ABSTRACT
Cryptosporidium spp. are apicomplexan parasites of global importance that cause human diarrheal disease. In vitro culture models that may be used to study this parasite and that have physiological relevance to in vivo infection remain suboptimal. Thus, the pathogenesis of cryptosporidiosis remains poorly characterized, and interventions for the disease are limited. In this study, we evaluated the potential of a novel bioengineered three-dimensional (3D) human intestinal tissue model (which we developed previously) to support long-term infection by Cryptosporidium parvum. Infection was assessed by immunofluorescence assays and confocal and scanning electron microscopy and quantified by quantitative reverse transcription-PCR. We found that C. parvum infected and developed in this tissue model for at least 17 days, the extent of the study time used in the present study. Contents from infected scaffolds could be transferred to fresh scaffolds to establish new infections for at least three rounds. Asexual and sexual stages and the formation of new oocysts were observed during the course of infection. Additionally, we observed ablation, blunting, or distortion of microvilli in infected epithelial cells. Ultimately, a 3D model system capable of supporting continuous Cryptosporidium infection will be a useful tool for the study of host-parasite interactions, identification of putative drug targets, screening of potential interventions, and propagation of genetically modified parasites.
KEYWORDS: 3D model, Cryptosporidium, in vitro culture, intestinal epithelial cells
INTRODUCTION
Cryptosporidium spp. are apicomplexan parasites of global importance that cause diarrheal disease in humans and animals worldwide (1, 2). Though immunocompetent individuals experience self-limiting or asymptomatic infection, immunocompromised hosts, such as untreated patients with HIV infection or AIDS (3) and malnourished children (1, 2) in resource-limited settings, may experience severe diarrhea, wasting, and death. In a recent case-control study, Cryptosporidium was one of four pathogens responsible for moderate to severe diarrhea in children under the age of 5 years and was associated with an increased risk of death in children from 1 to 2 years of age in seven countries in sub-Saharan Africa and South Asia (4). The only FDA-approved drug for the treatment of cryptosporidiosis is nitazoxanide, a drug with broad antiparasitic properties (5). However, this drug is not effective in immunocompromised patients (6) and has not been widely tested in malnourished children in resource-limited countries. Though significant improvements in water purification methods have occurred (7) since the outbreak of waterborne cryptosporidiosis in Milwaukee, WI, in 1993 (8), industrialized countries are seeing increased numbers of cases, largely due to recreational water outbreaks (9).
Unlike many other apicomplexan parasites which often require more than one host to complete their life cycles (10), Cryptosporidium completes its entire life cycle in a single host. Infection begins with the ingestion of oocysts in contaminated water or food by direct contact with infected humans or animals. In children and immunocompromised individuals in resource-limited settings, respiratory cryptosporidiosis may be transmitted by inhalation (11). Once oocysts reach the upper small intestine, excystation occurs, releasing four infectious sporozoites (12). These sporozoites glide along the surface of host cells using actin-dependent motility (13). Following attachment to and invasion of intestinal epithelial cells, two rounds of merogony (asexual reproduction) occur. During the first round, six to eight merozoites (14–16) are released into the gut and propagate infection by reinvading adjacent cells. The second round of merogony gives rise to four merozoites that continue on to gametogony (sexual replication) (17). As part of the sexual cycle, microgametes are formed; the microgametes fuse with macrogamonts during fertilization to form zygotes, which then mature into thick- or thin-walled oocysts. The thin-walled oocysts can excyst in the intestinal lumen and reinfect cells (16, 18, 19), while the thick-walled oocysts are excreted and released into the environment.
A major challenge in Cryptosporidium research is the lack of an in vitro culture system which supports continuous infection and completion of the life cycle (20). This has limited our understanding of the mechanisms involved in pathogenesis and identification of drug targets (21). A number of static two-dimensional (2D) in vitro culture systems employing primary cells or cell lines grown in monolayers have been reported to be permissive to infection (reviewed in references 20 and 22; 23, 24). However, in most of these systems, infection peaks by 3 days, followed by a sharp decline and incomplete replication (20). None of these systems support infection for more than 5 days. Until recently, the genetic manipulation of Cryptosporidium spp. had not been possible. However, even with new advances in the genetic modification of Cryptosporidium parvum, stable transgenic parasites cannot be selected or propagated in vitro (25).
Recently, we developed a novel bioengineered three-dimensional (3D) human intestinal tissue model using silk protein as the scaffolding biomaterial (26). This 3D porous protein scaffolding system contains a geometrically engineered hollow lumen with adaptability to both the large and small intestines. These intestinal tissues demonstrated representative human responses by permitting the continuous accumulation of mucous secretions on the epithelial surface, establishing low oxygen tension in the lumen, and interacting with gut-colonizing bacteria. The newly developed 3D intestinal model enabled months-long sustained access to these intestinal functions in vitro and could be integrated with a multitude of different organ mimics; therefore, it ensures a reliable ex vivo tissue system for studies in a broad context of human intestinal diseases and treatments.
Here, we demonstrate the use of this novel 3D bioengineered human intestinal tissue system as an improved in vitro model to support long-term Cryptosporidium infection.
RESULTS
C. parvum infection can be maintained in the 3D scaffold model for at least 15 to 17 days.
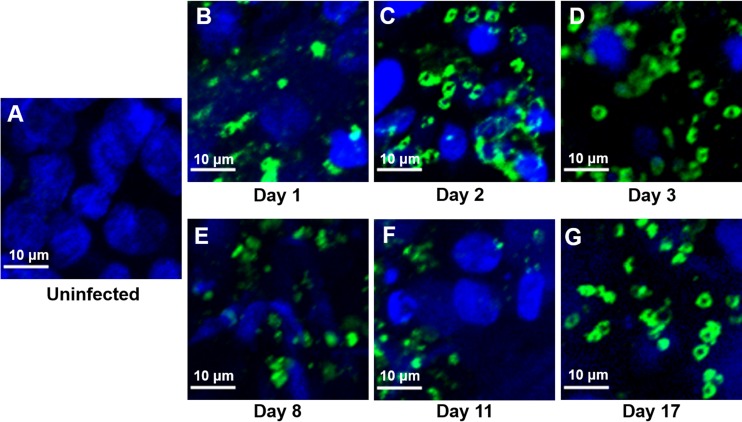
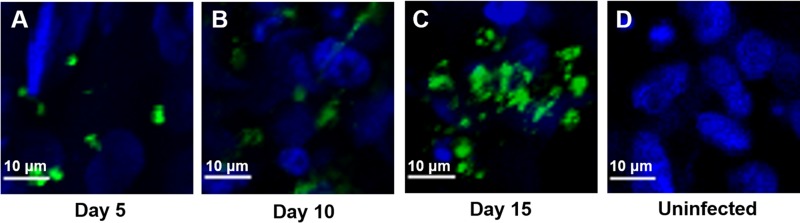
To determine whether the 3D scaffold model (Fig. 1) can support C. parvum infection, we infected Caco-2 and HT29-MTX cells in the scaffolds with oocysts and evaluated the presence of intracellular stages by immunofluorescence assay (IFA) and confocal microscopy using the 4E9 monoclonal antibody (MAb) at various time points. No staining in uninfected Caco-2 and HT29-MTX cells was observed in the scaffolds (Fig. 2A), confirming the specificity of the antibody. We detected staining (indicating intracellular C. parvum infection) for the entire duration of the time course of the experiments (Fig. 2B to G). We observed distinct areas of infection within the scaffolds rather than uniform infection throughout the tissue. When we examined Caco-2 and HT29-MTX cells infected with purified sporozoites in the scaffold model by IFA with MAb 4E9 and confocal microscopy, we detected staining for the duration of the 15-day time course (Fig. 3A to E). Although the infection patterns were similar to those of Caco-2 and HT29-MTX cells in scaffolds infected with oocysts, the levels of infection appeared to be less.
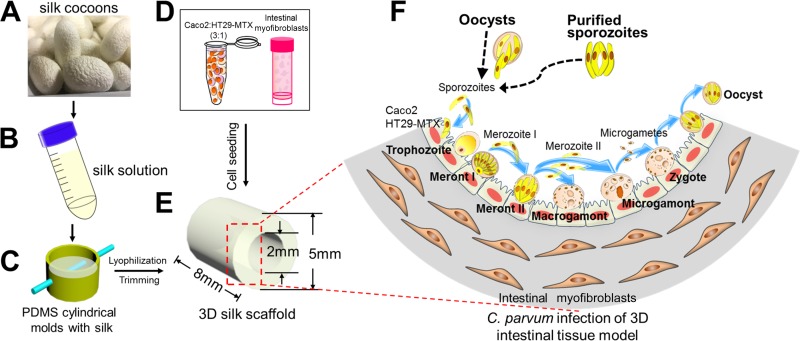
FIG 1.
Schematic of the fabrication process, cell seeding strategy, and C. parvum infection process for the 3D human intestinal model. (A and B) Silk cocoons (A) were regenerated into a 4 to 5% (wt/vol) viscous silk solution (B). (C) The silk solution was poured into PDMS molds, and a Teflon-coated stainless steel wire was inserted through the cross section of the cylinder to develop a hollow channel. (D and E) Caco-2 and HT29-MTX cells (D) were seeded into the hollow channel (E), while the porous bulk space was used to house H-InMyoFibs. (F) The Caco-2 and HT29-MTX cells in the scaffolds were infected with C. parvum oocysts or purified sporozoites, and intracellular development was allowed to proceed through asexual and sexual cycles to complete the life cycle with the formation of oocysts. (Republished from reference 21 with permission of the publisher as well as from Scientific Reports [26].)
FIG 2.
IFA and confocal microscopy of Caco-2 and HT29-MTX cells in scaffolds at various time points during infection with C. parvum oocysts. The infected scaffolds were fixed, permeabilized, and stained with MAb 4E9. Uninfected scaffolds (A) and infected scaffolds obtained at 1 day (B), 2 days (C), 3 days (D), 8 days (E), 11 days (F), and 17 days (G) postinfection are shown.
FIG 3.
IFA and confocal microscopy of Caco-2 and HT29-MTX cells in scaffolds infected with purified C. parvum sporozoites at various time points. Following infection, scaffolds were fixed, permeabilized, and stained with MAb 4E9. (A) Infected scaffolds at 5 days postinfection; (B) infected scaffolds at 10 days postinfection; (C) infected scaffolds at 15 days postinfection; (D) uninfected control scaffolds.
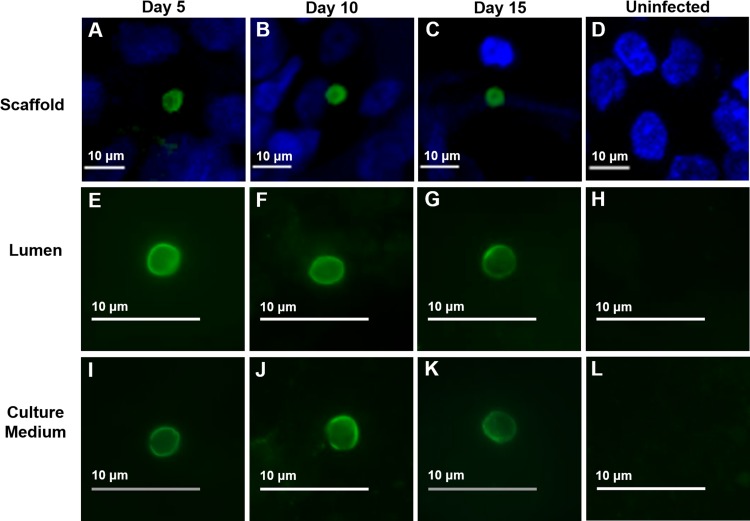
To determine whether C. parvum infection within this model system allowed completion of the full life cycle of the parasite, we examined sporozoite-infected scaffolds as well as luminal contents and the culture medium by IFA and confocal microscopy using an oocyst-specific MAb. Oocysts were not detected in purified sporozoites by IFA using the same MAb (see Fig. S1 in the supplemental material). We observed oocysts to be present in or adherent to cells within the scaffold lumen (Fig. 4A to C) as well as in the luminal contents of scaffolds (Fig. 4E to G) and in culture medium from the wells (Fig. 4I to K). No oocysts were observed in the purified sporozoite inoculum used for infection (Fig. S1) or in the lumen or medium of uninfected controls (Fig. 4D, H, and I), as determined using the same oocyst-specific MAb.
FIG 4.
IFA and confocal microscopy of oocyst production following infection with purified C. parvum sporozoites at various time points. (A to C) Oocysts within scaffolds; (E to G); oocysts within luminal material; (I to K) oocysts within culture medium; (D, H, L), uninfected controls. At each time point, scaffolds or material from the lumen or culture medium was fixed, permeabilized, and stained with an oocyst-specific MAb (Crypt-a-Glo).
C. parvum infection levels are maintained in the 3D scaffold model for at least 15 to 17 days.
Though microscopy can provide qualitative information about infection, a more precise means of quantification of live parasites is necessary to validate the model system. We quantified cDNA (derived from RNA) rather than DNA to obtain a more accurate representation of viable parasites, as C. parvum 18S rRNA degrades rapidly, within 3 h after parasite death (27, 28). The results for the controls, consisting of either no reverse transcriptase or no template, were negative.
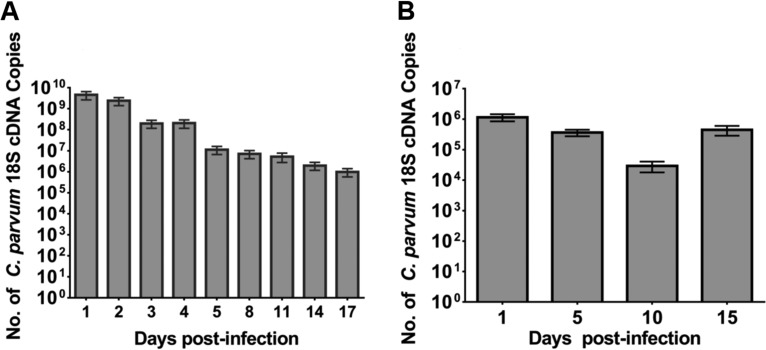
Although the C. parvum levels initially declined over the course of infection, infection levels remained relatively stable thereafter for the duration of the infection in the 3D scaffold model infected with oocysts (Fig. 5A). Although the infection levels in the model inoculated with purified sporozoites were less than those in the model infected with oocysts, the level of infection was relatively stable for the 15-day time course (Fig. 5B). It should be noted that these data only account for parasites present within the lumen and cells within the scaffold. They do not account for any parasites that may have been released into the culture medium.
FIG 5.
Quantification of C. parvum infection with oocysts (A) and purified sporozoites (B) in scaffolds by qRT-PCR. At the indicated times, the luminal contents and infected cells were harvested and infection was quantitated by qRT-PCR. Results are expressed as number of copies of C. parvum 18S cDNA obtained from a standard curve. Bars represent the means from six replicates with standard errors of the means. In panel A, P was <0.006 for day 1 compared to days 14 and 17 and P was <0.05 for day 2 compared to days 14 and 17. In panel B, P was <0.0005 for day 1 compared to day 10.
C. parvum asexual and sexual stages are visualized in the 3D scaffold model by SEM.
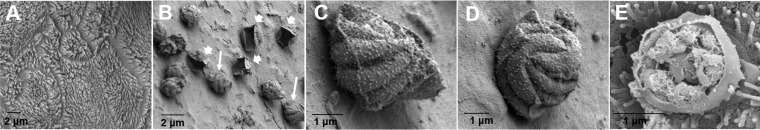
To examine which intracellular stages were present during infection, we analyzed infected Caco-2 and HT29-MTX cells in the 3D scaffold model by scanning electron microscopy (SEM). We observed densely packed microvilli with continuous brush borders in the uninfected Caco-2 and HT29-MTX cells (Fig. 6A). In contrast, we observed blunting, distortion, or ablation of the microvilli in infected areas (Fig. 6B to E), characteristic of infection in vivo (29). In the scaffold model infected with oocysts, intracellular stages, including those involved in asexual replication, were observed throughout infection. We were able to identify type I meronts (Fig. 6B to E) and empty parasitophorous vacuoles (Fig. 6B).
FIG 6.
Scanning electron micrographs of C. parvum oocyst-infected Caco-2 and HT29-MTX cells in scaffolds. (A) Uninfected cells; (B) type I meronts (thick arrows) and empty parasitophorous vacuoles (thin arrows) on day 3; (C and D) type I meronts containing eight merozoites on day 3 (the images are enlargements of the image in panel B); (E) a type I meront excysting on day 2.
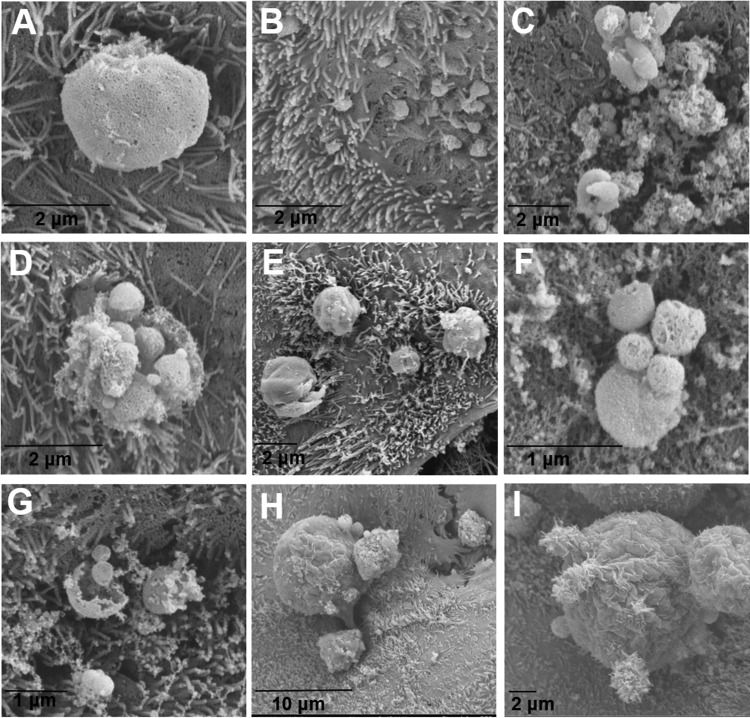
In sporozoite-infected scaffolds, intracellular stages, including those involved in asexual and sexual replication, were observed throughout infection by SEM. We were able to identify trophozoites (Fig. 7A), early type I merozoites invading host cells (Fig. 7B), type I meronts excysting (Fig. 7C to F), merozoites (Fig. 7F to G), and macrogamonts (Fig. 7H and I).
FIG 7.
Scanning electron micrographs of C. parvum sporozoite-infected scaffolds. (A) A trophozoite on day 5; (B) early penetration of type I merozoites on day 1; (C to E) type I meronts excysting on days 5 (C and D) and 2 (E); (F and G) free merozoites on day 5; (H and I) macrogamonts on day 3.
C. parvum can be passaged from an infected to a fresh 3D scaffold model.
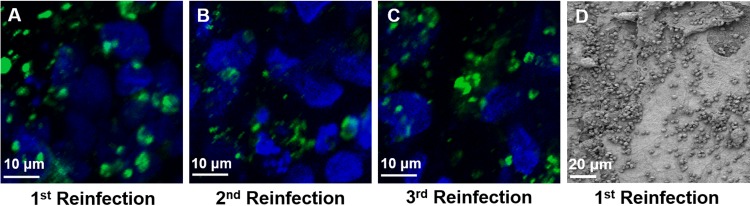
To determine whether this model system could support continuous infection by C. parvum, we inoculated Caco-2 and HT29-MTX cells in fresh scaffolds with luminal contents from infected cells in scaffolds after 3 days and analyzed the infection by IFA and confocal microscopy with MAb 4E9 staining as well as by SEM. We found that the luminal contents from infected Caco-2 and HT29-MTX cells in the scaffold model could be transferred to these cells in fresh scaffolds to establish new infections for at least three passages (Fig. 8).
FIG 8.
C. parvum can be passaged from an infected scaffold to an uninfected one to establish a new infection. (A to C) Following infection, scaffolds were fixed, permeabilized, and stained with MAb 4E9. Images are from 3 days after the first passage (A), second passage (B), or third passage (C). (D) Scanning electron micrograph of a C. parvum-infected scaffold at 3 days after the first passage.
DISCUSSION
The pathogenesis of cryptosporidiosis remains poorly characterized, in part due to the lack of in vitro, ex vivo, or in vivo models which recapitulate human intestinal structure, function, and disease. The need for such systems is paramount to advancing our understanding of Cryptosporidium-host interactions, which will aid in developing effective interventions.
In this study, we showed that the novel bioengineered 3D human intestinal model system was able to support C. parvum infections for at least 15 days. Since we have previously shown that this 3D human intestinal model system can be stably maintained for at least 8 weeks in culture (26), we anticipate that infections could be supported for durations longer than those demonstrated in the present work. We have also shown that it is possible to passage C. parvum from an infected scaffold to a fresh one for at least 3 cycles, something that has not been shown in previous studies (25). To our knowledge, this study demonstrates the longest continuous C. parvum infection in a static in vitro culture model. In addition, although this model has been shown to support infection by the bacterial pathogen Yersinia pseudotuberculosis and coculture with the commensal Lactobacillus rhamnosus (26), this is the first report of infection by a protozoan pathogen in this bioengineered 3D model.
This 3D model has specific features not present in previously reported 2D models which may contribute to the longer duration of infection. The 3D architecture of the silk scaffold system with a geometrically engineered hollow lumen allows seeding of Caco-2 and HT29-MTX cells circumferentially on the surface of the lumen and recapitulates the structure of the human small intestine. Two types of intestinal epithelial cells which are routinely used in coculture for intestinal model systems (30), cells of the Caco-2 and the HT29-MTX cell lines, which are differentiated for 2 weeks in situ, are included, resulting in enterocyte-like and goblet cell-like cells, respectively. These cells grow as highly polarized monolayers characterized by microvilli with continuous brush borders and the formation of tight junctions, identified by zonula occludens protein 1 (ZO-1) immunostaining (26), in the scaffold lumen. Biomarkers of the functional activity of enterocytes, such as alkaline phosphatase and sucrose isomaltase, as well as robust mucus secretion, identified by MUC-2 immunostaining and alcian blue staining, confirm that this model recapitulates human intestinal epithelial cell function. In addition, primary human intestinal myofibroblasts (H-InMyoFibs) expressing the Sm22α biomarker are injected via hollow channels into the porous bulk space of the scaffolds, where they secrete cytokines and growth factors which support the growth, differentiation, and expansion of human Caco-2 cells and HT29-MTX cells (31).
A major advantage of the 3D system over 2D models is the ability to generate depth-graded oxygen profiles in the luminal direction, with the lowest oxygen tensions being identified in the most distal regions, which recapitulates the proximal region-to-distal region decrease in luminal oxygen tension along the gastrointestinal tract (26). This is important, since Cryptosporidium is a microaerophilic/anaerobic organism (32) which preferentially colonizes the terminal ileum, which has a low-oxygen environment.
We identified both asexual and sexual stages of the parasite by SEM in Caco-2 and HT29-MTX cells in the 3D model system. We did not observe oocysts in the system by SEM, possibly because they were dislodged during processing. However, we observed oocysts present within the scaffold, in the luminal material, as well as in the culture medium by IFA and confocal microscopy utilizing an oocyst-specific MAb. This demonstrates that this model is capable of supporting the full life cycle of C. parvum.
The lower levels of infection obtained when the scaffolds were infected with isolated sporozoites versus oocysts is not surprising, given that this has been demonstrated previously, where a decrease in infectivity of 65% between infection with oocysts and infection with purified sporozoites was shown (24). It should also be noted that we analyzed only the luminal and scaffold contents and not the surrounding culture medium, which was changed every other day, for quantitative reverse transcription-PCR (qRT-PCR)-based quantification. Previous studies have observed free merozoites in culture as soon as 48 h postinfection (33).
Recently, there have been reports of improved in vitro culture systems for C. parvum. An organoid-type culture of HCT-8 cells in a low-shear microgravity environment was shown to support C. parvum infection (34). A culture system using primary intestinal epithelial cells isolated from jejunal tissue obtained during gastric bypass surgery was reported to support C. parvum infection for 5 days, with the production of oocyst-like structures being detected in the culture supernatant (23). While this was an improvement on previous in vitro methods, which have largely relied on the use of transformed cell lines grown in a monolayer (20), supplementation of the culture medium with both growth factors and antiapoptotic molecules was required. Additionally, the infectious dose of oocysts was restricted to small amounts, as a previous study showed that a larger inoculum resulted in poor infection levels and increased cell death (35). A non-carcinoma-derived human small intestinal cell line, FHs 74 Int, was also reported to support C. parvum infection at higher levels than other cell lines (24). However, these cells cannot be propagated for longer than 12 passages, and infection was carried out for only 48 h.
More recently, Morada et al. developed a culture system employing a hollow fiber technology with HCT-8 cells surrounding hollow fibers (36). This system allowed the in vitro production of C. parvum oocysts for 6 months but required specialized equipment to actively pump in nutrients and remove waste products. Although this system provides a unique method for continuous propagation, it is not easily scaled to use as an experimental system for testing a variety of conditions and/or time points or for drug screening. On the other hand, the small-scale 3D model that we used is not conducive to the propagation of large numbers of oocysts but does allow investigation of various host-parasite interactions and rapid drug screening.
There are a few limitations to the bioengineered silk 3D human intestinal model for C. parvum infection. The small scale of this 3D model system makes it more suited to investigation of C. parvum-host cell interactions, identification of putative drug targets, and, possibly, propagation of transgenic parasites than to large-scale propagation. Additionally, like most other in vitro culture models for C. parvum, this system employs transformed cancer-derived intestinal epithelial cells rather than primary human cells. Ongoing studies are directed at using primary human cells in the tissue systems and at improving the model using microfluidic approaches, incorporating peristaltic and flow conditions as well coculture with autologous immune cells and gut microbiota.
MATERIALS AND METHODS
Generation of 3D silk scaffolds.
3D silk scaffolds were prepared as described previously (26). Briefly, silk fibroin was extracted from Bombyx mori silkworm cocoons. To prepare silk scaffolds with hollow channels, special cylindrical molds were cast from polydimethylsiloxane (PDMS; Down Corning, Midland, MI). PDMS was prepared by mixing the base reagent with the curing reagent in a mass ratio of 10:1. The cylindrical PDMS molds consisted of a Teflon-coated stainless steel wire (diameter, 2 mm; McMaster-Carr) inserted through the cross section of the cylinder to develop a hollow channel in the silk scaffold. Finally, 4 to 5% (wt/vol) viscous silk solution was poured into the PDMS molds. The molds were frozen at −20°C overnight and then transferred to a lyophilizer for drying. The dried silk scaffolds were then autoclaved to induce the β-sheet conformation (insolubility in water), soaked in distilled water overnight, and trimmed along the axis of the hollow channel into a cuboid 5 by 5 by 8 mm. The fabrication method resulted in a scaffold consisting of a hollow channel space (diameter, 2 mm) and a bulk space around the channel that contained interconnected pores (Fig. 1).
Cell seeding on 3D silk scaffolds.
Cells of the human colon cancer-derived intestinal epithelial cell lines Caco-2 (CRL-2102) and HT29-MTX were obtained from ATCC (Manassas, VA) and the Public Health England Culture Collections (Salisbury, Great Britain), respectively. Both Caco-2 and HT29-MTX cells were grown in Dulbecco modified Eagle medium (DMEM; Gibco, Waltham, MA) supplemented with 10% fetal bovine serum, 10 μg/ml human transferrin (Gibco), 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B (Gibco). Primary human intestinal myofibroblasts (H-InMyoFibs) were purchased from Lonza (Walkersville, MD) and cultured in smooth muscle growth medium (SMGM; SmGM-2 BulletKit medium; Lonza, Walkersville, MD). Cells were maintained at 37°C in a humidified atmosphere with 5% CO2 and harvested with 0.25% trypsin-EDTA (Gibco) prior to seeding. The hollow channel of the 3D scaffolds was used to accommodate Caco-2 and HT29-MTX cells, while the porous bulk space was used to house the H-InMyoFibs. Gels containing a 10% cell suspension of H-InMyoFibs (in 10% DMEM [Gibco]), 10% 10× DMEM (Sigma-Aldrich, St. Louis, MO), and 80% type I rat tail collagen (2.01 mg/ml; First Link, Birmingham, UK) were delivered into the spongy silk scaffolds with the Teflon-coated stainless steel wires in place to leave the hollow channel open for the seeding of Caco-2 and HT29-MTX cells (Fig. 1D and E). After 20 to 25 min of gelation at 37°C, the Teflon-coated stainless steel wires were carefully removed from the scaffolds. After that, the hollow channels were seeded with the Caco-2/HT29-MTX cells (3:1) at a density of 4 × 106 cells/ml. Seeded scaffolds were incubated at 37°C for 1 h and then flipped down and incubated for an additional 1 h. During the incubation, a small amount of SMGM was dripped onto the scaffolds to keep them moistened. Seeded scaffolds were incubated in 12-well plates with culture medium (complete DMEM and SMGM at a 1:1 ratio) and incubated for 2 weeks (with the medium being changed every other day) at 37°C in 5% CO2 prior to infection with C. parvum.
C. parvum.
C. parvum (Iowa isolate) oocysts were obtained from Bunch Grass Farm, Deary, ID. Oocysts were stored in sterile phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) supplemented with 1,000 IU penicillin and 1,000 μg streptomycin at 4°C and used within 2 months of isolation.
Preparation of C. parvum oocysts for infection.
Prior to use, oocysts were surface sterilized with 10% (vol/vol) sodium hypochlorite on ice for 10 min. The oocysts were washed once by centrifugation with 1 ml sterile PBS and then incubated in 0.75% (vol/vol) sodium taurocholate solution in PBS at 15°C for 15 min and washed again in sterile PBS, prior to being resuspended in culture medium before infection. All centrifugation steps were performed at 10,000 × g for 2 min at 4°C.
Isolation of C. parvum sporozoites.
Purified sporozoites were obtained by excysting hypochlorite- and taurocholate-treated oocysts in sterile PBS for 1 h at 37°C, followed by filtration through a 3-μm-pore-size filter (EMD Millipore, Billerica, MA) to remove intact oocysts and oocyst shells. Routinely, excystation rates of ∼80% were observed. Purified sporozoites were centrifuged at 16,000 × g for 5 min at 4°C and resuspended in culture medium. Aliquots were spotted on poly-l-lysine-coated 8-well slides (Sigma, Waltham, MA). Dried spots were fixed with ice-cold methanol for 30 min at room temperature, washed with PBS, and then stained with an oocyst-specific MAb (Crypt-a-Glo; Waterborne, Inc., New Orleans, LA) for 30 min at room temperature or an MAb specific for sporozoites and intracellular stages (Sporo-Glo; Waterborne, Inc.) for 30 min at room temperature. A drop of No-Fade mounting medium (Waterborne, Inc., New Orleans, LA) was placed on each well before it was sealed with a coverslip. Slides were imaged by differential interference contrast (DIC) or fluorescence using a Zeiss Axio Imager Z.1 fluorescence microscope at ×100 magnification.
C. parvum infection of 3D scaffold model with oocysts.
The Caco-2 and HT29-MTX cells in the scaffolds were infected by inoculating 106 hypochlorite- and taurocholate-treated oocysts into the lumen and incubated at 37°C in 5% CO2 for 2 h in 10-cm plates without culture medium to allow the excystation of oocysts and invasion by sporozoites. The scaffolds were then placed in 12-well plates with culture medium and incubated for 1 to 17 days at 37°C in 5% CO2. For passaging, at 3 days postinfection, the luminal contents from infected scaffolds were harvested by aspiration using a pipette, concentrated by centrifugation at 16,000 × g for 2 min at 4°C, and used to infect new scaffolds. The medium was changed every other day for continuous infection and for passaging.
C. parvum infection of 3D scaffold model with purified sporozoites.
Purified sporozoites (4 × 106) were inoculated into the lumen of the scaffolds, which were incubated at 37°C in 5% CO2 for 2 h in 10-cm plates without culture medium to allow invasion. After incubation, the scaffolds were placed in 12-well plates with culture medium and incubated for 1 to 15 days at 37°C in 5% CO2.
Immunofluorescence assay (IFA) and confocal microscopy.
At each time point, the scaffolds were removed from the medium and fixed in 4% paraformaldehyde for 30 min at room temperature. The scaffolds were cut in half along the longitudinal axis to allow better exposure of the luminal surfaces. Fixed scaffolds were permeabilized in 0.1% Triton X-100 in PBS at room temperature for 15 min, and nonspecific binding was blocked with 5% normal goat serum (NGS; Atlanta Biologicals, Flowery Branch, GA) in PBS or 1% bovine serum albumin (BSA; Thermo Fisher, Waltham, MA) in PBS at 4°C overnight. The scaffolds were incubated with 4E9, an MAb to a glycan epitope on C. parvum invasive and intracellular stages (37), at room temperature for 1.5 h, followed by incubation with Alexa Fluor 488-conjugated goat anti-mouse IgM (Molecular Probes/Thermo Fisher Scientific, Waltham, MA) at room temperature for 1.5 h or with Crypt-a-Glo at room temperature for 1.5 h. The scaffolds were then counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) at room temperature for 30 min. Just prior to imaging, each half was cut in half again and placed luminal side down (facing the objective) on a glass coverslip. All scaffolds were imaged using a Nikon Eclipse Ti confocal microscope at ×20 magnification.
IFA of luminal contents and culture medium.
At each time point, the luminal contents and culture medium from wells containing scaffolds were collected by aspiration and concentrated by centrifugation at 16,000 × g for 3 min at 4°C. The resultant pellet was resuspended in sterile PBS, and aliquots were spotted on poly-l-lysine-coated 8-well slides. Dried spots were fixed with ice-cold methanol for 30 min at room temperature, stained with DAPI for 5 min, washed with PBS, and then stained with Crypt-a-Glo for 30 min at room temperature. The slides were mounted as described above for purified sporozoites and imaged using a Zeiss Axio Imager Z.1 fluorescence microscope at ×100 magnification.
Scanning electron microscopy (SEM).
Infected and uninfected scaffolds were fixed in 0.5% glutaraldehyde (Sigma, Waltham, MA), followed by progressive dehydration in a graded series of ethanol (30%, 50%, 75%, and 95% ethanol and twice in 100% ethanol for 30 min at each concentration). The samples were subsequently dried by critical point drying with a liquid CO2 dryer (AutoSamdri-815; Tousimis Research Corp.), coated with 5 nm platinum-palladium using a sputter coater (208HR; Cressington Scientific Instruments Inc., Cranberry Township, PA), and imaged using a Zeiss UltraPlus or Zeiss Ultra 55 scanning electron microscope (Carl Zeiss SMT Inc.) at a voltage of 2 to 3 kV. Previously published studies showing scanning electron micrographs of C. parvum were used as a reference (14, 38) to identify the various developmental stages.
Extraction of RNA from scaffolds.
At each time point, the luminal contents were aspirated from each scaffold and reserved. A 0.25% trypsin-EDTA solution was added to the lumen, and the scaffold was incubated at 37°C for 10 min. Luminal contents were collected, the scaffold was flipped 180°, and trypsinization was repeated. All reserved contents were combined, and the mixture was centrifuged at 10,000 × g for 2 min at 4°C. RNA was extracted from the pellet using an RNeasy Plus minikit (Qiagen, Valencia, CA). RNA was extracted (from three infected scaffolds for infection with oocysts and two infected scaffolds for infection with sporozoites) at each time point and pooled. Contaminating genomic DNA was removed by treatment with a Turbo DNA-free kit (Ambion, Waltham, MA).
Quantification of C. parvum infection by qRT-PCR.
Five nanograms of RNA, quantified by use of a NanoDrop spectrophotometer (Thermo Fisher, Waltham, MA), was used to synthesize cDNA using a high-capacity reverse transcription kit (Applied Biosystems, Foster City, CA). Quantitative reverse transcription-PCR (qRT-PCR) was then performed using C. parvum 18S rRNA gene-specific primers (27) and QuantiTect SYBR green master mix (Qiagen). Quantitative PCRs (qPCRs) were performed in 96-well plates using an Mx3000P qPCR system (Agilent Technologies, Santa Clara, CA). Briefly, the reaction mixtures were heated to 95°C for 15 min and then subjected to 40 thermal cycles (94°C for 30 s, 52°C for 30 s, and 72°C for 30 s) of PCR amplification. After amplification, melting curve analysis was performed at temperatures of between 55 and 95°C to assess the specificity of the reactions. Three replicate reactions were performed for each sample, and the qPCR assay was repeated twice for a total of 6 technical replicates. Assays with a coefficient of variation (CV) of greater than 15% between replicates were repeated. 18S cDNA copy numbers were determined by comparison with a standard curve obtained by qPCR of DNA extracted from 102 to 107 oocysts using a QIAamp DNA minikit (Qiagen) as described previously (33, 39). Since each C. parvum genome contains 5 copies of the 18S rRNA gene (40) and each oocyst contains 4 sporozoites, the oocyst number was multiplied by 20 to obtain the approximate total number of 18S ribosomal cDNA copies.
Statistical analysis.
Statistical analyses were performed in GraphPad Prism software (version 7 for Windows; GraphPad Software, San Diego, CA) using the Kruskal-Wallace test followed by the Dunn's multiple-comparison test. P values of ≤0.05 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. C. Andrew Thompson, School of Veterinary and Life Sciences, Murdoch University, Western Australia, for help with the identification of C. parvum life cycle stages in SEM images.
This work was supported by a grant from the Tufts Innovation Institute, Tufts University, Medford, MA, and by NIH (P41 EB002520). M.A.D.R. was supported in part by NIH training grant T32 AI 7077-30 to the Tufts University Sackler School of Graduate Biomedical Sciences.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00731-16.
REFERENCES
- 1.Shirley DA, Moonah SN, Kotloff KL. 2012. Burden of disease from cryptosporidiosis. Curr Opin Infect Dis 25:555–563. doi: 10.1097/QCO.0b013e328357e569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Checkley W, White AC Jr, Jaganath D, Arrowood MJ, Chalmers RM, Chen X, Fayer R, Griffiths JK, Guerrant RL, Hedstrom L, Huston CD, Kotloff KL, Kang G, Mead JR, Miller M, Petri WA Jr, Priest JW, Roos DS, Striepen B, Thompson RC, Ward HD, Van Voorhis WA, Xiao L, Zhu G, Houpt ER. 2015. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect Dis 15:85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Connor RM, Shaffie R, Kang G, Ward HD. 2011. Cryptosporidiosis in patients with HIV/AIDS. AIDS 25:549–560. doi: 10.1097/QAD.0b013e3283437e88. [DOI] [PubMed] [Google Scholar]
- 4.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque ASG, Zaidi AKM, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 5.Rossignol JF, Ayoub A, Ayers MS. 2001. Treatment of diarrhea caused by Cryptosporidium parvum: a prospective randomized, double-blind, placebo-controlled study of nitazoxanide. J Infect Dis 184:103–106. doi: 10.1086/321008. [DOI] [PubMed] [Google Scholar]
- 6.Abubakar I, Aliyu SH, Arumugam C, Usman NK, Hunter PR. 2007. Treatment of cryptosporidiosis in immunocompromised individuals: systematic review and meta-analysis. Br J Clin Pharmacol 63:387–393. doi: 10.1111/j.1365-2125.2007.02873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shannon MA, Bohn PW, Elimelech M, Georgiadis JG, Marinas BJ, Mayes AM. 2008. Science and technology for water purification in the coming decades. Nature 452:301–310. doi: 10.1038/nature06599. [DOI] [PubMed] [Google Scholar]
- 8.Mac Kenzie WR, Hoxie NJ, Proctor ME, Gradus MS, Blair KA, Peterson DE, Kazmierczak JJ, Addiss DG, Fox KR, Rose JB, Davis JP. 1994. A massive outbreak in Milwaukee of cryptosporidium infection transmitted through the public water supply. N Engl J Med 331:161–167. doi: 10.1056/NEJM199407213310304. [DOI] [PubMed] [Google Scholar]
- 9.Hlavsa MC, Roberts VA, Kahler AM, Hilborn ED, Mecher TR, Beach MJ, Wade TJ, Yoder JS. 2015. Outbreaks of illness associated with recreational water—United States, 2011-2012. MMWR Morb Mortal Wkly Rep 64:668–672. [PMC free article] [PubMed] [Google Scholar]
- 10.Esch KJ, Petersen CA. 2013. Transmission and epidemiology of zoonotic protozoal diseases of companion animals. Clin Microbiol Rev 26:58–85. doi: 10.1128/CMR.00067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sponseller JK, Griffiths JK, Tzipori S. 2014. The evolution of respiratory cryptosporidiosis: evidence for transmission by inhalation. Clin Microbiol Rev 27:575–586. doi: 10.1128/CMR.00115-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol 124:80–89. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Wetzel DM, Schmidt J, Kuhlenschmidt MS, Dubey JP, Sibley LD. 2005. Gliding motility leads to active cellular invasion by Cryptosporidium parvum sporozoites. Infect Immun 73:5379–5387. doi: 10.1128/IAI.73.9.5379-5387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borowski H, Thompson RC, Armstrong T, Clode PL. 2010. Morphological characterization of Cryptosporidium parvum life-cycle stages in an in vitro model system. Parasitology 137:13–26. doi: 10.1017/S0031182009990837. [DOI] [PubMed] [Google Scholar]
- 15.Current WL, Haynes TB. 1984. Complete development of Cryptosporidium in cell culture. Science 224:603–605. doi: 10.1126/science.6710159. [DOI] [PubMed] [Google Scholar]
- 16.Hijjawi NS, Meloni BP, Morgan UM, Thompson RC. 2001. Complete development and long-term maintenance of Cryptosporidium parvum human and cattle genotypes in cell culture. Int J Parasitol 31:1048–1055. doi: 10.1016/S0020-7519(01)00212-0. [DOI] [PubMed] [Google Scholar]
- 17.Bouzid M, Hunter PR, Chalmers RM, Tyler KM. 2013. Cryptosporidium pathogenicity and virulence. Clin Microbiol Rev 26:115–134. doi: 10.1128/CMR.00076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Donoghue PJ. 1995. Cryptosporidium and cryptosporidiosis in man and animals. Int J Parasitol 25:139–195. doi: 10.1016/0020-7519(94)E0059-V. [DOI] [PubMed] [Google Scholar]
- 19.Current WL, Reese NC. 1986. A comparison of endogenous development of three isolates of Cryptosporidium in suckling mice. J Protozool 33:98–108. doi: 10.1111/j.1550-7408.1986.tb05567.x. [DOI] [PubMed] [Google Scholar]
- 20.Karanis P, Aldeyarbi HM. 2011. Evolution of Cryptosporidium in vitro culture. Int J Parasitol 41:1231–1242. doi: 10.1016/j.ijpara.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Lendner M, Daugschies A. 2014. Cryptosporidium infections: molecular advances. Parasitology 141:1511–1532. doi: 10.1017/S0031182014000237. [DOI] [PubMed] [Google Scholar]
- 22.Arrowood MJ. 2002. In vitro cultivation of Cryptosporidium species. Clin Microbiol Rev 15:390–400. doi: 10.1128/CMR.15.3.390-400.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castellanos-Gonzalez A, Cabada MM, Nichols J, Gomez G, White AC Jr. 2013. Human primary intestinal epithelial cells as an improved in vitro model for Cryptosporidium parvum infection. Infect Immun 81:1996–2001. doi: 10.1128/IAI.01131-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varughese EA, Bennett-Stamper CL, Wymer LJ, Yadav JS. 2014. A new in vitro model using small intestinal epithelial cells to enhance infection of Cryptosporidium parvum. J Microbiol Methods 106:47–54. doi: 10.1016/j.mimet.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 25.Vinayak S, Pawlowic MC, Sateriale A, Brooks CF, Studstill CJ, Bar-Peled Y, Cipriano MJ, Striepen B. 2015. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature 523:477–480. doi: 10.1038/nature14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Lin Y, Davis KM, Wang Q, Rnjak-Kovacina J, Li C, Isberg RR, Kumamoto CA, Mecsas J, Kaplan DL. 2015. Robust bioengineered 3D functional human intestinal epithelium. Sci Rep 5:13708. doi: 10.1038/srep13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai X, Woods KM, Upton SJ, Zhu G. 2005. Application of quantitative real-time reverse transcription-PCR in assessing drug efficacy against the intracellular pathogen Cryptosporidium parvum in vitro. Antimicrob Agents Chemother 49:4437–4442. doi: 10.1128/AAC.49.11.4437-4442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fontaine M, Guillot E. 2003. Study of 18S rRNA and rDNA stability by real-time RT-PCR in heat-inactivated Cryptosporidium parvum oocysts. FEMS Microbiol Lett 226:237–243. doi: 10.1016/S0378-1097(03)00538-X. [DOI] [PubMed] [Google Scholar]
- 29.Okhuysen PC, Chappell CL. 2002. Cryptosporidium virulence determinants—are we there yet? Int J Parasitol 32:517–525. doi: 10.1016/S0020-7519(01)00356-3. [DOI] [PubMed] [Google Scholar]
- 30.Mahler GJ, Shuler ML, Glahn RP. 2009. Characterization of Caco-2 and HT29-MTX cocultures in an in vitro digestion/cell culture model used to predict iron bioavailability. J Nutr Biochem 20:494–502. doi: 10.1016/j.jnutbio.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 31.Lahar N, Lei NY, Wang J, Jabaji Z, Tung SC, Joshi V, Lewis M, Stelzner M, Martin MG, Dunn JC. 2011. Intestinal subepithelial myofibroblasts support in vitro and in vivo growth of human small intestinal epithelium. PLoS One 6:e26898. doi: 10.1371/journal.pone.0026898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makiuchi T, Nozaki T. 2014. Highly divergent mitochondrion-related organelles in anaerobic parasitic protozoa. Biochimie 100:3–17. doi: 10.1016/j.biochi.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Zhu G. 2015. Quantitative RT-PCR assay for high-throughput screening (HTS) of drugs against the growth of Cryptosporidium parvum in vitro. Front Microbiol 6:991. doi: 10.3389/fmicb.2015.00991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alcantara Warren C, Destura RV, Sevilleja JE, Barroso LF, Carvalho H, Barrett LJ, O'Brien AD, Guerrant RL. 2008. Detection of epithelial-cell injury, and quantification of infection, in the HCT-8 organoid model of cryptosporidiosis. J Infect Dis 198:143–149. doi: 10.1086/588819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dann SM, Wang HC, Gambarin KJ, Actor JK, Robinson P, Lewis DE, Caillat-Zucman S, White AC Jr. 2005. Interleukin-15 activates human natural killer cells to clear the intestinal protozoan cryptosporidium. J Infect Dis 192:1294–1302. doi: 10.1086/444393. [DOI] [PubMed] [Google Scholar]
- 36.Morada M, Lee S, Gunther-Cummins L, Weiss LM, Widmer G, Tzipori S, Yarlett N. 2016. Continuous culture of Cryptosporidium parvum using hollow fiber technology. Int J Parasitol 46:21–29. doi: 10.1016/j.ijpara.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 37.Cevallos AM, Bhat N, Verdon R, Hamer DH, Stein B, Tzipori S, Pereira ME, Keusch GT, Ward HD. 2000. Mediation of Cryptosporidium parvum infection in vitro by mucin-like glycoproteins defined by a neutralizing monoclonal antibody. Infect Immun 68:5167–5175. doi: 10.1128/IAI.68.9.5167-5175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edwards H, Thompson RC, Koh WH, Clode PL. 2012. Labeling surface epitopes to identify Cryptosporidium life stages using a scanning electron microscopy-based immunogold approach. Mol Cell Probes 26:21–28. doi: 10.1016/j.mcp.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 39.Fontaine M, Guillot E. 2003. An immunomagnetic separation-real-time PCR method for quantification of Cryptosporidium parvum in water samples. J Microbiol Methods 54:29–36. doi: 10.1016/S0167-7012(03)00005-8. [DOI] [PubMed] [Google Scholar]
- 40.Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Iyer L, Anantharaman V, Aravind L, Kapur V. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.