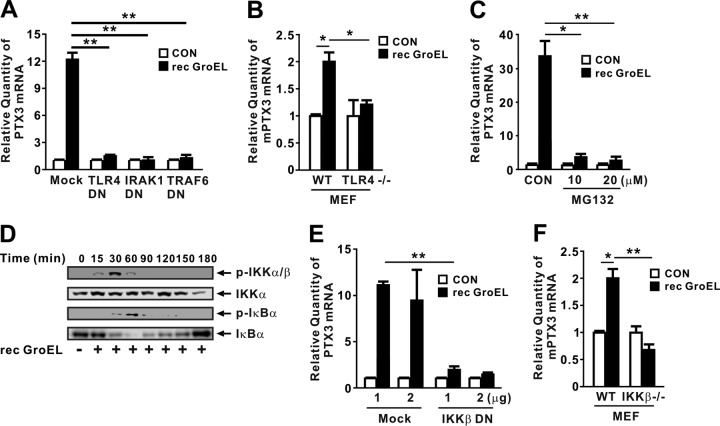
Given that HSP60 is a ligand of TLR4 (16), it is likely that GroEL induces PTX3 expression via TLR4 signaling. To investigate the involvement of the TLR4 pathway, we transfected THP-1 cells with dominant-negative (DN) variants that disrupt the activity of proteins involved in the TLR4 signaling cascade. As shown in Fig. 4A, cells transfected with DN variants of TLR4, IRAK1, or TRAF6 did not exhibit induction in response to recombinant GroEL, indicating that induction of PTX3 expression is under the control of the TLR4 pathway. The involvement of TLR4 was further confirmed by examining PTX3 expression in mouse embryonic fibroblast (MEF) Tlr4−/− cells (Fig. 4B). TLR4 leads to the activation of nuclear factor-κB (NF-κB) (17). To determine whether recombinant GroEL induces the transcription of PTX3 through the activation of NF-κB, THP-1 cells were pretreated with MG132, a chemical inhibitor of NF-κB activity, prior to treatment with recombinant GroEL. Expression of PTX3 was significantly reduced by pretreatment with MG132 (Fig. 4C), suggesting that NF-κB is involved in induction of GroEL-mediated PTX3 expression. We verified the activation of NF-κB in response to the treatment with recombinant GroEL by detecting phosphorylation of IKKα/β and IκBα, as well as the degradation of IκBα (Fig. 4D). The involvement of NF-κB was further confirmed by more specific approaches, i.e., transfection of an IKKβ DN overexpression construct (Fig. 4E) and IKKβ−/− MEFs (Fig. 4F), in which the absence of IKKβ protein was confirmed as previously reported (18). Taken together, these results suggest that recombinant GroEL induces the expression of PTX3 via the TLR4 and NF-κB signaling pathways.
FIG 4.
GroEL-induced expression of PTX3 is mediated by the TLR4/NF-κB signaling pathways. (A) Overexpression of dominant-negative (DN) TLR4, IRAK1, and TRAF6 inhibited induction of PTX3 mRNA expression by recombinant P. aeruginosa GroEL (1 μg/ml) in THP-1 cells. (B) Recombinant P. aeruginosa GroEL (1 μg/ml) induced mouse PTX3 (mPTX3) gene transcription in wt MEFs but not in Tlr4−/− MEFs. (C) Pretreatment for 1 h with MG132 inhibited induction of PTX3 mRNA expression by recombinant P. aeruginosa GroEL (1 μg/ml) in THP-1 cells after 4 h of incubation. (D) Recombinant P. aeruginosa GroEL (1 μg/ml) potently induced phosphorylation of IKKα/β and IκBα, as well as degradation of IκBα, in THP-1 cells, as assessed by Western blotting. (E) Overexpression of DN IKKβ inhibited induction of PTX3 mRNA expression by recombinant P. aeruginosa GroEL (1 μg/ml) in THP-1 cells after 4 h of incubation. (F) Recombinant P. aeruginosa GroEL (1 μg/ml) induced Ptx3 transcription in wt MEFs, but not in IKKβ−/− MEFs, after 4 h of incubation. The PTX3 mRNA was measured by qRT-PCR. Data in panels A to C and panels E and F are expressed as means ± the SD (n = 3). Data in panel D are representative of three separate experiments. *, P < 0.05; **, P < 0.01 versus Mock (A and E), CON (B, C, and F), or wt (B and F). A control extract (A to C, E, and F) was used as a control (CON).
GroEL-induced PTX3 expression is mediated by downregulation of miR-9.
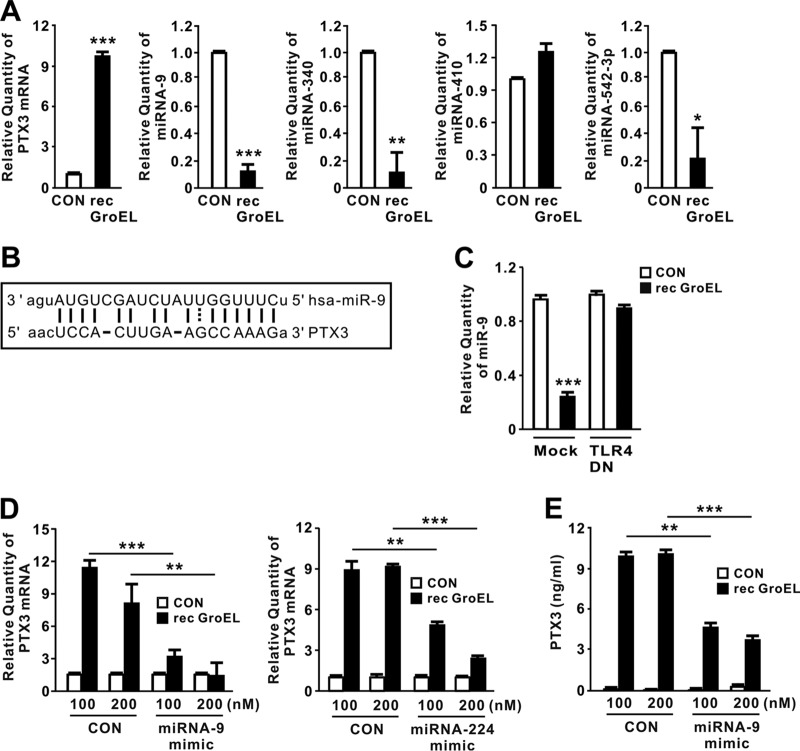
MicroRNAs (miRNAs) are small noncoding RNAs engaged in posttranscriptional control of gene expression (19). Recent work showed that miR-224 is involved in downregulation of PTX3 production (20). To expand our knowledge of miRNAs involved in GroEL-mediated PTX3 induction, we used a target prediction tool (http://www.microrna.org/microrna/home.do) to obtain a list of miRNA candidates (miR-9, miR-542-3p, miR-181d, miR-410, miR-204, miR-211, miR-340, miR-224, and miR-29b) that aligned with PTX3 mRNA. We then performed miRNA qRT-PCR analysis to examine the expression levels of each candidate in response to GroEL. This analysis revealed that levels of miR-9, miR-340, and miR-542-3p decreased after GroEL treatment, whereas levels of the remaining miRNAs were statistically insignificant (Fig. 5A). As predicted by the target prediction tool, miR-9, which showed the most significant decrease, contains a highly conserved binding site within PTX3 mRNA; an alignment is provided in Fig. 5B. Next, we investigated whether miR-9 expression in response to recombinant GroEL treatment is influenced by the TLR4 pathway. As shown in Fig. 5C, cells transfected with TLR4 DN did not show reduced miR-9 expression, indicating that the reduction in miR-9 expression is under the control of the TLR4 pathway. To verify the biological effects of miR-9 on PTX3 expression, we next transfected cells with miR-9 mimics. Since miR-224 targets the transcript of PTX3 (20, 21), we transfected cells with miR-224 mimics as a positive control. Similar to the inhibitory effect observed for miR-224 mimics, miR-9 mimics abrogated recombinant GroEL-mediated induction of PTX3 in a dose-dependent manner (Fig. 5D and E), supporting the idea that GroEL-mediated downregulation of miR-9 plays an important role in inducing PTX3 expression.
FIG 5.
GroEL-induced PTX3 expression is mediated by downregulation of miR-9. (A) THP-1 cells were treated with recombinant P. aeruginosa GroEL (1 μg/ml) for 4 h. After treatment, levels of PTX3 mRNA, miRNA-9, miRNA-340, miRNA-410, and miRNA-542-3p were measured by qRT-PCR. (B) miR-9 was aligned with PTX3 mRNA (predicted by a target prediction tool [http://www.microrna.org/microrna/home.do]). (C) Overexpression of TLR4 DN in THP-1 cells did not reduce miR-9 expression in response to recombinant P. aeruginosa GroEL (1 μg/ml). (D and E) THP-1 cells were transfected with either miRNA-9 or miRNA-224 mimic. At 24 h posttransfection, the transfected cells were treated with recombinant P. aeruginosa GroEL (1 μg/ml) for either 4 h (D) or 24 h (E). After treatment, the PTX3 mRNA level was measured by qRT-PCR (D), and the level of PTX3 protein in supernatant was measured by ELISA (E). Data in panels A, C, D, and E are expressed as means ± the SD (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus CON (A, C to E). A control extract (A, C to E) was used as a control (CON).
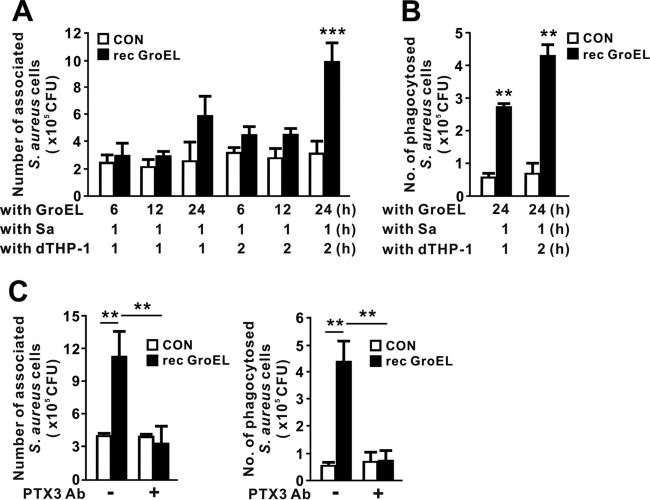
GroEL-mediated production of PTX3 enhances the phagocytosis of S. aureus.
P. aeruginosa and S. aureus are the leading causes of nosocomial infections, and their drug resistance profiles are an increasing cause for concern in the medical community (22). Although PTX3 is essential for protection against P. aeruginosa (13, 23), its role against S. aureus remains unclear. Given that recombinant GroEL was capable of inducing PTX3, we next investigated the effect of GroEL-induced production of PTX3 in clearance of S. aureus via phagocytosis. We first treated THP-1 cells with recombinant GroEL for 6, 12, or 24 h to obtain conditioned media, which were assumed to contain secreted PTX3, and then incubated S. aureus with the conditioned media for 1 h to allow opsonization of bacteria by PTX3. We then transferred opsonized S. aureus to dishes containing activated macrophage differentiated THP-1 (dTHP-1) cells, incubated the samples for 1 or 2 h, and counted the S. aureus organisms associated with and phagocytosed into cells. When we used conditioned media from cells treated with GroEL for 24 h, macrophage-associated bacteria appeared, and their number increased with incubation time (Fig. 6A). Consistent with this, the number of bacteria phagocytosed by dTHP-1 cells increased in a time-dependent manner (Fig. 6B). This observation suggests that facilitation of phagocytosis could be mediated by an increase of opsonization via PTX3 induced by GroEL treatment. To determine whether the bacterial association and phagocytosis were specifically caused by the PTX3, we used a PTX3-specific polyclonal antibody to neutralize the PTX3. As shown in Fig. 6C, the addition of antibody clearly decreased the numbers of associated and phagocytosed S. aureus organisms, suggesting that the increase in phagocytosis was mediated by secreted PTX3.
FIG 6.
GroEL-mediated production of PTX3 promotes phagocytosis of S. aureus. (A and B) THP-1 cells were pretreated with recombinant P. aeruginosa GroEL (1 μg/ml) for 6, 12, or 24 h. After pretreatment, supernatants were collected and incubated with S. aureus (Sa) for 1 h. The supernatants containing S. aureus were transferred onto PMA-induced differentiated THP-1 (dTHP-1) cells and incubated for 1 or 2 h. The numbers of associated (A) or phagocytosed (B) S. aureus organisms were calculated by counting CFU as described in Materials and Methods. (C) THP-1 cells were pretreated with recombinant P. aeruginosa GroEL (1 μg/ml) for 24 h. After pretreatment, supernatants were mixed with 4 μg of anti-PTX3 polyclonal antibody (Ab)/ml for 1 h and then incubated with S. aureus for 1 h. The supernatants containing S. aureus were transferred onto PMA-induced dTHP-1 cells and incubated for 2 h. The numbers of associated or phagocytosed S. aureus organisms were calculated by counting the CFU. Data in panels A to C are expressed as means ± the SD (n = 3). **, P < 0.01; ***, P < 0.001 versus CON (A to C) or Mock (C). A control extract (A to C) was used as a control (CON).
DISCUSSION
PTX3, a nonredundant soluble PRR, plays an important role in preventing pulmonary infection by promoting P. aeruginosa clearance through opsonization followed by facilitation of phagocytosis (13). In this study, in response to P. aeruginosa infection, PTX3 expression increased in a dose-dependent manner (Fig. 1A), suggesting that microbial moieties are involved in induction. Upregulation of PTX3 is induced by proinflammatory cytokines, such as interleukin-1β (IL-1β) and tumor necrosis factor alpha (TNF-α), as well as microbial moieties, such as outer membrane component LPS and the mycobacterial cell wall component lipoarabinomannan (24–27). However, the contribution of soluble molecules from P. aeruginosa to induction remained unclear. As shown in Fig. 1B, supernatants obtained from cultures of P. aeruginosa increased PTX3 expression in an incubation time-dependent manner, indicating the presence of potent soluble molecules in the culture supernatants. Although the induction of PTX3 was observed in diverse cell types, including A549 and BEAS-2B lung epithelial cells, in response to IL-1β and TNF-α (24), supernatant-mediated induction was observed only in THP-1 monocytes and not in the epithelial cells (Fig. 1D). This indicates that regulation of PTX3 expression in diverse cell types depends on specific stimuli; the observations here support the notion that monocytes are the major cell type in which expression of PTX3 is induced in response to components in the culture supernatant.
P. aeruginosa expresses numerous virulence factors, including membrane-associated PAMPs such as LPS and flagella, as well as soluble molecules, such as extracellular secreted molecules and effector molecules translocated into host cells through the type III secretion system (1). Size fractionation and proteomic analysis identified GroEL, a homolog of HSP60 that facilitates protein folding, as a potent inducing molecule in the culture supernatant. Therefore, we examined the ability of recombinant GroEL protein to induce expression of PTX3 after excluding the effects of contaminants such as LPS and nonspecific E. coli-derived proteins. To neutralize the effect of LPS, the preparations were first treated with 20 μg of PMB/ml; the concentration of PMB could be sufficient based on previous results showing that PTX3 expression induced by PMB-treated GroEL was much higher than that induced by the PMB-treated control extract (Fig. 3B). In addition, treatment of cells with Triton X-114-pretreated recombinant GroEL still resulted in expression of PTX3 (Fig. 3C). Triton X-114 pretreatment eliminated most of the residual LPS present in the purified proteins; an LAL endotoxin assay (Pierce Thermo, Rockford, IL) revealed contamination levels of 0.07 U/ml. Furthermore, the fact that a mutant GroEL (D398A) protein obtained using the same purification procedure had a weaker effect than the wild-type protein supports the idea that GroEL itself, rather than contaminating LPS, was responsible for the induction of PTX3 (Fig. 3D to F). Therefore, we conclude that GroEL is indeed involved in inducing the expression of PTX3.
GroEL is located primarily in the cytosolic fraction, but we detected it in the extracellular fraction of P. aeruginosa, an observation consistent with a previous report that identified GroEL in an extracellular fraction by two-dimensional gel electrophoresis analysis (28). Interestingly, HSP60 homologs have no leader sequence or other recognizable motifs to direct their secretion into extracellular space. Thus, these proteins are nonclassical secreted proteins (29). The mechanisms by which P. aeruginosa releases GroEL have yet to be elucidated. Given that the human ortholog HSP60 induced PTX3 expression (Fig. 3G), we postulated that induction of PTX3 expression could be mediated by cytotoxic damage, which causes release of the HSP60. However, we did not observe any signs of cytotoxicity in response to treatment with supernatants or recombinant GroEL in a lactate dehydrogenase (LDH) release assay.
GroEL requires ATP binding and hydrolysis activities to function properly as a chaperonin (30). Our data suggest that GroEL is involved in the induction of PTX3 expression and that its ATP hydrolysis activity is at least partly required for this induction based on results obtained with a GroEL D398A protein defective in ATP hydrolysis but not ATP binding (14) (Fig. 3D to F). This suggests that the mutation impairing ATP hydrolysis causes a slight conformational change in GroEL and that this structural change makes GroEL less effective at inducing PTX3 expression. In addition to aspartate 398, a previous report suggests that aspartate 52 is also important for ATP hydrolysis (14). Therefore, we expect that GroEL (D52A/D398A) double mutants could be less effective at inducing PTX3 expression. However, the precise role of ATP hydrolysis in induction of PTX3 expression remains unclear. NCBI Protein BLAST revealed that the amino acid sequence of P. aeruginosa-derived GroEL shares 51% identity with human-derived HSP60, encoded by HSPD1, and 80% with E. coli-derived GroEL, encoded by groL. All of these homologs were confirmed to induce PTX3 (Fig. 3G). Similarly, human and chlamydial HSP60 activate JNK and IKK signaling via TLR2 and TLR4 (31), implying that the underlying mechanisms of PTX3 induction are shared by all HSP60 homologs.
Although TLR4 is a PRR involved in sensing of the membrane-associated moiety LPS of Gram-negative bacteria (32), we observed that GroEL could also signal through TLR4 to induce expression of PTX3 (Fig. 4A and B). GroEL derived from Porphyromonas gingivalis upregulates the expression of PTX3 via TLR4 signaling (33), and TLR4-deficient mice exhibit reduced expression of PTX3 (17). A549 and BEAS-2B cells have lower levels of TLR4 activity (24, 34), which could lead to lower levels of PTX3 expression. NF-κB also acts as a key transcription regulator that initiates inflammatory responses during airway infections (35). As expected, expression of PTX3 was controlled by the NF-κB signaling cascade (Fig. 4C to F). Consistent with this, studies show that a regulatory region within the human PTX3 promoter contains an NF-κB-binding site (36) and that P. gingivalis GroEL can stimulate the transcriptional activity of NF-κB (33). AKT activation is also involved in PTX3 expression, as determined by pretreatment of cells with the chemical inhibitor LY294002 (data not shown), an observation consistent with a previous report demonstrating that PI3K/AKT is essential for PTX3 activation (37). According to a previous report that IL-1 signaling phosphorylates AKT, followed by activation of downstream NF-κB signaling (37), GroEL could activate the AKT signaling cascade, which subsequently activates the NF-κB pathway. We also investigated whether mitogen-activated protein kinases (MAPKs) are involved by pretreatment of chemical inhibitors, but the results revealed that MAPKs have no significant effect on PTX3 induction (data not shown).
miRNAs play major roles in posttranscriptional regulation in many biological systems (38, 39). miRNAs function by directly binding to the 3′ untranslated region (3′UTR) of specific target mRNAs, thereby suppressing protein expression (20, 21). By applying a target prediction tool (http://www.microrna.org/microrna/home.do), we obtained a list of miRNA candidates and ultimately identified miR-9. Transfection of cells with a miR-9 mimic reduced the level of PTX3 transcripts (as observed for a miR-224 mimic) (Fig. 5D and E), whereas transfection with a miR-340 mimic did not (data not shown), even though its expression was also clearly reduced in response to GroEL (as shown in Fig. 5A). miR-224 targets the transcript of PTX3, and the expression of miR-224 is inhibited by transforming growth factor β1 treatment (20, 21). However, GroEL did not show significant suppression of miR-224 expression (data not shown).
To promote clearance of infected microbes, hosts must have mechanisms for modulation of innate immunity in response to microbial moieties. Here, we show that P. aeruginosa-derived GroEL can increase the production of PTX3, which contributes to host innate immune responses by protecting against certain infections. GroEL treatment induced PTX3 expression as early as 1 h posttreatment. This peaked by 4 h, before gradually decreasing again, indicating that responses may occur relatively quickly. GroEL is recognized by TLR4, resulting in phosphorylation of IKKαβ and subsequent phosphorylation of IκBα via the TLR signaling cascade (Fig. 4D). The resulting free NF-κB transcription factors move to the nucleus and increase expression of PTX3 at the mRNA level. In response to GroEL, the level of miRNA-9, which targets the PTX3 transcript, is decreased as early as 4 h posttreatment (Fig. 5A), implying that GroEL is sufficient to stabilize the PTX3 transcript by reducing the activity of miRNA-9. PTX3, produced as a soluble recognition receptor, was released and showed effects after as little as 6 h of incubation (Fig. 6A). During initial infection of the airway, P. aeruginosa may be recognized by macrophages via interaction with membrane-associated PAMPs such as LPS. In addition to known PAMPs, the immune system stimulates effective defense responses by detecting diverse secreted factors such as GroEL. In this way, the immune system can respond to fluctuations in the number of bacterial pathogens. Moreover, P. aeruginosa and S. aureus are the leading causes of nosocomial infections, and their intrinsic and acquired antibiotic resistance is of increasing concern to the medical community (22) because the pathogens are becoming more difficult to treat. The potential therapeutic effect of PTX3 is essential for protection against P. aeruginosa (13). Here, we demonstrated that PTX3 is effective against S. aureus (Fig. 6). The findings imply that GroEL might be a useful therapeutic tool against such infections. In addition, a detailed understanding of the regulatory mechanisms underlying GroEL-induced production of PTX3, especially GroEL-mediated suppression of miR-9, is an important goal for the future.
MATERIALS AND METHODS
Reagents.
MG132 and PMB were purchased from A.G. Scientific (San Diego, CA) and Sigma-Aldrich (St. Louis, MO), respectively.
Bacterial strains and culture conditions.
P. aeruginosa wild-type (wt) strains (PAO1, PAK, PA103, and PA14) (40–42) and an S. aureus wt strain (ATCC 25923) were used in this study. Unless specified otherwise, PAO1 was used to treat cells. P. aeruginosa was grown in Luria broth rich medium (yeast extract, 0.5%; tryptone, 1%; and NaCl, 1%; all wt/vol) or minimal medium A [MMA; 1.05% K2HPO4, 0.45% KH2PO4, 0.1% (NH4)2SO4, 0.05% sodium citrate·2H2O, 0.845% glutamate, and 1% glycerol (all wt/vol percentages except for glycerol, which was vol/vol)] at 37°C. S. aureus was cultured in tryptic soy broth (Difco) medium at 37°C. Whole bacterial cells cultured in MMA were harvested at 10,000 × g for 20 min at 4°C to obtain the supernatant (Sup) and the pellet after overnight incubation. The bacterial culture supernatant of P. aeruginosa cultivated in MMA was filtered through either a low-protein-binding 0.2-μm-pore-size membrane (Corning Star, Cambridge, MA) to completely remove the bacteria or a low-protein-binding 50-kDa-pore-size Unltracel-50 membrane (EMD Millipore, Darmstadt, Germany) for size fractionation. For the preparation of live bacteria, the bacterial pellet was suspended in Dulbecco phosphate-buffered saline (HyClone, Rockford, IL).
Cell culture.
All media described below were supplemented with 10% heat-inactivated fetal bovine serum (FBS; HyClone, Rockford, IL), penicillin (100 U/ml), and streptomycin (0.1 mg/ml). A549 cells (human alveolar epithelial cells), THP-1 cells (human monocytes), and U937 cells (human monocytes) were cultured in Roswell Park Memorial Institute 1640 (RPMI 1640; HyClone, Rockford, IL). wt MEFs, IκB kinase β-deficient (IKKβ−/−) MEFs, and BEAS-2B (human bronchial epithelial) cells were cultured in Dulbecco modified Eagle medium (HyClone, Rockford, IL). Unless specified otherwise, THP-1 cells were exposed to bacteria for 4 h at various MOIs or recombinant protein for 4 h at 1 μg/ml. Cells were maintained at 37°C in a humidified 5% CO2 air-jacketed incubator.
Construction of His-tagged GroEL plasmid and purification of recombinant protein.
The coding regions of groEL (PA4385), groL, and HSPD1 loci were amplified from genomic DNA derived from P. aeruginosa strain PAO1 and E. coli strain W3110 and from cDNA from human THP-1 cells, respectively, by PCR using the following primers containing restriction enzyme recognition sites for BamHI, SacI, and HindIII: P. aeruginosa groEL, 5′-GCCGGATCCTATGGCTGCCAAAGAAGTTAAG-3′ and 5′-CCGGAGCTCTCCAACCACAGGGGCCGG-3′; E. coli groL, 5′-CAGGAGCTCAATGGCAGCTAAAGACGTAAAATTCG-3′ and 5′-CAGAAGCTTTTGTTTATTTCTGCGAGGTGCAG-3′; and human HSPD1, 5′-CCGGAGCTCAATGCTTCGGTTACCCACAG-3′ and 5′-CCGAAGCTTGGGCTTCCTGTCACAGTTC-3′ (restriction sites are underlined). The resultant 1.6-, 1.6-, and 1.7-kb PCR products were cloned into a pETDuet-1 vector (Novagen, Germany), and the construction was confirmed by sequence analysis with pET Upstream primer (69214-3) and DuetDOWN1 primer (71179-3). To stimulate the expression of recombinant GroEL protein in E. coli strain DE3-BL21, bacteria were cultivated for 16 h at 20°C in the presence of IPTG (isopropyl-β-d-thiogalactopyranoside) and then harvested prior to suspension in phosphate-buffered saline (PBS). The bacterial cell suspension was sonicated on ice 600 times at 150 W for 1 s at 5-s intervals using a Digital Sonifier (Branson Ultrasonics, Danbury, CT). Residual intact cells were removed by centrifugation at 10,000 × g for 30 min at 4°C. The bacterial lysate was incubated with cobalt resin (Sigma-Aldrich) at 4°C for 1 h and then washed three times with 10 mM imidazole in PBS, followed by elution with 200 mM imidazole in PBS. To obtain a control extract, E. coli strain DE3-BL21 harboring a pETDuet-1 vector was subjected to the same procedures. The control extract was used to evaluate the effect of recombinant GroEL throughout the study. The recombinant protein and control extract were treated with PMB prior to use. GroEL proteins were quantitated by using a protein assay (Bio-Rad, Hercules, CA). Purity was further confirmed using a Coomassie blue-stained SDS-polyacrylamide gel. In addition, LPS was removed by phase-separation treatment with Triton X-114 as described previously (43), and the amount of residual LPS was examined using an LAL chromogenic endotoxin quantitation kit.
Transfection of plasmids and miRNA mimic.
Expression plasmids for IKKβ dominant negative (DN), TLR4 DN, IL-1 receptor-associated kinases 1 (IRAK1) DN, and TNF receptor-associated factor 6 (TRAF6) DN were described previously (44). The plasmids were prepared using an Endo-Free Plasmid Maxi kit (Qiagen, Valencia, CA), and miRNA mimic was purchased in AccuTarget (Bioneer, South Korea). Cells were transfected using a neon electroporation system (Invitrogen, Carlsbad, CA). Briefly, 5 × 106 cells in 100 μl of R buffer were electroporated in 100-μl tips with 6 or 12 μg of plasmid and plated into 1 ml of RPMI 1640 supplemented with 10% FBS in a 12-well plate. The electroporation parameters of THP-1 were as follows: 1,400 V, 20 mA, and two pulses. At 24 h posttransfection, 106 cells per ml were plated into each well of a 12-well plate (45, 46).
Quantitative real-time PCR for PTX3 mRNA.
Total RNA was isolated using TRIzol reagent (Invitrogen, Grand Island, NY). cDNA was synthesized from total RNA using a ReverTra Ace qRT-PCR kit (Toyobo, Japan). Quantitative reverse transcription-PCR (qRT-PCR) was performed using SYBR green PCR master mix (Kapa Biosystems, Woburn, MA). The primer sequences were as follows: human PTX3 (5′-TTGGACAACGAAATAGACAATGGA-3′ and 5′-GTCGTCCGTGGCTTGCA-3′) and mouse Ptx3 (5′-GTCGTCGGTGGCCTGCA-3′ and 5′-TTGGACAACGAAATAGACAATGGA-3′). Reactions were performed on a CFX96 real-time PCR system (Bio-Rad) under the following thermal conditions: stage 1, 50°C for 2 min and 95°C for 10 min, and stage 2, 95°C for 15 s and 60°C for 1 min. Stage 2 was repeated for 40 cycles. The relative quantities of PTX3 mRNA were calculated using the comparative threshold cycle (CT) method and normalized against glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The primer sequences were as follows: human GAPDH (5′-CCCTCCAAAATCAAGTGG-3′ and 5′-CCATCCACAGTCTTCTGG-3′) and mouse Gapdh (5′-CTCATGACCACAGTCCATGC-3′ and 5′-CACATTGGGGGTAGGAACAC-3′).
Quantitative real-time PCR for miRNA.
Total RNA was isolated using the mirVana kit (Invitrogen). Expression of microRNA (miRNA) was examined as described previously (47). Briefly, 1 μg of total RNA was polyadenylated for 1 h at 37°C using a polyadenylation kit (Invitrogen). cDNA was synthesized from polyadenylated RNA using primer RT1 (5′-GCGAGCACAGAATTAATACGACTCCTGGGCAATTTTTTTTTTTTVN-3′) and the ReverTra Ace qRT-PCR kit (Toyobo, Japan). Quantitative reverse transcription-PCR (qRT-PCR) was performed using SYBR green PCR master mix. Primer sequences were as follows: miR-9, 5′-TCTTTGGTTATCTAGCTGTATGA-3′; miR-340, 5′-TTATAAAGCAATGAGACTGATT-3′; miR-410, 5′-AATATAACACAGATGGCCTGT-3′; miR-542-3p, 5′-TGTGACAGATTGATAACTGAAA-3′; and universal primer RACE, 5′-GCGAGCACAGAATTAATACGAC-3′. Reactions were performed on a CFX96 real-time PCR system. The relative quantities of miR-9 were calculated using the comparative CT method and normalized against U6 (5′-ATGACACGCAAATTCGTGAAGC-3′) and RACE (rapid amplification of cDNA ends).
Immunoblotting analysis.
Cells were lysed on ice for 10 min in 20 mM Tris-HCl (pH 7.4), 50 mM NaCl, 50 mM sodium pyrophosphate, 30 mM NaF, 5 μM zinc chloride, 2 mM iodoacetic acid, and 1% Triton X-100 in distilled water supplemented with 1 mM phenylmethylsulfonyl fluoride (Thermo Scientific) and 0.1 mM sodium orthovanadate (Sigma-Aldrich). The lysates were centrifuged at 10,000 × g for 15 min at 4°C, and the protein concentration was measured using the bicinchoninic acid method (Pierce, Rockford, IL). Proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Membranes were blocked in TBS (10 mM Tris-HCl [pH 7.5], 150 mM NaCl) with 5% nonfat dry milk for 1 h and then incubated for 16 h at 4°C with primary antibodies against IκBα, phosphorylated IκBα (p-IκBα), IKKα, and p-IKKα/β (Cell Signaling Technology, Danvers, MA). Blots were washed and incubated with appropriate secondary antibodies and visualized using WEST-ZOLplus chemiluminescent substrate (Intron, South Korea) on an ImageQuant LAS 4000 (GE Healthcare Life Sciences).
Enzyme-linked immunosorbent assay (ELISA).
The amount of PTX3 released into supernatants was assayed using a Quantikine Human Pentraxin 3/TSG-14 immunoassay kit (R&D Systems, Minneapolis, MN).
CFU quantitation of associated and phagocytosed bacteria.
To obtain differentiated THP-1 (dTHP-1) cells, THP-1 monocytes were seeded in a 12-well plate at 106 cells per well in the presence of phorbol 12-myristate 13-acetate (PMA; 5 ng/ml) (48). The treatment was continued at 37°C for 24 h in RPMI 1640 supplemented with serum, and then the cells were washed and incubated for 4 h in serum-free RPMI 1640. To collect supernatants from recombinant GroEL-treated THP-1 cells, the cells were seeded in a 12-well plate at 106 cells per well, followed by incubation at 37°C for 4 h in serum-free RPMI 1640. The cultured cells were then treated with recombinant GroEL and incubated at 37°C for 24 h. Supernatants were collected by centrifugation and filtered through a low-protein-binding 0.2-μm-pore-size membrane (Corning Star) to remove the THP-1 cells. To neutralize the effect of PTX3 present in supernatants, the supernatants were treated with anti-PTX3 antibody (4 μg/ml; R&D Systems) for 1 h at room temperature. For analysis of the associated and phagocytosed S. aureus, prepared supernatants were incubated with bacteria at an MOI of 10 for 1 h and then transferred to dTHP-1 cells, followed by incubation for an additional 1 or 2 h. To determine the number of S. aureus organisms, the treated dTHP-1 cells were washed five times with 1 ml of PBS to remove nonadherent bacteria and then incubated with 1 ml of 0.2% Triton X-100 in PBS for 10 min. To determine the number of phagocytosed S. aureus organisms, dTHP-1 cells were treated with 1 ml of serum-free RPMI 1640 supplemented with 200 μg of gentamicin/ml for 1 h to remove membrane-associated S. aureus. After a washing step, the cells were incubated with 1 ml of 0.2% Triton X-100 in PBS for 10 min. The number of associated or phagocytosed S. aureus organisms was calculated from the CFU counts.
Statistical analysis.
Statistical analyses were performed with Student t test or one-way analysis of variance, followed by Tukey's post hoc multiple-range test using the Instat package from GraphPad (GraphPad Software, Inc., San Diego, CA). A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
This study was supported by the Korea-China Joint Research Program (NRF-2014K1A3A1A20034794), the BK21 Plus Program of the Ministry of Education, and the Ministry of Science and Technology of China (2015DFG32500).
REFERENCES
- 1.Gellatly SL, Hancock RE. 2013. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 67:159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 2.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. 2013. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci U S A 110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Kawai T, Akira S. 2007. TLR signaling. Semin Immunol 19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Calderwood SK, Mambula SS, Gray PJ Jr. 2007. Extracellular heat shock proteins in cell signaling and immunity. Ann N Y Acad Sci 1113:28–39. doi: 10.1196/annals.1391.019. [DOI] [PubMed] [Google Scholar]
- 6.Quintana FJ, Cohen IR. 2011. The HSP60 immune system network. Trends Immunol 32:89–95. doi: 10.1016/j.it.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Fayet O, Ziegelhoffer T, Georgopoulos C. 1989. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J Bacteriol 171:1379–1385. doi: 10.1128/jb.171.3.1379-1385.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabona P, Reddi K, Khan S, Nair SP, Crean SJ, Meghji S, Wilson M, Preuss M, Miller AD, Poole S, Carne S, Henderson B. 1998. Homogeneous Escherichia coli chaperonin 60 induces IL-1β and IL-6 gene expression in human monocytes by a mechanism independent of protein conformation. J Immunol 161:1414–1421. [PubMed] [Google Scholar]
- 9.Galdiero M, de l'Ero GC, Marcatili A. 1997. Cytokine and adhesion molecule expression in human monocytes and endothelial cells stimulated with bacterial heat shock proteins. Infect Immun 65:699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garlanda C, Bottazzi B, Bastone A, Mantovani A. 2005. Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu Rev Immunol 23:337–366. doi: 10.1146/annurev.immunol.23.021704.115756. [DOI] [PubMed] [Google Scholar]
- 11.Bottazzi B, Doni A, Garlanda C, Mantovani A. 2010. An integrated view of humoral innate immunity: pentraxins as a paradigm. Annu Rev Immunol 28:157–183. doi: 10.1146/annurev-immunol-030409-101305. [DOI] [PubMed] [Google Scholar]
- 12.Garlanda C, Hirsch E, Bozza S, Salustri A, De Acetis M, Nota R, Maccagno A, Riva F, Bottazzi B, Peri G, Doni A, Vago L, Botto M, De Santis R, Carminati P, Siracusa G, Altruda F, Vecchi A, Romani L, Mantovani A. 2002. Nonredundant role of the long pentraxin PTX3 in anti-fungal innate immune response. Nature 420:182–186. doi: 10.1038/nature01195. [DOI] [PubMed] [Google Scholar]
- 13.Moalli F, Paroni M, Veliz Rodriguez T, Riva F, Polentarutti N, Bottazzi B, Valentino S, Mantero S, Nebuloni M, Mantovani A, Bragonzi A, Garlanda C. 2011. The therapeutic potential of the humoral pattern recognition molecule PTX3 in chronic lung infection caused by Pseudomonas aeruginosa. J Immunol 186:5425–5434. doi: 10.4049/jimmunol.1002035. [DOI] [PubMed] [Google Scholar]
- 14.Koike-Takeshita A, Mitsuoka K, Taguchi H. 2014. Asp-52 in combination with Asp-398 plays a critical role in ATP hydrolysis of chaperonin GroEL. J Biol Chem 289:30005–30011. doi: 10.1074/jbc.M114.593822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin FY, Hsiao FP, Huang CY, Shih CM, Tsao NW, Tsai CS, Yang SF, Chang NC, Hung SL, Lin YW. 2014. Porphyromonas gingivalis GroEL induces osteoclastogenesis of periodontal ligament cells and enhances alveolar bone resorption in rats. PLoS One 9:e102450. doi: 10.1371/journal.pone.0102450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim SC, Stice JP, Chen L, Jung JS, Gupta S, Wang Y, Baumgarten G, Trial J, Knowlton AA. 2009. Extracellular heat shock protein 60, cardiac myocytes, and apoptosis. Circ Res 105:1186–1195. doi: 10.1161/CIRCRESAHA.109.209643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian J, Guo X, Liu XM, Liu L, Weng QF, Dong SJ, Knowlton AA, Yuan WJ, Lin L. 2013. Extracellular HSP60 induces inflammation through activating and up-regulating TLRs in cardiomyocytes. Cardiovasc Res 98:391–401. doi: 10.1093/cvr/cvt047. [DOI] [PubMed] [Google Scholar]
- 18.Shin HS, Ha UH. 2011. Up-regulation of human bradykinin B1 receptor by secreted components of Pseudomonas aeruginosa via a NF-κB pathway in epithelial cells. FEMS Immunol Med Microbiol 63:418–426. doi: 10.1111/j.1574-695X.2011.00868.x. [DOI] [PubMed] [Google Scholar]
- 19.Ambros V. 2004. The functions of animal microRNAs. Nature 431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 20.Yao G, Liang M, Liang N, Yin M, Lu M, Lian J, Wang Y, Sun F. 2014. MicroRNA-224 is involved in the regulation of mouse cumulus expansion by targeting Ptx3. Mol Cell Endocrinol 382:244–253. doi: 10.1016/j.mce.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 21.Rudnicki A, Shivatzki S, Beyer LA, Takada Y, Raphael Y, Avraham KB. 2014. microRNA-224 regulates pentraxin 3, a component of the humoral arm of innate immunity, in inner ear inflammation. Hum Mol Genet 23:3138–3146. doi: 10.1093/hmg/ddu023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowy FD. 2003. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest 111:1265–1273. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaillon S, Mancuso G, Hamon Y, Beauvillain C, Cotici V, Midiri A, Bottazzi B, Nebuloni M, Garlanda C, Fremaux I, Gauchat JF, Descamps P, Beninati C, Mantovani A, Jeannin P, Delneste Y. 2013. Prototypic long pentraxin PTX3 is present in breast milk, spreads in tissues, and protects neonate mice from Pseudomonas aeruginosa lung infection. J Immunol 191:1873–1882. doi: 10.4049/jimmunol.1201642. [DOI] [PubMed] [Google Scholar]
- 24.Han B, Mura M, Andrade CF, Okutani D, Lodyga M, dos Santos CC, Keshavjee S, Matthay M, Liu M. 2005. TNFα-induced long pentraxin PTX3 expression in human lung epithelial cells via JNK. J Immunol 175:8303–8311. doi: 10.4049/jimmunol.175.12.8303. [DOI] [PubMed] [Google Scholar]
- 25.Doni A, Peri G, Chieppa M, Allavena P, Pasqualini F, Vago L, Romani L, Garlanda C, Mantovani A. 2003. Production of the soluble pattern recognition receptor PTX3 by myeloid, but not plasmacytoid, dendritic cells. Eur J Immunol 33:2886–2893. doi: 10.1002/eji.200324390. [DOI] [PubMed] [Google Scholar]
- 26.Imamura M, Kawasaki T, Savchenko AS, Ohashi R, Jiang S, Miyamoto K, Ito Y, Iwanari H, Sagara M, Tanaka T, Hamakubo T, Kodama T, Uchiyama M, Naito M. 2007. Lipopolysaccharide induced expression of pentraxin 3 in human neutrophils and monocyte-derived macrophages. Cell Immunol 248:86–94. doi: 10.1016/j.cellimm.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Vouret-Craviari V, Matteucci C, Peri G, Poli G, Introna M, Mantovani A. 1997. Expression of a long pentraxin, PTX3, by monocytes exposed to the mycobacterial cell wall component lipoarabinomannan. Infect Immun 65:1345–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nouwens AS, Willcox MD, Walsh BJ, Cordwell SJ. 2002. Proteomic comparison of membrane and extracellular proteins from invasive (PAO1) and cytotoxic (6206) strains of Pseudomonas aeruginosa. Proteomics 2:1325–1346. doi:. [DOI] [PubMed] [Google Scholar]
- 29.Bendtsen JD, Kiemer L, Fausboll A, Brunak S. 2005. Non-classical protein secretion in bacteria. BMC Microbiol 5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clare DK, Saibil HR. 2013. ATP-driven molecular chaperone machines. Biopolymers 99:846–859. doi: 10.1002/bip.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vabulas RM, Ahmad-Nejad P, da Costa C, Miethke T, Kirschning CJ, Hacker H, Wagner H. 2001. Endocytosed HSP60s use Toll-like receptor 2 (TLR2) and TLR4 to activate the toll/interleukin-1 receptor signaling pathway in innate immune cells. J Biol Chem 276:31332–31339. doi: 10.1074/jbc.M103217200. [DOI] [PubMed] [Google Scholar]
- 32.Ospelt C, Gay S. 2010. TLRs and chronic inflammation. Int J Biochem Cell Biol 42:495–505. doi: 10.1016/j.biocel.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Argueta JG, Shiota S, Yamaguchi N, Masuhiro Y, Hanazawa S. 2006. Induction of Porphyromonas gingivalis GroEL signaling via binding to Toll-like receptors 2 and 4. Oral Microbiol Immunol 21:245–251. doi: 10.1111/j.1399-302X.2006.00286.x. [DOI] [PubMed] [Google Scholar]
- 34.Perros F, Lambrecht BN, Hammad H. 2011. TLR4 signaling in pulmonary stromal cells is critical for inflammation and immunity in the airways. Respir Res 12:125. doi: 10.1186/1465-9921-12-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatada EN, Krappmann D, Scheidereit C. 2000. NF-κB and the innate immune response. Curr Opin Immunol 12:52–58. doi: 10.1016/S0952-7915(99)00050-3. [DOI] [PubMed] [Google Scholar]
- 36.Basile A, Sica A, d'Aniello E, Breviario F, Garrido G, Castellano M, Mantovani A, Introna M. 1997. Characterization of the promoter for the human long pentraxin PTX3: role of NF-κB in tumor necrosis factor-alpha and interleukin-1beta regulation. J Biol Chem 272:8172–8178. [DOI] [PubMed] [Google Scholar]
- 37.Chang WC, Wu SL, Huang WC, Hsu JY, Chan SH, Wang JM, Tsai JP, Chen BK. 2015. PTX3 gene activation in EGF-induced head and neck cancer cell metastasis. Oncotarget 6:7741–7757. doi: 10.18632/oncotarget.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Connell RM, Rao DS, Baltimore D. 2012. microRNA regulation of inflammatory responses. Annu Rev Immunol 30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 39.Zhang B, Pan X, Cobb GP, Anderson TA. 2007. microRNAs as oncogenes and tumor suppressors. Dev Biol 302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 40.Ha U, Jin S. 2001. Growth phase-dependent invasion of Pseudomonas aeruginosa and its survival within HeLa cells. Infect Immun 69:4398–4406. doi: 10.1128/IAI.69.7.4398-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holloway BW. 1955. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol 13:572–581. [DOI] [PubMed] [Google Scholar]
- 42.Kaufman MR, Jia J, Zeng L, Ha U, Chow M, Jin S. 2000. Pseudomonas aeruginosa mediated apoptosis requires the ADP-ribosylating activity of ExoS. Microbiology 146:2531–2541. doi: 10.1099/00221287-146-10-2531. [DOI] [PubMed] [Google Scholar]
- 43.Aida Y, Pabst MJ. 1990. Removal of endotoxin from protein solutions by phase separation using Triton X-114. J Immunol Methods 132:191–195. doi: 10.1016/0022-1759(90)90029-U. [DOI] [PubMed] [Google Scholar]
- 44.Ha U, Lim JH, Jono H, Koga T, Srivastava A, Malley R, Pages G, Pouyssegur J, Li JD. 2007. A novel role for IκB kinase (IKK) alpha and IKKβ in ERK-dependent up-regulation of MUC5AC mucin transcription by Streptococcus pneumoniae. J Immunol 178:1736–1747. doi: 10.4049/jimmunol.178.3.1736. [DOI] [PubMed] [Google Scholar]
- 45.Khare S, Dorfleutner A, Bryan NB, Yun C, Radian AD, de Almeida L, Rojanasakul Y, Stehlik C. 2012. An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity 36:464–476. doi: 10.1016/j.immuni.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plank TD, Whitehurst JT, Kieft JS. 2013. Cell type specificity and structural determinants of IRES activity from the 5′ leaders of different HIV-1 transcripts. Nucleic Acids Res 41:6698–6714. doi: 10.1093/nar/gkt358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reichenstein I, Aizenberg N, Goshen M, Bentwich Z, Avni YS. 2010. A novel qPCR assay for viral encoded microRNAs. J Virol Methods 163:323–328. doi: 10.1016/j.jviromet.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 48.Park EK, Jung HS, Yang HI, Yoo MC, Kim C, Kim KS. 2007. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm Res 56:45–50. doi: 10.1007/s00011-007-6115-5. [DOI] [PubMed] [Google Scholar]