LETTER
After the pde1 gene was found to be essential for growth in an experimental meningitis model (1), Cron et al. further showed in a 2011 study that Streptococcus pneumoniae mutants with pde1 (SP2205 in TIGR4; SPD2032 in D39) and its paralogue pde2 (SP1298 in TIGR4; SPD1153 in D39) knocked out exhibited reduced host cell adherence and attenuated virulence in a mouse model of meningitis (2). Following work confirmed that Pde1 acts as a phosphodiesterase, cleaving c-di-AMP into pApA (3, 4). These signaling molecules are known to have broad effects on the cell (5) and were again shown to affect growth and virulence in a mouse model of pneumonia. In both studies, the authors suggested that these proteins are promising vaccine targets; however, further evidence of their importance in human infection is needed to bolster these claims.
In a recent study of 674 adults with culture-proven pneumococcal meningitis (6), we searched concurrently sampled bacterial genomes from the blood and cerebrospinal fluid (CSF) for adaptation to either niche occurring postinvasion (7). Here we present results of additional analysis performed using this study that support the conclusions of Cron et al. with respect to a natural population.
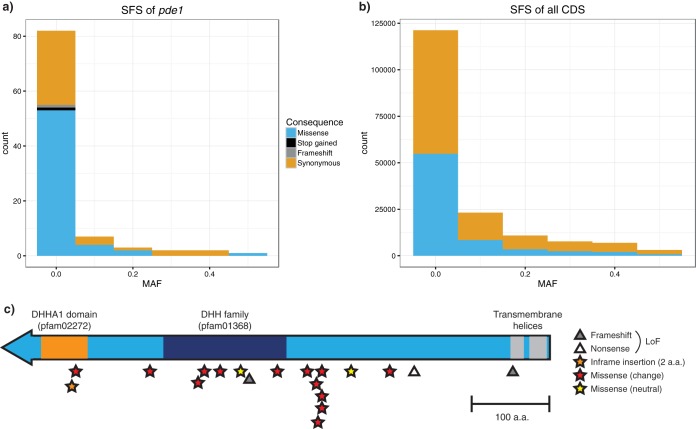
First, we observed that pde1 did not appear to be under selection in the sampled population, as the ratio of nonsynonymous to synonymous mutations was neutral (dN/dS = 0.89) and contained variants with a site frequency spectrum similar to that of other genes (Fig. 1a and b; Tajima's D = −1.69; P = 0.94). However, comparing the variations between samples taken from the same patient during meningitis and given the overall small number of mutations occurring during the rapid progression of disease, pde1 showed a significant enrichment of mutations (P < 10−10). As all these mutations were nonsynonymous, this strongly implies that selection acts on pde1 during the course of invasive disease.
FIG 1.
Evidence of selection on pde1 during meningitis. Panels a and b show the site frequency spectra (SFS; histograms of minor allele frequency) of mutations in just pde1 and in all coding regions (CDS), respectively. Variants are colored according to the predicted effect. Panel c shows the positions and predicted effects of mutations observed in pde1 during cases of meningitis and pfam predicted domains. MAF, minor allele frequency; a.a., amino acids.
We computationally predicted (8, 9) the effect of the 19 mutations observed to occur in pde1 during meningitis and have plotted these along with the predicted functional domains in Fig. 1c. Of these mutations, 14 are predicted to change protein function, without causing a loss of function (LoF). The mutations are not evenly distributed across the gene and are mostly clustered in the DHH family domain or just before it. While this does not allow a singular interpretation of the effect of these variants on gene function, we are able to conclude that selection appears to be operating on pde1 during meningitis.
This corollary from our study therefore strongly supports the conclusion of Cron et al. that pde1 is essential for virulence and additionally shows variation to be important in specific regions of pde1 which should be considered in follow-up work. Together, these studies give good evidence that Pde1 might be an important component of a pneumococcal protein vaccine.
Ed. Note: The author of the published article did not feel that a response was necessary.
REFERENCES
- 1.Molzen TE, Burghout P, Bootsma HJ, Brandt CT, van der Gaast-de Jongh CE, Eleveld MJ, Verbeek MM, Frimodt-Møller N, Østergaard C, Hermans PWM. 2011. Genome-wide identification of Streptococcus pneumoniae genes essential for bacterial replication during experimental meningitis. Infect Immun 79:288–297. doi: 10.1128/IAI.00631-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cron LE, Stol K, Burghout P, van Selm S, Simonetti ER, Bootsma HJ, Hermans PWM. 2011. Two DHH subfamily 1 proteins contribute to pneumococcal virulence and confer protection against pneumococcal disease. Infect Immun 79:3697–3710. doi: 10.1128/IAI.01383-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai Y, Yang J, Eisele LE, Underwood AJ, Koestler BJ, Waters CM, Metzger DW, Bai G. 2013. Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di-AMP phosphodiesterase activity and affect bacterial growth and virulence. J Bacteriol 195:5123–5132. doi: 10.1128/JB.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuipers K, Gallay C, Martínek V, Rohde M, Martínková M, van der Beek SL, Jong WS, Venselaar H, Zomer A, Bootsma H, Veening JW, de Jonge MI. 2016. Highly conserved nucleotide phosphatase essential for membrane lipid homeostasis in Streptococcus pneumoniae. Mol Microbiol 101:12–26. doi: 10.1111/mmi.13312. [DOI] [PubMed] [Google Scholar]
- 5.Tamayo R, Pratt JT, Camilli A. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol 61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bijlsma MW, Brouwer MC, Kasanmoentalib ES, Kloek AT, Lucas MJ, Tanck MW, van der Ende A, van de Beek D. 2016. Community-acquired bacterial meningitis in adults in the Netherlands, 2006–14: a prospective cohort study. Lancet Infect Dis 16:339–347. doi: 10.1016/S1473-3099(15)00430-2. [DOI] [PubMed] [Google Scholar]
- 7.Lees JA, Kremer PHC, Manso AS, Croucher NJ, Ferwerda B, Serón MV, Oggioni MR, Parkhill J, Brouwer MC, van der Ende A, van de Beek D, Bentley SD. 2017. Large scale genomic analysis shows no evidence for pathogen adaptation between the blood and cerebrospinal fluid niches during bacterial meningitis. Microb Genom 3:1–12. doi: 10.1099/mgen.0.000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. 2012. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]