ABSTRACT
MicroRNAs (miRNAs) are short, noncoding RNAs involved in the regulation of several processes associated with inflammatory diseases and infection. Bacterial infection modulates miRNA expression to subvert any innate immune response. In this study we analyzed, using microarray analysis, the bacterial modulation of miRNAs in bone marrow-derived macrophages (BMMs) in which activity was induced by infection with Porphyromonas gingivalis. The expression of several miRNAs was modulated 3 h postinfection (at a multiplicity of infection of 25). A bioinformatic analysis was performed to further identify pathways related to the innate immune host response under the influence of selected miRNAs. To assess the effects of the miRNAs identified on cytokine secretion (tumor necrosis factor alpha [TNF-α] and interleukin-10 [IL-10]), BMMs were transfected with selected miRNA mimics and inhibitors. Transfection with mmu-miR-155 and mmu-miR-2137 did not modify TNF-α secretion, while their inhibitors increased it. Inhibitors of mmu-miR-2137 and mmu-miR-7674 increased the secretion of the anti-inflammatory factor IL-10. In P. gingivalis-infected BMMs, mmu-miR-155-5p significantly decreased TNF-α secretion while inhibitor of mmu-miR-2137 increased IL-10 secretion. In vivo, in a mouse model of P. gingivalis-induced calvarial bone resorption, injection of mmu-miR-155-5p or anti-mmu-miR-2137 reduced the size of the lesion significantly. Furthermore, anti-mmu-miR-2137 significantly reduced inflammatory cell infiltration, osteoclast activity, and bone loss. Bioinformatic analysis demonstrated that pathways related to cytokine- and chemokine-related pathways but also osteoclast differentiation may be involved in the effects observed. This study contributes further to our understanding of P. gingivalis-induced modulation of miRNAs and their physiological effects. It highlights the potential therapeutic merits of targeting mmu-miR-155-5p and mmu-miR-2137 to control inflammation induced by P. gingivalis infection.
KEYWORDS: Porphyromonas gingivalis, inflammation, innate immunity, macrophages, miRNA
INTRODUCTION
Periodontitis (PD) is a common chronic inflammatory disease that can induce the destruction of tooth-supporting tissues (1, 2) and has been linked to several systemic diseases, such as cardiovascular disease (3, 4). The pathogenesis of PD is associated with a dysbiosis of the periodontal microbiota. This dysbiosis is characterized by a shift from a symbiotic microbial community to a pathogenic one that is composed mainly of anaerobic bacteria, resulting in alteration of the host-microbe cross talk (4). Porphyromonas gingivalis is a Gram-negative anaerobic bacterium that is considered a keystone pathogen (4). It produces several virulence factors such as lipopolysaccharide (LPS) and fimbriae in order to activate macrophages (5, 6) and invade different cell types, such as endothelial (7) and epithelial cells (8, 9). Several mechanisms, such as modulation of gene and protein expression, are used by P. gingivalis to compromise immune function at the periodontal level (1, 3). For instance, in epithelial cells, P. gingivalis, through its LPS, increases the secretion not only of several proinflammatory cytokines, including tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6), but also of anti-inflammatory cytokines such as IL-10, demonstrating the complexity of the cell's response to infection (10). These inflammatory processes and immune responses activated after bacterial infection are also responsible for tissue destruction.
Macrophages are pivotal cells in the host response because of their direct contact with infectious agents or their by-products (5, 11). These cells are often found in chronic-PD-diseased tissues (12) and represent an important fraction of the total inflammatory cell population (13). P. gingivalis is known to be able to invade macrophages (14) and to induce an important modulation of gene expression, resulting in the synthesis of several mRNAs related to signaling, apoptosis, cytokine receptors, and chemokine pathways (3, 6). Because posttranscriptional regulation may occur specifically through microRNAs (miRNAs), it is important to note that it is not known if these mRNA modulations have biological consequences.
Recognized as critical mediators in gene expression, miRNAs are short, noncoding RNAs involved in many physiological and pathological processes (1, 3, 15). In recent years, several roles for miRNAs in numerous inflammatory diseases, such as cardiovascular diseases, diabetes, obesity, arthritis, and PD, have been described (16). Partially complementary to the sequence of mRNAs in the 3′ untranslated region, miRNAs can downregulate gene expression at the posttranscriptional level (17). However, the identification of miRNA expression and the roles of miRNAs in inflammatory and infectious diseases are not fully understood. Analysis of the modulation of miRNA expression in infectious and inflammatory diseases may lead to the identification of new potential biomarkers and therapeutics (16).
In the context of PD, the impact of miRNA expression induced by infection with common periodontal pathogens such as P. gingivalis has been evaluated mostly in gingival tissue samples. Several miRNAs that can be differentially expressed in healthy and diseased tissues have been described (18). However, a precise correlation between specific pathogens and the different cell types involved in periodontal destruction has not been made.
The aims of this study were to identify miRNAs differentially expressed by macrophages in response to P. gingivalis infection and to assess their impact at the cytokine level. Furthermore, we wanted to evaluate, in vitro and in vivo, the ability of miRNAs with anti-inflammatory properties to reduce inflammatory effects associated with P. gingivalis infection. A bioinformatic pathway analysis revealed signal transduction targets associated with the most promising miRNAs.
RESULTS
miRNAs differentially expressed in control and infected cells.
To evaluate the effects of P. gingivalis infection on the miRNA expression profile in BMMs, the expression profile of 1,908 mouse miRNAs was subjected to microarray analysis. A total of 203 miRNAs showed differential expression with a false discovery rate-corrected P (q) value of <0.25, and clustering trends appeared between the groups (see Fig. S1 in the supplemental material). The analysis was restricted to the most differentially expressed miRNAs identified with higher-stringency parameters (a threshold of a ≥2- or ≤2-fold change and P and q values of ≤0.001). As a result, eight miRNAs were identified as differentially expressed between BMMs and the P. gingivalis-infected BMMs. Five miRNAs were upregulated (mmu-miR-7674-5p, mmu-miR-6975-5p, mmu-miR-155-5p, mmu-miR-347-3b, mmu-miR-347-3e), and three were downregulated (mmu-miR-2137, mmu-miR-8109, mmu-miR-211-3p) (Table 1). The results obtained with the microarray analysis were validated by reverse transcription (RT)-quantitative PCR (qPCR) for all of the miRNAs identified except mmu-miR-6975-5p and mmu-miR-8109 because of primer GC content and technical issues (Table 1).
TABLE 1.
Differential expression of miRNAs between BMMs infected with P. gingivalis and control
| miRNA differentially expressed upon P. gingivalis infectiona | Fold change |
|---|---|
| Upregulated | |
| mmu-miR-7674-5pb | 27.3 |
| mmu-miR-6975-5p | 4.4 |
| mmu-miR-3473bb | 2.7 |
| mmu-miR-3473eb | 2.7 |
| mmu-miR-155-5pb | 5.4 |
| Downregulated | |
| mmu-miR-8109 | −3.3 |
| mmu-miR-2137b | −3.2 |
| mmu-miR-211-3pb | −2.4 |
These genes were identified by a threshold of a ≥2- or ≤2-fold change and P and q values of ≤0.001.
miRNA expression validated by RT-qPCR.
Predicting the targets of differentially expressed miRNAs.
To assist in the selection of miRNAs with potential impact on the host response and identify the biological pathways that may be affected by these miRNAs, a bioinformatic analysis was performed. Therefore, after having analyzed the putative mRNA targets of the miRNAs differentially expressed in naive BMMs and P. gingivalis-infected BMMs, 3,796 potential target genes were predicted. The number of predicted target genes varied from 44 (mmu-miR-8109) to 1,262 (mmu-miR-6975-5p). Interactions of target genes were extracted from the STRING database, and the resulting protein-protein interaction (PPI) network was visualized with Cytoscape. To facilitate analysis, only proteins with at least two interactions with identified targets were considered. The network contains 6,746 nodes and 51,470 edges, where nodes represent target genes and edges represent interactions between target genes. Topological modules (densely connected regions) were identified by the Markov clustering (MCL) algorithm. A total of 639 modules were identified, 211 of which contained >5 nodes, and the largest module contained 613 nodes. Pathway analysis led to the identification of miRNA target genes in several pathways related to the inflammatory response, including chemokine signaling pathways and cytokine-cytokine receptor interactions (Table 2). Interestingly, even if several pathways related to a cell's life were identified, such as the cell cycle, ribosome, spliceosome, from the 10th largest identified topological modules, three of them were linked to pathways related to the inflammatory response highlighting the potential involvement of identified miRNAs in the modulation of the inflammatory response in P. gingivalis-infected BMMs.
TABLE 2.
Predicted pathways of the identified targets gene of selected miRNAsa
| Topological module and predicted pathways | Benjamini-corrected P value |
|---|---|
| 1 Olfactory transduction | 0.0E0 |
| 2 | |
| Neuroactive ligand-receptor interaction | 1.2E-107 |
| Taste transduction | 4.9E-36 |
| Calcium signaling pathway | 1.5E-19 |
| Chemokine signaling pathway | 2.7E-17 |
| Cytokine-cytokine receptor interaction | 4.3E-8 |
| Complement and coagulation cascades | 5.7E-6 |
| CyclicAMP signaling pathway | 6.6E-6 |
| 3 | |
| Ribosome | 1.2E-141 |
| Protein export | 1.3E-19 |
| 4 | |
| Cell cycle | 3.0E-86 |
| Progesterone-mediated oocyte maturation | 1.7E-25 |
| Oocyte meiosis | 1.7E-24 |
| HTLV-I infection | 1.8E-13 |
| p53 signaling pathway | 6.8E-12 |
| Viral carcinogenesis | 20.7E-11 |
| Ubiquitin-mediated proteolysis | 8.7E-11 |
| Small-cell lung cancer | 2.3E-9 |
| Hepatitis B | 1.2E-8 |
| FoxO signaling pathway | 3.5E-8 |
| Pathways in cancer | 2.3E-7 |
| 5 | |
| PPAR signaling pathway | 2.4E-21 |
| Thyroid hormone signaling pathway | 1.2E-19 |
| Adipocytokine signaling pathway | 3.6E-7 |
| Bile secretion | 4.3E-5 |
The predicted pathways of the five largest topological modules are presented. Only pathways with a Benjamini-corrected P value of <0.00001 were considered. Boldface type indicates pathways related to immune and inflammatory response.
Impact of identified miRNAs on TNF-α and IL-10 secretion in BMMs.
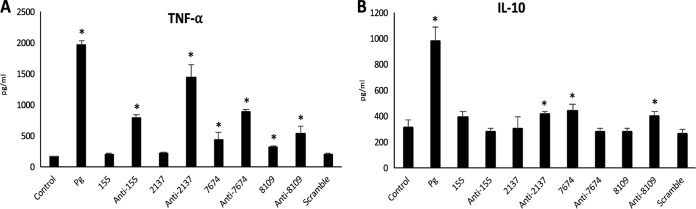
To evaluate the effects of the miRNAs identified on the inflammatory response, BMMs were transfected with mimics and inhibitors of the most differentially expressed miRNAs (mmu-miR-155-5p, mmu-miR-2137, mmu-miR-7674, mmu-miR-8109). This experiment was aimed at evaluating the specific effect of each miRNA to assess its role in the inflammatory response to P. gingivalis infection and especially in proinflammatory TNF-α and anti-inflammatory IL-10 secretion.
Transfection of BMMs with miR-155 and miR-2137 did not significantly modify TNF-α secretion, while their respective inhibitors significantly increased its secretion level. Regarding miR-7674 and miR-8109, transfection induced an increase in TNF-α to a lesser extent than transfection with the respective inhibitors (Fig. 1A).
FIG 1.
(A) TNF-α secretion in transfected BMMs with selected miRNAs and their inhibitors (*, P < 0.05). (B) IL-10 secretion in transfected BMMs with selected miRNAs and their inhibitors (*, P < 0.05). Pg, P. gingivalis.
Regarding IL-10 secretion, only a few of the miRNAs tested modified its secretion profile significantly. Mimics of mmu-miR-7674 and inhibitors of mmu-miR-2137 and mmu-miR-8109 induced a significant increase in IL-10 release in comparison with that in the control (P < 0.05) (Fig. 1B). Therefore, it appears that the miRNAs identified are able to modulate the secretion rate of these two cytokines.
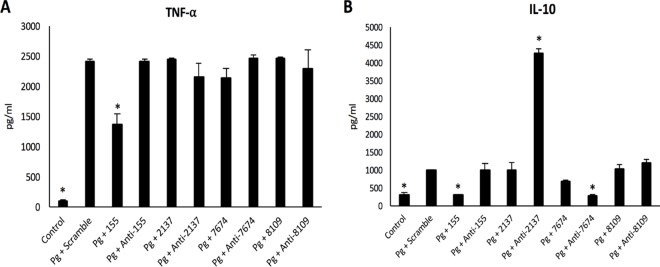
BMM transfection with selected miRNAs modulates cytokine secretion induced by P. gingivalis infection.
To evaluate if concomitant exposure to selected miRNAs is able to modulate a cell's response to P. gingivalis infection, transfected BMMs were infected and TNF-α/IL-10 secretion was evaluated. Interestingly, transfection with miR-155 significantly decreased TNF-α secretion by 30% (Fig. 2A). However, this effect was not observed with other mimics or inhibitors. Regarding IL-10 secretion, the transfection of anti-miR-2137 significantly increased IL-10 secretion (Fig. 2B) while miR-155 and anti-miR-7674 decreased it significantly (P < 0.05).
FIG 2.
(A) Dosages of TNF-α in supernatants of transfected BMMs infected with P. gingivalis (Pg) at 24 h. *, P < 0.05 versus P. gingivalis plus scramble. (B) Dosages of IL-10 in supernatants of transfected BMMs infected with P. gingivalis at 24 h. *, P < 0.05 versus P. gingivalis plus scramble.
Taken together, these data show that transfection with miRNAs, especially miR-155 or anti-miR-2137, modulates the host response to P. gingivalis infection.
Effects of miR-155 and anti-miR-2137 in vivo.
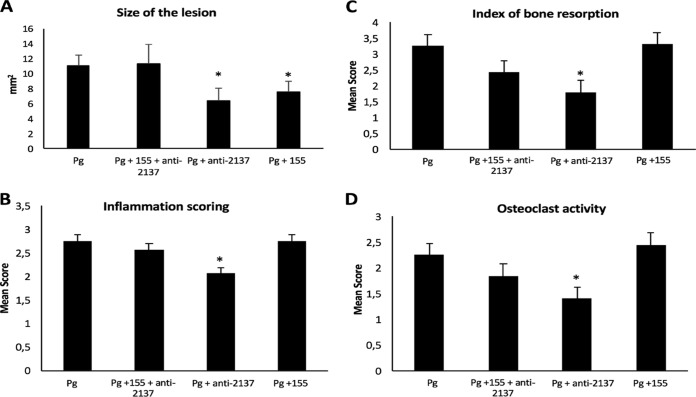
In this context, an in vivo proof-of-concept experiment was conducted to assess the potential effects of miR-155 and anti-miR-2137 in a mouse model of calvarial bone resorption induced by P. gingivalis infection. To determine their potential roles in the inflammatory response and bone resorption, selected miRNAs were injected concomitantly with P. gingivalis into mouse calvaria. After 7 days, the area of the induced lesion in each mouse was measured (Fig. 3A), and the mean area was 11.08 ± 1.47 mm2. The injection of miR-155 or anti-miR-2137 alone concomitantly with P. gingivalis significantly decreased the size of the lesion (7.55 ± 1.6 and 6.42 ± 1.4 mm2, respectively), demonstrating a modulation of the inflammatory process in vivo.
FIG 3.
(A) Sizes of calvarial lesions 7 days after P. gingivalis (Pg; 5 × 108 CFU) infection and the injection of miR-155 and anti-miR-2137. *, P < 0.05, n = 4. (B) Quantitative analysis of the index of ICI in each of the following groups: P. gingivalis-only infected, combination group (P. gingivalis and anti-miR-2137 plus miR-155), P. gingivalis and anti-miR-2137, and P. gingivalis and miR-155. *, P < 0.05. (C) Quantitative analysis of calvarial bone resorption in each of the following groups: P. gingivalis-only infected, combination group (P. gingivalis and anti-miR-2137 plus miR-155), P. gingivalis and anti-miR-2137, and P. gingivalis and miR-155. *, P < 0.05. (D) Quantitative analysis of osteoclast activity in each of the following groups: P. gingivalis-only infected, combination group (P. gingivalis and anti-miR-2137 plus miR-155), P. gingivalis and anti-miR-2137, and P. gingivalis and miR-155. *, P < 0.05.
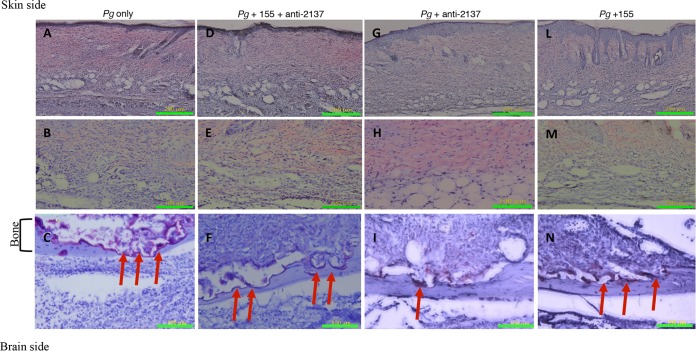
Histological examinations showed some degree of inflammatory cell infiltration (ICI; leukocytes, lymphocytes, and macrophages) in all of the groups (Fig. 4) in comparison with untreated mice (see Fig. S2). However, animals infected with P. gingivalis and treated with anti-miR-2137 showed a reduced amount of inflammation (Fig. 4G and H), in comparison with the P. gingivalis-infected group (Fig. 4A and B), supported by our quantitative analysis (Fig. 3B). P. gingivalis infection induced bone resorption (Fig. 4C) correlated with increased osteoclast activity, as evidenced by resorption lacunae. When anti-miR-2137 was injected concomitantly with the infection, a significant reduction of calvarial bone loss and osteoclast activity (Fig. 3C and D) was observed, while mice injected with miR-155 did not show significantly reduced ICI, bone loss, or osteoclast activity. However, while no significant reduction of ICI was observed in the combination group, a trend toward reduction of bone loss and osteoclast activity was observed (Fig. 3C and D).
FIG 4.
Histological sections. Shown are representative samples of skin and underlying calvarial bone at the middle of the lesion from each of the following groups: P. gingivalis (Pg) (A to C), combination (P. gingivalis and miR-155 plus anti-miR-2137) (D to F), P. gingivalis and anti-miR-2137 (G to I), and P. gingivalis and miR-155 (L to N). TRAP staining of bone (bottom row, ×200 magnification) and hematoxylin-and-eosin staining of skin (top row, ×100 magnification; middle row, ×200 magnification) are shown. Arrows indicate TRAP-stained multinucleated osteoclasts attached to bone and bone resorption lacunae.
Identification of potential pathways targeted by mmu-miR-155 and mmu-miR-2137.
As some biological effects have been observed in vitro and in vivo, a new set of bioinformatic analyses regarding mmu-miR-155 or mmu-miR-2137 were performed to determine which molecular pathway can be linked to the results observed.
Regarding mmu-miR-155, 130 topological modules were identified by the MCL algorithm, 52 of which contained more than five nodes. When the largest topological modules (see Fig. S3) were considered, several pathways related to the inflammatory response but also pathogen recognition were identified as potential targets, including cytokine- and chemokine-related pathways and the Toll-like receptor (TLR) signaling pathway but also osteoclast differentiation. Furthermore, gene ontology analysis confirmed the potential involvement of this miRNA in the genes associated with the inflammatory response (Benjamini-corrected P value, 1.3E-24). Interestingly, one other topological module (see Fig. S4) was related to the Jak-STAT pathway, a well-described pathway related to apoptosis but also to proinflammatory cytokine production, notably through targeting of the TNF and NF-κB signaling pathways. Furthermore, associations with pathways related to epithelial cell invasion and osteoclast differentiation have also been underlined for this module, especially through direct targeting of the phosphatidylinositol 3-kinase–AKT pathway.
Regarding mmu-miR-2137, 10 topological modules were identified, 4 of which contained more than five nodes. The largest module was associated with the cell cycle (Benjamini-corrected P value, 2.1E-12). However, no pathway related to inflammation has been identified yet.
DISCUSSION
This study aimed to evaluate the effects of P. gingivalis infection on miRNA expression in BMMs and to evaluate their role in the inflammatory host response in vitro and in vivo. This wide-scale analysis led to the identification of several miRNAs that are differentially expressed in infected cells. To determine the effects associated with this modulation and potential implications for the innate immune response, a focus was placed on miRNAs that have a high level of significance. Therefore, analyses at the gene level, through bioinformatic prediction, and at the protein level of cytokine secretion were conducted that suggest roles for mmu-miR-155-5p, mmu-miR-2137, mmu-miR-7674, and mmu-miR-8109 in P. gingivalis-induced host response modulation. Interestingly, the present study also shows the potential of using specific miRNAs as a therapeutic approach to control the P. gingivalis-induced inflammatory response in a mouse model of inflammation and bone resorption.
The capacity for modulation of the host response to P. gingivalis infection is part of the bacterial survival strategy. Several mechanisms have been developed by such bacteria in order to invade and proliferate within the host cell, whether at the gene (6) or the protein (19, 20) level. Regulation of protein expression is critically important in homeostasis and pathology. Because miRNAs have been shown to control 30% of all protein-coding genes and also to participate in the regulation of almost every cellular process (17, 21), miRNAs could be considered important posttranscriptional regulators. Their expression has been related to the dysregulation of several biological processes in both innate and adaptive immune responses (5). The impact of infection on miRNA expression has already been described for several bacteria in different cell models. This is especially true for macrophages, which play an important role in cytokine secretion, the immune response, and early recognition and clearance of bacteria (22). For instance, in a model of human macrophages infected with Mycobacterium tuberculosis, it has been observed that the infection leads to the specific expression of several miRNAs that play key roles in immune and inflammatory pathways (23). In the context of PD, up- or downregulation of the expression of several miRNAs, such as miR-155, miR-200, and miR-146, that are involved in the regulation of several cellular processes, including the control of TLR sensitivity, the cell cycle, apoptosis, and autophagy, has also been observed (5). More specifically, effects on miRNA regulation induced by keystone pathogens such as P. gingivalis have also been investigated. In macrophages, stimulation by P. gingivalis LPS modulates the expression of miR-24, a negative regulator of macrophage classical activation (24). In our model, P. gingivalis infection also downregulated mmu-miR-211-3p expression; this observation was also visible in a Candida albicans infection model, contributing to suppression of the host immune response (19). Interestingly some other miRNAs have already been described as potential regulators of the host immune response induced by P. gingivalis or its virulence factors, such as miR-146a, which downregulates proinflammatory cytokine secretion by blocking TLR pathways in vitro (25, 26) or in vivo (27). Several other pathways have been identified as putative targets of P. gingivalis leading to uncontrolled secretion of cytokines such as SOCS3 (28).
In our study, the effects of P. gingivalis infection on 1,908 miRNAs were evaluated. We observed that infection led to specific modulation of the expression of eight miRNAs (mmu-miR-155-5p, mmu-miR-211-3p, mmu-miR-2137, mmu-miR-3473b, mmu-miR-3473e, mmu-miR-6975-5p, mmu-miR-7674-5p, and mmu-miR-8109) when high-stringency criteria were used.
miR-155 expression is upregulated by P. gingivalis infection. This miRNA is abundantly expressed in immune cells such as macrophages and active B and T cells and in dendritic cells. It is involved in the immune response, as well as in protection against the harmful effects of inflammation through the modulation of pathways related to apoptosis, proliferation, and host-pathogen interactions (29). Interestingly, in human gingival tissues affected by PD, an increase in miR-155 expression has already been observed (18), with several pathogens seemingly able to increase it. In macrophages infected with Helicobacter pylori, the increased expression of miR-155 acts as an inflammatory promoter, inducing proinflammatory cytokine (TNF-α, IL-23, IL-10, IL-8) secretion (30). However, in our in vitro model of transfection, miR-155 did not modify the secretion of TNF-α, while its inhibition increased TNF-α levels significantly, demonstrating a potentially differential effect dependent on the type of pathogen, the cell type, or the method of miRNA delivery.
Surprising results have been obtained regarding the effects of miR-7674 and miR-8109 on TNF-α secretion. Transfection with either mimics or inhibitors of these miRNAs induced an increase in TNF-α secretion. Precise targets associated with TNF-α have not been identified for these miRNAs yet, and at this point, the precise significance of these results remains unclear. Additional experiments are required to fill this knowledge gap.
Furthermore, some of the miRNA mimics or inhibitors selected increased the secretion of IL-10 in macrophages, highlighting their potential role in modulation of the innate immune response. The increase in anti-inflammatory IL-10 secretion has already been observed during P. gingivalis infection and is considered part of the bacterial survival strategy (31, 32). Therefore, modulation of the secretion of this cytokine through miRNA expression may be one of the mechanisms involved in the host response associated with P. gingivalis infection, as suggested by our pathway bioinformatic analysis. However, it is important to take into account that the observed increase in IL-10 secretion in transfected BMMs was small, emphasizing the need for confirmatory experiments regarding their impact on specific molecular pathways associated with the secretion of this cytokine.
Since PD is a chronic disease, infection may occur in already activated cells. Accordingly, the effect of P. gingivalis infection on pretreated macrophages was evaluated. Our results show that the cell response to infection is modulated by such pretreatment. For instance, a decrease in TNF-α secretion was observed in cells transfected with miR-155 and infected with P. gingivalis. Therefore, we hypothesized that modulation of miR-155 expression by P. gingivalis is one of the mechanisms developed by the bacterium to restrain the host immune response to P. gingivalis infection. An anti-inflammatory effect was also observed when macrophages were transfected with anti-miR-2137 and infected with P. gingivalis, resulting in amplification of anti-inflammatory IL-10 secretion. From a therapeutic point of view, their ability to reduce the proinflammatory aspects and promote anti-inflammatory aspects of the host response may be an interesting strategy to control the adverse effects of P. gingivalis infection.
Furthermore, our in vitro results were confirmed in vivo in a mouse calvarial model. This model has often been used to study inflammation and bone loss in response to infection. We elected to use the calvarial model to assess the impact of identified miRNA mimics or inhibitors on the modulation of P. gingivalis-induced inflammation and bone resorption (33) for two reasons. The first is easy delivery of the miRNA. With easy access, we were confident in our abilities to deliver the miRNA to the proper location and accurately test for efficacy. Furthermore, it allows us to deliver a precise dose of miRNAs. The second is specificity to P. gingivalis pathogenicity. We wanted to test the effects of the miRNAs identified on P. gingivalis pathogenicity only and avoid the oral cavity, as this would have introduced additional confounding factors (i.e., microorganisms). Therefore, we elected to use the calvarial model in this study, as the murine PD model (i.e., oral gavage or infected ligatures) would introduce an additional unknown factor (host oral flora), which is preferably avoided in proof-of-concept experiments.
At the histological level, P. gingivalis infection induced bone loss, as reported previously (34). Interestingly, coinjection with miR-155 or anti-miR-2137 significantly decreased some aspects of the induced lesion, notably its size. Through their modulation of macrophage polarization, as for miR-155 (35), the therapeutic potential of miRNAs in the treatment of several inflammation-related diseases has already been proposed. Previously, it was shown that miR-155 was able to affect the levels of expression of other miRNAs (5), which may explain the observed effect in vivo. Such a role was already suggested for miR-155 in a previous study (36), where this miRNA was described as having modulated osteoclastogenesis by targeting SOCS1 and MITF. Interestingly, our in vivo model showed that the reduction of TNF-α by miR-155 was not enough to significantly decrease ICI, bone loss, or osteoclast activity. Further investigation of the role of miR-155 in inflammation might reveal an alternative pathway. Regarding mmu-miR-2137, in vitro results were substantiated in vivo when anti-miR-2137 was coinjected with P. gingivalis, leading to a reduction in osteoclast activity and bone loss. This indicated a potential role for anti-miR-2137 in bone metabolism and revealed that the increase in IL-10 production caused by miR-2137 inhibition was sufficient to produce a robust reduction in P. gingivalis-induced ICI, bone loss, or osteoclast activity. However, the injection of the combination group did not affect the size of the lesion associated with P. gingivalis infection as we had hypothesized. This may be a consequence of the ability of miR-155 to affect the expression of other miRNAs (5). This could also be attributed to the fact that we used a lower concentration of each miRNA because of the inherent toxicity of miRNA seen at a higher concentration in our in vivo model. More importantly, quantitative histological examination revealed that the effect of miRNA-155 on TNF-α was not sufficient to reduce the strong inflammation produced by P. gingivalis, which in turn could reverse the robust effect of anti-miR-2137. However, osteoclast activity was significantly reduced by this combination (miRNA-155 and anti-miR-2137).
Cognizant of the calvarial model's limitations, the present study has demonstrated proof of principle in the identification and confirmation of the roles of specific miRNAs in the inflammatory process associated with P. gingivalis pathogenicity. The present results should be confirmed in a specific model of experimental PD such as an oral-gavage- or infected-ligature-induced model (33) to evaluate precisely the potential therapeutic interest of these miRNAs in the context of a more complex lesion induced by a multispecies complex biofilm such as in PD.
Recently, the roles and functions of miRNAs have been more appreciated in the context of PD. However, available studies analyzed miRNA expression in healthy and diseased gingival biopsy specimens without taking the differences in cell populations into account, leading to biased results (5). To prevent skewed results due to preactivation of macrophage cells, our study focused on BMMs, which can be considered a naive cell population. Indeed, the macrophage response to infection is certainly under the influence of several factors, including preactivation (37), ruling out the use of peritoneal macrophages.
miRNA research in the periodontal field is a work in progress, as knowledge increases continuously. However, the identification of new miRNA sequences calls for attention. In our study, some of the miRNAs identified were poorly described in the literature and the potential molecular pathways involved in their action could only be approached through bioinformatic analysis. Therefore, further research with other types of immune and periodontal cells but also specific in vivo models of PD (33) should pave the way to a better understanding of their role in periodontal disease development and to identify their potential use as therapeutics. Their pivotal role in a network of cellular mechanisms may allow us to use them to target well-identified pathways specifically with the aim to control infection-related processes. The development of specific carriers is mandatory to develop highly selective miRNA-based therapies.
MATERIALS AND METHODS
Bacterial strain.
P. gingivalis ATCC 33277 (kindly provided by R. Lamont, University of Louisville, Louisville, KY) was cultured and maintained on enriched tryptic soy agar plates containing a mixture of defibrinated sheep blood, 5 mg/ml hemin, and 1 mg/ml menadione at 37°C under anaerobic conditions. Bacterial colonies where then transferred into brain heart infusion medium (Becton, Dickinson and Company, Sparks, MD) supplemented with the same materials. On the day of infection, bacteria were centrifuged and washed with phosphate-buffered saline (PBS) and the number of bacteria was determined by measuring the optical density at 600 nm.
Macrophage cultures.
BMMs were harvested as described previously (38). Briefly, bone marrow cells from femora of donor mice were incubated for 7 days at 37°C in RPMI medium supplemented with L929. BMMs were cultured in RPMI medium supplemented with 10,000 U/liter penicillin, 100 mg/liter streptomycin, and 10% fetal bovine serum in a humidified atmosphere with 5% CO2.
Infection of BMMs with P. gingivalis.
A total of 1 × 106 BMMs were plated in six-well plates. Adherent BMMs were infected with live P. gingivalis at a multiplicity of infection (MOI) of 25 (3). Cells and supernatants were harvested 3 h after infection with live bacteria.
RNA extraction and microarray analysis.
BMMs were washed with cold PBS 3 h after infection, and the total RNAs were extracted with the miRNeasy minikit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Sample integrity was verified with RNA 6000 Nano Assay RNA chips run in an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). The RNAs (300 μg) were then labeled with the FlashTag HSR kit (Genisphere Inc., Hatfield, PA) according to the manufacturer's protocol. The labeled RNAs were hybridized with miRNA 4.0 arrays (Affymetrix, Santa Clara, CA) for 16 h in GeneChip Hybridization Oven 640 at 48°C with rotation (60 rpm). The hybridized samples were washed and stained with Affymetrix Fluidics Station 450. The first staining with streptavidin–R-phycoerythrin was followed by signal amplification with a biotinylated goat anti-streptavidin antibody and another streptavidin–R-phycoerythrin staining (Hybridization, Washing and Staining kit; Affymetrix, Santa Clara, CA). Microarrays were immediately scanned with Affymetrix GeneArray Scanner 3000 7G Plus (Affymetrix, Santa Clara, CA). The Affymetrix Expression console software was used for summarization, normalization, and quality control of the resulting CEL files. All experiments were performed in triplicate.
Data processing.
Raw Affymetrix CEL files (GeneChip miRNA version 4.0 array) were normalized to produce probe set level expression values for all mouse and control probe sets with the Affymetrix Expression Console (version 1.4.1), Robust Multiarray Average (39), and Detection Above BackGround. The analysis was limited to 1,908 mature mouse miRNAs assessed by the array. A principal-component analysis was performed with the prcomp R function with expression values that had been normalized across all of the samples to a mean of 0 and a standard deviation of 1. Analyses of variance was performed with the f.pvalue function in the sva package (version 3.4.0). Pairwise differential miRNA expression was assessed with the moderated (empirical Bayesian) t test implemented in the limma package (version 3.14.4) (i.e., creation of simple linear models with lmFit, followed by empirical Bayesian adjustments with eBayes). Correction for multiple-hypothesis testing was accomplished with the Benjamini-Hochberg false-discovery rate (40). All statistical analyses were performed in the R environment for statistical computing (version 2.15.1).
RT and real-time PCR validation.
Three micrograms of total RNA was used for RT-qPCR with the miScript II RT kit according to the manufacturer's instructions. Real-time PCR was conducted with the miScript SYBR green PCR kit and the Bio-Rad RT-PCR system (Bio-Rad, Hercules, CA). All reagents and primers were purchased from Qiagen, Valencia, CA.
Identification and analysis of predicted target genes of differentially expressed miRNAs.
miRWalk 2.0 (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/) (41) was used to identify putative target genes from eight identified miRNAs with a predicted P value of <0.05. Interactomic data were extracted from the STRING 10 database (42), which contains known and predicted physical and functional PPIs from diverse sources. Only interactions with the highest confidence were selected (combined score, ≥0.9). The open-source software platform Cytoscape 3.2.1 (43) was used to visualize and analyze PPI networks. The clusterMaker2 0.9.5 plugin (44) from Cytoscape, and in particular the MCL algorithm (45) (granularity parameter of 2.5), was applied to identify topological modules. KEGG pathway enrichment analysis was performed with DAVID release 6.8 (46) to study the functional significance of the modules.
Transient miRNA transfections.
The miScript miRNA mimics used were syn-mmu-miR-155-5p (miR-155), syn-mmu-miR-2137 (miR-2137), syn-mmu-miR-7674-5p (miR-7674), and syn-mmu-miR-8109 (miR-8109), and the miScript miRNA inhibitors used were anti-mmu-miR-155-5p (anti-miR-155), anti-mmu-miR-2137 (anti-miR-2137), anti-mmu-miR-7674 (anti-miR-7674), and anti-mmu-miR-8109 (anti-miR-8109). All were purchased from Qiagen (Valencia, CA). As a control, AllStars Neg. siRNA AF 546 (Qiagen) was used. One day prior to the transfection, 4 × 105 cells were seeded into a 24-well plate. Transfection was performed with HiPerfect Transfection Reagent (Qiagen) according to the manufacturer's protocol. BMMs were transfected with miRNA mimics or inhibitors at a final concentration of 50 nM and infected with P. gingivalis (MOI of 25).
Cytokine analysis.
The supernatants of the P. gingivalis-infected BMMs that were transfected with the miRNA mimics or inhibitors were collected after 24 h of treatment. The levels of TNF-α and IL-10 were evaluated with enzyme-linked immunosorbent assay kits (Life Technologies Corporation, Carlsbad, CA) according to the manufacturer's protocol.
Mouse calvarial bone resorption model.
The 8- to 12-week-old wild-type C57BL/6J mice used in this study were purchased from The Jackson Laboratory (Bar Harbor, ME). All experiments involving animals were approved by the Boston University Institutional Animal Care and Use Committee and were performed in compliance with relevant animal care and use laws. Regular mouse chow and water were provided ad libitum. The mice (n = 4) were separated into the following four groups: (i) P. gingivalis-infected mice as a control; (ii) P. gingivalis, 8 μg of miR-155, and 8 μg of anti-miR-2137 inhibitor (here, this group is referred to as the combination group); (iii) P. gingivalis and 16 μg of miR-155; and (iv) P. gingivalis and 16 μg of anti-miR-2137. The final volume of each individual injection was 100 μl in PBS.
All of the procedures were done with mice under anesthesia as previously described (47). Anesthesia was obtained by the intraperitoneal injection of ketamine-xylazine. First, the heads of mice were shaved, and then P. gingivalis (5 × 108 CFU in 100 μl of PBS) and the mmu-miR 155 mimic, the mmu-miR 2137 inhibitor, or both were injected subcutaneously at the same site. Injections were carried out with a 30.5-gauge needle at a point on the midline of the skull between the ears and eyes. Mice were sacrificed 7 days after the injection. The size of the lesion resulting from the injection in each animal (area in square millimeters) was analyzed with ImageJ.
Histological analysis.
The calvarium of each mouse was fixed in 2% paraformaldehyde in PBS (Affymetrix, Inc., Cleveland, OH) for 2 h at room temperature and refrigerated in PBS until ready for further processing. Samples were embedded in an optimal cutting temperature compound (Fisher HealthCare, Houston, TX) and frozen at the time of sectioning. Sections of 5 to 8 μm were stained with hematoxylin and eosin and prepared for further analysis. To analyze osteoclast activity, a tartrate-resistant acid phosphatase (TRAP) assay was performed according to an already established protocol (48). To quantify the histological sections, samples were evaluated by double-blinded examiners according to three different scoring protocols. (i) Calvarial bone resorption was scored on a scale of 0 through 4, with 0 corresponding to no calvarial bone resorption, 1 meaning that one-fourth of the calvarial bone was affected but there was no through-and-through penetration, 2 meaning that half of the calvarial bone was affected but there was no through-and-through penetration, 3 meaning that half of the calvarial bone was affected with through-and-through bone penetration <1 mm in width, and 4 meaning that more than half of the calvarial bone was affected with through-and-through bone penetration >1 mm wide. (ii) Osteoclast activity was scored on a scale of 0 to 3, with 0 corresponding to no osteoclast activity with no sign of bone resorption; 1 corresponding to the presence of some osteoclast lacunae with minimum bone resorption; 2 corresponding to the presence of 5 to 10 osteoclast lacunae with bone resorption, and 3 corresponding to the presence of >10 osteoclast lacunae and severe bone resorption. (iii) ICI was scored on a scale of 0 through 5, with 0 corresponding to no sign of inflammation, 1 corresponding to incipient ICI in the derma/subderma, 2 corresponding to mild ICI in the derma/subderma, 3 corresponding to moderate ICI in the derma/subderma and scarce inflammation in the surrounding bone, 4 corresponding to advanced ICI in the derma/subderma and mild inflammation in the surrounding bone, and 5 corresponding to severe ICI in the derma/subderma and advanced inflammation in the surrounding bone.
Statistical analysis.
Data were analyzed for statistical significance with XLStat (Addinsoft, New York, NY). P values were calculated with the Mann-Whitney t test.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants RO1HL076801 and RO1DE014079 to S.A.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00771-16.
REFERENCES
- 1.Hajishengallis G, Lamont RJ. 2014. Breaking bad: manipulation of the host response by Porphyromonas gingivalis. Eur J Immunol 44:328–338. doi: 10.1002/eji.201344202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pihlstrom BL, Michalowicz BS, Johnson NW. 2005. Periodontal diseases. Lancet 366:1809–1820. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Q, Amar S. 2007. Identification of signaling pathways in macrophage exposed to Porphyromonas gingivalis or to its purified cell wall components. J Immunol 179:7777–7790. doi: 10.4049/jimmunol.179.11.7777. [DOI] [PubMed] [Google Scholar]
- 4.Hajishengallis G. 2015. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol 15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kebschull M, Papapanou PN. 2015. Mini but mighty: microRNAs in the pathobiology of periodontal disease. Periodontol 2000 69:201–220. doi: 10.1111/prd.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu W-H, Hu H, Zhou Q, Xia Y, Amar S. 2010. Bioinformatics analysis of macrophages exposed to porphyromonas gingivalis: implications in acute vs. chronic infections. PLoS One 5:e15613. doi: 10.1371/journal.pone.0015613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodrigues PH, Reyes L, Chadda AS, Bélanger M, Wallet SM, Akin D, Dunn W, Progulske-Fox A. 2012. Porphyromonas gingivalis strain specific interactions with human coronary artery endothelial cells: a comparative study. PLoS One 7:e52606. doi: 10.1371/journal.pone.0052606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morandini AC, Ramos-Junior ES, Potempa J, Nguyen K-A, Oliveira AC, Bellio M, Ojcius DM, Scharfstein J, Coutinho-Silva R. 2014. Porphyromonas gingivalis fimbriae dampen P2X7-dependent interleukin-1β secretion. J Innate Immun 6:831–845. doi: 10.1159/000363338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato Y, Hagiwara M, Ishihara Y, Isoda R, Sugiura S, Komatsu T, Ishida N, Noguchi T, Matsushita K. 2014. TNF-α augmented Porphyromonas gingivalis invasion in human gingival epithelial cells through Rab5 and ICAM-1. BMC Microbiol 14:229. doi: 10.1186/s12866-014-0229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kocgozlu L, Elkaim R, Tenenbaum H, Werner S. 2009. Variable cell responses to P. gingivalis lipopolysaccharide. J Dent Res 88:741–745. doi: 10.1177/0022034509341166. [DOI] [PubMed] [Google Scholar]
- 11.Giannobile WV. 2008. Host response therapeutics for periodontal diseases. J Periodontol 79:1592–1600. doi: 10.1902/jop.2008.080174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gemmell E, McHugh GB, Grieco DA, Seymour GJ. 2001. Costimulatory molecules in human periodontal disease tissues. J Periodont Res 36:92–100. doi: 10.1034/j.1600-0765.2001.360205.x. [DOI] [PubMed] [Google Scholar]
- 13.Okada H, Murakami S. 1998. Cytokine expression in periodontal health and disease. Crit Rev Oral Biol Med 9:248–266. doi: 10.1177/10454411980090030101. [DOI] [PubMed] [Google Scholar]
- 14.Olczak T, Sosicka P, Olczak M. 2015. HmuY is an important virulence factor for Porphyromonas gingivalis growth in the heme-limited host environment and infection of macrophages. Biochem Biophys Res Commun 467:748–753. doi: 10.1016/j.bbrc.2015.10.070. [DOI] [PubMed] [Google Scholar]
- 15.Hosin AA, Prasad A, Viiri LE, Davies AH, Shalhoub J. 2014. MicroRNAs in atherosclerosis. J Vasc Res 51:338–349. doi: 10.1159/000368193. [DOI] [PubMed] [Google Scholar]
- 16.Marques-Rocha JL, Samblas M, Milagro FI, Bressan J, Martínez JA, Marti A. 2015. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J 29:3595–3611. doi: 10.1096/fj.14-260323. [DOI] [PubMed] [Google Scholar]
- 17.Simo G, Lueong S, Grebaut P, Guny G, Hoheisel JD. 2015. Micro RNA expression profiles in peripheral blood cells of rats that were experimentally infected with Trypanosoma congolense and different Trypanosoma brucei subspecies. Microbes Infect 17:596–608. doi: 10.1016/j.micinf.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Stoecklin-Wasmer C, Guarnieri P, Celenti R, Demmer RT, Kebschull M, Papapanou PN. 2012. MicroRNAs and their target genes in gingival tissues. J Dent Res 91:934–940. doi: 10.1177/0022034512456551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X-Y, Zhang K, Jiang Z-Y, Cai L-H. 2014. MiR-204/miR-211 downregulation contributes to candidemia-induced kidney injuries via derepression of Hmx1 expression. Life Sci 102:139–144. doi: 10.1016/j.lfs.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Huck O, Elkaim R, Davideau J-L, Tenebaum H. 2015. Porphyromonas gingivalis-impaired innate immune response via NLRP3 proteolysis in endothelial cells. Innate Immun 21:65–72. doi: 10.1177/1753425914523459. [DOI] [PubMed] [Google Scholar]
- 21.Filipowicz W, Bhattacharyya SN, Sonenberg N. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 22.Silva N, Abusleme L, Bravo D, Dutzan N, Garcia-Sesnich J, Vernal R, Hernandez M, Gamonal J. 2015. Host response mechanisms in periodontal diseases. J Appl Oral Sci 23:329–355. doi: 10.1590/1678-775720140259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng L, Leung E, Lee N, Lui G, To K-F, Chan RCY, Ip M. 2015. Differential microrna expression in human macrophages with mycobacterium tuberculosis infection of beijing/w and non-beijing/w strain types. PLoS One 10:e0126018. doi: 10.1371/journal.pone.0126018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fordham JB, Naqvi AR, Nares S. 2015. miR-24 regulates macrophage polarization and plasticity. J Clin Cell Immunol 6:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang S-Y, Xue D, Xie Y-F, Zhu D-W, Dong Y-Y, Wei C-C, Deng J-Y. 2015. The negative feedback regulation of microRNA-146a in human periodontal ligament cells after Porphyromonas gingivalis lipopolysaccharide stimulation. Inflamm Res 64:441–451. doi: 10.1007/s00011-015-0824-y. [DOI] [PubMed] [Google Scholar]
- 26.Benakanakere MR, Li Q, Eskan MA, Singh AV, Zhao J, Galicia JC, Stathopoulou P, Knudsen TB, Kinane DF. 2009. Modulation of TLR2 protein expression by miR-105 in human oral keratinocytes. J Biol Chem 284:23107–23115. doi: 10.1074/jbc.M109.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nahid MA, Rivera M, Lucas A, Chan EKL, Kesavalu L. 2011. Polymicrobial infection with periodontal pathogens specifically enhances microRNA miR-146a in ApoE−/− mice during experimental periodontal disease. Infect Immun 79:1597–1605. doi: 10.1128/IAI.01062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moffatt CE, Lamont RJ. 2011. Porphyromonas gingivalis induction of microRNA-203 expression controls suppressor of cytokine signaling 3 in gingival epithelial cells. Infect Immun 79:2632–2637. doi: 10.1128/IAI.00082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng F-R, Tang L-J, He Y, Garcia RC. 2015. An update on the role of miRNA-155 in pathogenic microbial infections. Microbes Infect 17:613–621. doi: 10.1016/j.micinf.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Yao Y, Li G, Wu J, Zhang X, Wang J. 2015. Inflammatory response of macrophages cultured with Helicobacter pylori strains was regulated by miR-155. Int J Clin Exp Pathol 8:4545–4554. [PMC free article] [PubMed] [Google Scholar]
- 31.Martin M, Schifferle RE, Cuesta N, Vogel SN, Katz J, Michalek SM. 2003. Role of the phosphatidylinositol 3 kinase-Akt pathway in the regulation of IL-10 and IL-12 by Porphyromonas gingivalis lipopolysaccharide. J Immunol 171:717–725. doi: 10.4049/jimmunol.171.2.717. [DOI] [PubMed] [Google Scholar]
- 32.Yilmaz O, Jungas T, Verbeke P, Ojcius DM. 2004. Activation of the phosphatidylinositol 3-kinase/Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infect Immun 72:3743–3751. doi: 10.1128/IAI.72.7.3743-3751.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graves DT, Kang J, Andriankaja O, Wada K, Rossa C Jr. 2012. Animal models to study host-bacteria interactions involved in periodontitis. Front Oral Biol 15:117–132. doi: 10.1159/000329675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meka A, Bakthavatchalu V, Sathishkumar S, Lopez MC, Verma RK, Wallet SM, Bhattacharyya I, Boyce BF, Handfield M, Lamont RJ, Baker HV, Ebersole JL, Kesavalu L. 2010. Porphyromonas gingivalis infection-induced tissue and bone transcriptional profiles. Mol Oral Microbiol 25:61–74. doi: 10.1111/j.2041-1014.2009.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Essandoh K, Li Y, Huo J, Fan G-C. 2016. Mirna-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock 46:122–131. doi: 10.1097/SHK.0000000000000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Zhao H, Chen J, Xia B, Jin Y, Wei W, Shen J, Huang Y. 2012. Interferon-β-induced miR-155 inhibits osteoclast differentiation by targeting SOCS1 and MITF. FEBS Lett 586:3255–3262. doi: 10.1016/j.febslet.2012.06.047. [DOI] [PubMed] [Google Scholar]
- 37.Papadopoulos G, Shaik-Dasthagirisaheb YB, Huang N, Viglianti GA, Henderson AJ, Kantarci A, Gibson FC. 27 June 2016. Immunologic environment influences macrophage response to porphyromonas gingivalis. Mol Oral Microbiol doi: 10.1111/omi.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Goncalves R, Mosser DM. 2008. The isolation and characterization of murine macrophages. Curr Protoc Immunol Chapter 14:Unit 14.1. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 40.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc Ser B 57:289–300. [Google Scholar]
- 41.Dweep H, Gretz N, Sticht C. 2014. miRWalk database for miRNA-target interactions. Methods Mol Biol 1182:289–305. doi: 10.1007/978-1-4939-1062-5_25. [DOI] [PubMed] [Google Scholar]
- 42.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, Mering von C. 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris JH, Apeltsin L, Newman AM, Baumbach J, Wittkop T, Su G, Bader GD, Ferrin TE. 2011. clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics 12:436. doi: 10.1186/1471-2105-12-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 47.Graves DT, Naguib G, Lu H, Desta T, Amar S. 2005. Porphyromonas gingivalis fimbriae are pro-inflammatory but do not play a prominent role in the innate immune response to P. gingivalis. J Endotoxin Res 11:13–18. doi: 10.1179/096805105225006722. [DOI] [PubMed] [Google Scholar]
- 48.Chiang CY, Kyritsis G, Graves DT, Amar S. 1999. Interleukin-1 and tumor necrosis factor activities partially account for calvarial bone resorption induced by local injection of lipopolysaccharide. Infect Immun 67:4231–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.