ABSTRACT
Bacterial oligoribonuclease (Orn) is a conserved 3′-to-5′ exonuclease. In Pseudomonas aeruginosa, it has been demonstrated that Orn plays a major role in the hydrolysis of pGpG, which is required for cyclic-di-GMP homeostasis. Meanwhile, Orn is involved in the degradation of nanoRNAs, which can alter global gene expression by serving as transcription initiation primers. Previously, we found that Orn is required for the type III secretion system and pathogenesis of P. aeruginosa, indicating a role of Orn in the bacterial response to environmental stimuli. Here we report that Orn is required for the tolerance of P. aeruginosa to ciprofloxacin. Transcriptome analysis of an orn mutant revealed the upregulation of pyocin biosynthesis genes. Mutation of genes involved in pyocin biosynthesis in the background of an orn mutant restored bacterial tolerance to ciprofloxacin. We further demonstrate that the upregulation of pyocin biosynthesis genes is due to RecA-mediated autoproteolysis of PrtR, which is the major negative regulator of pyocin biosynthesis genes. In addition, the SOS response genes were upregulated in the orn mutant, indicating a DNA damage stress. Therefore, our results revealed a novel role of Orn in bacterial tolerance to ciprofloxacin.
KEYWORDS: oligoribonuclease, pyocin, Pseudomonas aeruginosa, ciprofloxacin
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic, pathogenic Gram-negative bacterium which causes acute and chronic infections in humans. P. aeruginosa is intrinsically resistant to antibiotics largely owing to multiple chromosomally encoded multidrug efflux systems as well as low membrane permeability (1). Interestingly, numerous genes that sensitize bacteria to antibiotics have also been identified in the chromosome of P. aeruginosa (2–4). For example, pyocin biosynthesis genes contribute to bacterial susceptibility to quinolones, as mutations in those genes increase bacterial resistance to quinolones. Pyocins are bacteriocins that mainly target other P. aeruginosa strains (5). The release of pyocins is mediated by holin-like PA0614 and lysozyme-like PA0629, and pyocin release results in the lysis of the producer cells (6, 7). Expression of pyocin genes is repressed by a DNA binding protein, PrtR. Genotoxic agents, such as mitomycin C and quinolones, induce DNA damage and subsequently activate RecA. The activated RecA then induces the autoproteolysis of PrtR, resulting in the derepression of PrtN, which directly activates the expression of pyocin biosynthesis genes and the subsequent release of pyocins through cell lysis (7–9).
Besides individual genes, bacterial growth modes affect antibiotic resistance levels. Bacteria growing in sessile biofilms are much more resistant to various antibiotics and environmental stresses than planktonic bacteria (10–12). Bacteria inside biofilms are shielded from host phagocytes and antibiotics by the biofilm matrix, which is mainly composed of polysaccharide, DNA, and proteins (13–15). In addition, multidrug efflux systems are upregulated in bacteria embedded in biofilms (16), and the slow growth of bacteria inside biofilms further enhances bacterial resistance to antibiotics (17).
Biofilm formation is regulated by a second messenger, cyclic-di-GMP (c-di-GMP) (18, 19), which is synthesized by enzymes containing glycine-glycine-aspartate-glutamate-phenylalanine (GGDEF) domains and degraded by enzymes containing histidine-aspartate-glycine-tyrosine-proline (HD-GYP) or glutamate-alanine-leucine (EAL) domains (20). EAL domain-containing enzymes hydrolyze c-di-GMP to 5′-phosphoguanylyl-(3′, 5′)-guanosine (pGpG) (21), which is degraded primarily by the oligoribonuclease (Orn). Mutation of orn leads to the accumulation of pGpG, which inhibits the function of EAL domain-containing enzymes, resulting in increased levels of c-di-GMP and the consequent hyperbiofilm phenotype (22, 23).
Orn is a highly conserved 3′-to-5′ exonuclease (24). Other than degrading pGpG, Orn plays a major role in the hydrolysis of 2- to 5-nucleotide (nt) RNAs, namely, nanoRNAs (25). NanoRNAs can serve as primers for transcription initiation. The aberrant accumulation of nanoRNAs due to defective Orn alters global gene expression (26). Previously, we found that Orn is required for the expression of the type III secretion system (T3SS) genes in P. aeruginosa (27). Whether Orn plays other roles during infection or in the bacterial response to environmental stresses is not known.
In this study, we investigated the role of Orn in bacterial resistance to antibiotics. Mutation of orn drastically increased bacterial susceptibility to quinolones but not to tetracycline, aminoglycoside, or β-lactam antibiotics. Gene expression profile analysis and genetic experimentation demonstrated that the upregulation of pyocin biosynthesis genes contributed to the hypersusceptibility to quinolones. We further found that RecA-mediated PrtR autoproteolysis was responsible for the upregulation of pyocin biosynthesis genes. In addition, SOS response genes were found to be upregulated in the orn mutant. Thus, our results revealed a novel role of Orn in genome integrity and bacterial resistance to quinolones.
RESULTS
Oligoribonuclease is required for bacterial resistance to fluoroquinolones.
In P. aeruginosa, oligoribonuclease controls the intracellular levels of c-di-GMP and nanoRNA, both of which affect the expression of multiple genes (22, 26, 28, 29). To test the overall influence of oligoribonuclease on bacterial antibiotic resistance, we examined the MICs of various antibiotics for a Δorn mutant. Mutation of orn rendered the bacteria slightly more susceptible to tetracycline and carbenicillin (Table 1). Unexpectedly, the Δorn mutant was much more susceptible to quinolones and showed 8- and 4-fold decreases in the MICs of ciprofloxacin and ofloxacin, respectively. Complementation with an orn gene (Δorn/orn) restored bacterial resistance (Table 1). These results suggest that oligoribonuclease plays an essential role in bacterial resistance to quinolones.
TABLE 1.
Bacterial susceptibilities to antibiotics
| Strain | MIC (μg/ml)a |
||||||
|---|---|---|---|---|---|---|---|
| TET | TOB | GM | CAR | MEM | CIP | OFL | |
| PA14 | 12.5 | 0.625 | 1.25 | 100 | 2.5 | 0.25 | 1 |
| PA14 Δorn | 6.25 | 0.625 | 1.25 | 50 | 2.5 | 0.03 | 0.25 |
| PA14 Δorn/orn | 25 | 0.625 | 1.25 | 100 | 2.5 | 0.25 | 1 |
TET, tetracycline; TOB, tobramycin; GM, gentamicin; CAR, carbenicillin; MEM, meropenem; CIP, ciprofloxacin; OFL, ofloxacin.
Upregulation of pyocin genes contributes to increased susceptibility to ciprofloxacin in the orn mutant.
To understand the mechanism of increased susceptibility to ciprofloxacin in the Δorn mutant, we performed transcriptome analyses. No significant decrease in the levels of expression of multidrug efflux system genes in the Δorn mutant compared to those in wild-type strain PA14 was observed (see Table S1 in the supplemental material). Of note, all the genes involved in pyocin biosynthesis were upregulated (Table 2). Real-time PCR assays confirmed the upregulation of genes involved in the production of F-, R-, and S-type pyocins (Fig. 1). Complementation with an orn gene restored the expression levels of those genes (Fig. 1).
TABLE 2.
mRNA levels of pyocin biosynthesis genes in the Δorn mutant compared to those in wild-type strain PA14
| PA14 locus tag | PAO1 locus tag | Product | Fold changea | P value |
|---|---|---|---|---|
| PA14_07970 | PA0612 | Repressor, PtrB | 3.053 | 1.892E−03 |
| PA14_07980 | PA0613 | Hypothetical protein | 3.238 | 6.263E−11 |
| PA14_08000 | PA0615 | Hypothetical protein | 3.585 | 2.459E−12 |
| PA14_08010 | PA0616 | Hypothetical protein | 3.695 | 1.222E−08 |
| PA14_08020 | PA0617 | Probable bacteriophage protein | 3.709 | 2.966E−09 |
| PA14_08030 | PA0618 | Probable bacteriophage protein | 3.664 | 1.997E−08 |
| PA14_08040 | PA0619 | Probable bacteriophage protein | 3.626 | 3.419E−13 |
| PA14_08050 | PA0620 | Probable bacteriophage protein | 2.924 | 2.020E−10 |
| PA14_08060 | PA0621 | Conserved hypothetical protein | 2.376 | 4.971E−05 |
| PA14_08070 | PA0622 | Probable bacteriophage protein | 3.234 | 5.329E−12 |
| PA14_08090 | PA0623 | Probable bacteriophage protein | 2.999 | 1.919E−10 |
| PA14_08100 | PA0624 | Hypothetical protein | 3.342 | 8.559E−11 |
| PA14_08120 | PA0625 | Hypothetical protein | 3.299 | 8.352E−09 |
| PA14_08130 | PA0626 | Hypothetical protein | 3.377 | 3.302E−05 |
| PA14_08140 | PA0627 | Conserved hypothetical protein | 3.108 | 3.297E−08 |
| PA14_08150 | PA0628 | Conserved hypothetical protein | 3.385 | 5.495E−08 |
| PA14_08160 | PA0629 | Conserved hypothetical protein | 3.689 | 6.944E−07 |
| PA14_08180 | PA0630 | Hypothetical protein | 4.223 | 2.695E−07 |
| PA14_08200 | PA0632 | Hypothetical protein | 3.000 | 4.495E−06 |
| PA14_08210 | PA0633 | Hypothetical protein | 2.824 | 4.475E−09 |
| PA14_08220 | PA0634 | Hypothetical protein | 2.740 | 1.271E−08 |
| PA14_08230 | PA0635 | Hypothetical protein | 2.783 | 3.407E−06 |
| PA14_08240 | PA0636 | Hypothetical protein | 3.454 | 8.654E−11 |
| PA14_59220 | PA0985 | Pyocin S5 | 2.337 | 6.858E−07 |
The data represent the fold change in PA14 Δorn compared with that in PA14, which was given a value of 1.
FIG 1.
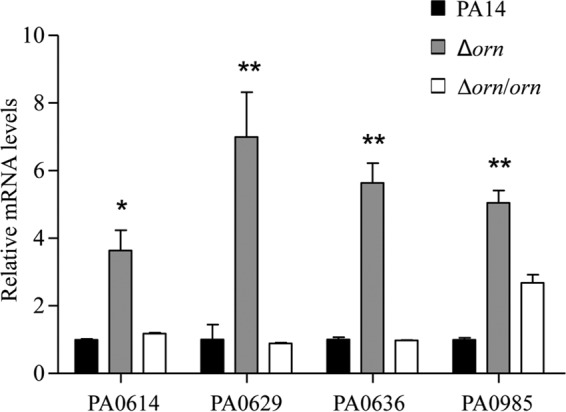
Expression levels of PA0614, PA0629, PA0636, and PA0985. Total RNA was isolated from the indicated strains, and the mRNA levels were determined by real-time PCR by use of PA0668.1 as an internal control. The results shown represent data from three independent experiments with similar results. Error bars represent standard deviations. *, P < 0.05, and **, P < 0.01, compared to PA14 or the complemented strain by Student's t test.
In P. aeruginosa, pyocin biosynthesis is induced by genotoxic agents, such as fluoroquinolones, mitomycin C, and UV light. The production and release of pyocins result in cell lysis (7, 9). Mutation of pyocin biosynthesis genes increases bacterial resistance to ciprofloxacin (2, 3). To test the role of pyocin biosynthesis genes in the hypersusceptibility of the Δorn mutant, we knocked out prtN and PA0629, which encode a positive regulator of pyocin biosynthesis genes and a putative lysozyme, respectively. Consistent with previous reports, mutation of prtN or PA0629 in wild-type strain PA14 increased the level of bacterial resistance to ciprofloxacin by 2-fold (2, 3). However, mutation of prtN or PA0629 in the Δorn mutant increased the level of resistance up to 8-fold (Table 3). These results suggest that the upregulation of pyocin biosynthesis genes in the orn mutant contributes to the increased susceptibility to ciprofloxacin.
TABLE 3.
Bacterial susceptibilities to ciprofloxacin
| Strain | Ciprofloxacin MIC (μg/ml) |
|---|---|
| PA14 | 0.25 |
| Δorn | 0.03 |
| PA14 ΔprtN | 0.5 |
| PA14 ΔPA0629 | 0.5 |
| PA14 Δorn ΔprtN | 0.25 |
| PA14 Δorn ΔPA0629 | 0.25 |
| PA14/pMMB67EH | 0.25 |
| PA14 Δorn/pMMB67EH | 0.03 |
| PA14/pMMB67EH-prtRS162A-His | 0.5 |
| PA14 Δorn/pMMB67EH-prtRS162A-His | 0.1 |
| PA14 ΔrecA::Gm | 0.03 |
| PA14 Δorn ΔrecA::Gm | 0.01 |
PrtR stability is reduced by the mutation of orn.
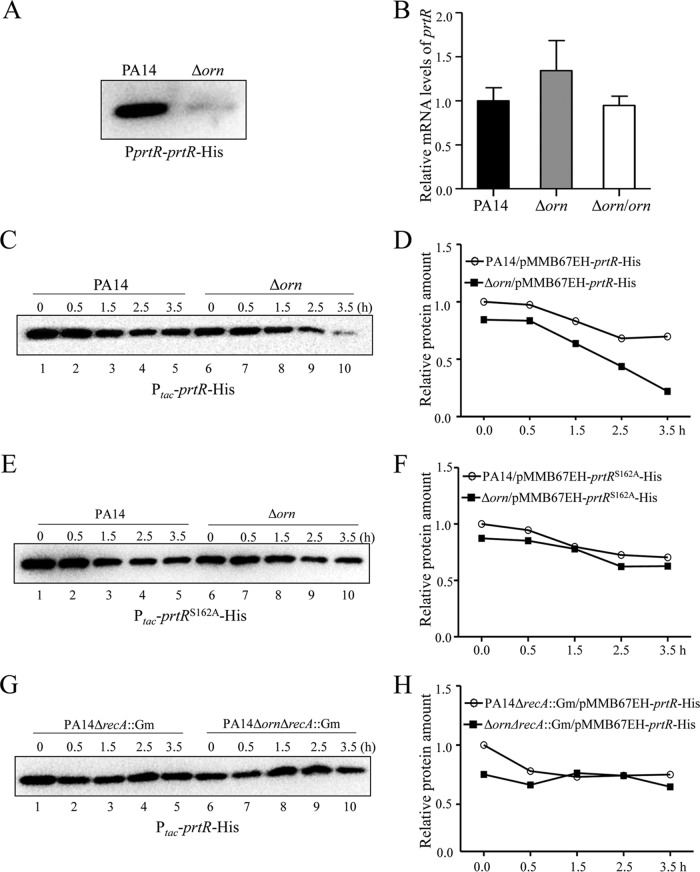
The expression of prtN is directly repressed by PrtR (8). Previously, we demonstrated that the PA0612-PA0613 operon is directly regulated by PrtR (30). Real-time PCR assays revealed higher levels of PrtN and PA0613 mRNA in the Δorn mutant (Fig. 2A and B), suggesting a reduced PrtR protein level.
FIG 2.
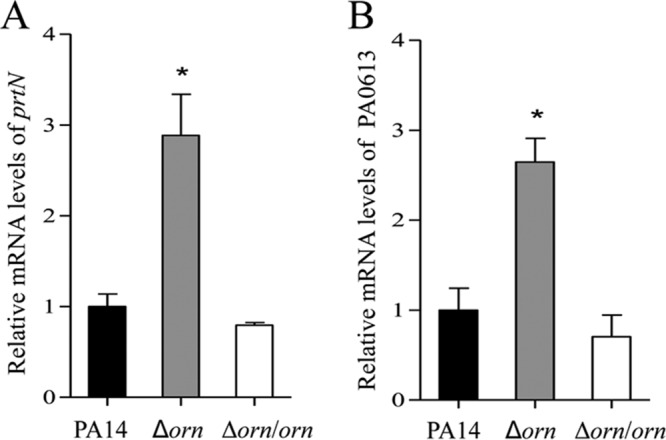
Expression levels of prtN and PA0613. The relative levels of prtN (A) and PA0613 (B) mRNA in the indicated strains were determined by real-time PCR, with PA0668.1 serving as an internal control. The results shown represent data from three independent experiments with similar results. Error bars represent standard deviations. *, P < 0.05 compared to PA14 or the complemented strain by Student's t test.
To test the level of the PrtR protein, a prtR gene that had a His tag at its C terminus and whose expression was driven by its native promoter (PprtR-prtR-His) (31) was transferred into wild-type strain PA14 and the Δorn mutant. Indeed, the PrtR-His level was lower in the Δorn mutant than the PA14 parent strain (Fig. 3A). However, the PrtR mRNA level in the Δorn mutant was 1.3-fold of that in wild-type strain PA14 (Fig. 3B), indicating posttranscriptional regulation. Next, we constructed a prtR gene that had a His tag at its C terminus and whose expression was driven by an exogenous tac promoter (Ptac-prtR-His). Transcription of the prtR-His was induced by IPTG (isopropyl-β-d-thiogalactopyranoside). To monitor the stability of PrtR, chloramphenicol was added to the culture to block protein translation and the PrtR-His levels were subsequently determined. Right after chloramphenicol was added, the PrtR-His level in the Δorn mutant was slightly lower than that in wild-type strain PA14 (Fig. 3C, lanes 1 and 6, and D). However, the PrtR-His level dropped faster in the Δorn mutant (Fig. 3C and D), indicating decreased stability.
FIG 3.
Expression levels of PrtR. (A) Protein levels of PrtR in PA14 or the Δorn mutant with PprtR-prtR-His integrated into the chromosome. Bacteria were grown to an OD600 of 1.0, and bacterial samples were collected. The levels of PrtR-His were determined by Western blotting. (B) The relative levels of prtR mRNA in wild-type strain PA14 and the Δorn mutant were determined by real-time PCR, with PA0668.1 serving as an internal control. Bacteria of the indicated strains carrying pMMB67EH-prtR-His (C, D, G, H) or pMMB67EH-prtRS162A-His (E, F) were grown to an OD600 of 0.4 to 0.6, followed by addition of IPTG to 0.01 mM. After 1 h, 500 μg/ml chloramphenicol was added to the medium. Then, bacteria of each strain were collected at the indicated time points. Samples from equivalent numbers of bacterial cells were loaded onto an SDS-polyacrylamide gel, and the levels of PrtR-His were determined by Western blotting. The band intensities were quantified with ImageJ software.
Degradation of PrtR is mediated by its autoproteolytic activity (31). Replacement of the serine residue at codon 162 with an alanine (PrtRS162A) inactivates the autoprotease domain, which increases the stability of the protein (32, 33). To test whether the accelerated degradation of PrtR is due to autoproteolysis, we constructed a prtR gene with the S162A mutation (prtRS162A) that had a His tag at the C terminus and whose expression was driven by a tac promoter (Ptac-prtRS162A-His) and monitored the stability of the protein. The initial PrtRS162A-His level in the Δorn mutant was slightly lower than that in the wild-type strain PA14. Nevertheless, the PrtRS162A-His proteins exhibited similar stabilities in the two strains (Fig. 3E and F). These results suggest that the orn mutation leads to enhanced autoproteolysis of PrtR.
Overexpression of PrtRS162A increases the resistance of the Δorn mutant to ciprofloxacin.
If the enhanced autoproteolysis of PrtR plays a major role in the increased susceptibility to ciprofloxacin, the overexpression of stable PrtRS162A should increase bacterial resistance to ciprofloxacin. Indeed, overexpression of PrtRS162A in the Δorn mutant increased the MIC of ciprofloxacin by 4-fold, whereas in the wild-type strain, the MIC was increased by 2-fold (Table 3). These results suggest that overexpression of PrtRS162A has a more profound effect on the Δorn mutant than on wild-type strain PA14.
Overall, our results shown above demonstrate that mutation of orn leads to enhanced autoproteolysis of PrtR and, subsequently, the upregulation of pyocin genes, which results in increased bacterial susceptibility to ciprofloxacin.
RecA plays a role in the enhanced autoproteolysis of PrtR in the Δorn mutant.
The autoproteolytic activity of PrtR is activated by RecA (31). To assess the role of RecA in the Δorn mutant, we replaced the coding region of recA with an aacC1 cassette (ΔrecA::Gm) and monitored the stability of PrtR-His. Mutation of recA resulted in similar stabilities of PrtR-His in the two strains (Fig. 3G and H). Replacement of the recA gene with an aacC1 cassette (ΔrecA::Gm) increased bacterial susceptibility to ciprofloxacin. Nevertheless, the difference in the MIC of ciprofloxacin between PA14 and the Δorn mutant was reduced from 8-fold to 2-fold by the mutation of recA (Table 3), indicating a role of RecA in the increased susceptibility of the Δorn mutant.
RecA senses DNA damage and initiates an SOS response by inducing the autoproteolysis of LexA (34). Our results so far implied an impairment of DNA integrity in the Δorn mutant. We then measured the expression levels of SOS response genes with a LexA box in the promoter regions. Indeed, the mRNA levels of recN, uvrA, uvrD, and lexA were higher in the Δorn mutant, indicating a stronger SOS response (Fig. 4). These results suggest that mutation of orn might impair DNA integrity, which promotes the RecA-mediated autoproteolysis of PrtR and the subsequent upregulation of pyocin biosynthesis genes, resulting in increased bacterial susceptibility to ciprofloxacin.
FIG 4.
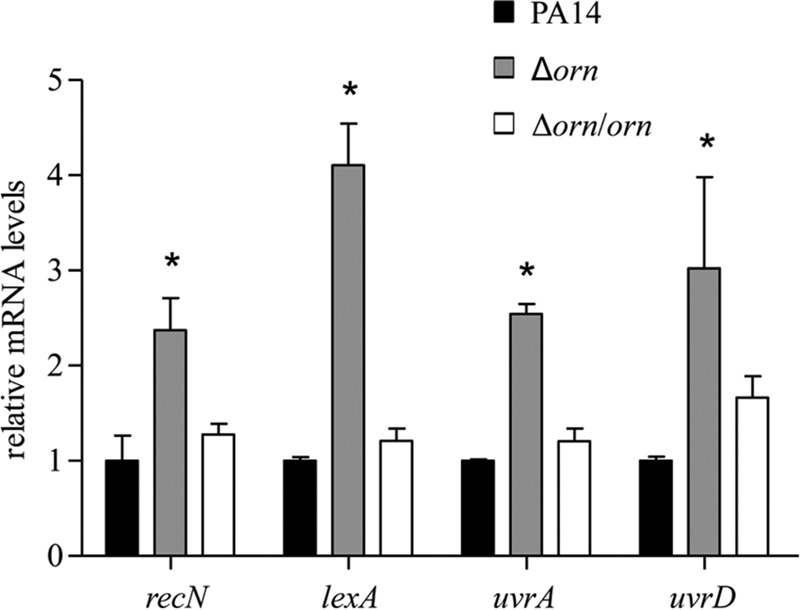
Expression levels of SOS response genes. Total RNA was isolated from wild-type strain PA14, the Δorn mutant, and the complemented strain. The relative levels of recN, lexA, uvrA and uvrD mRNA were determined by real-time PCR, with PA0668.1 serving as an internal control. Error bars indicate standard deviations. *, P < 0.05 compared to PA14 or the complemented strain by Student's t test.
DISCUSSION
In this study, we found that Orn is required for the stability of PrtR. PrtR is homologous to λ phage CI (7). It binds to and represses the promoter of prtN, which encodes the positive regulator for pyocin biosynthesis genes (8). Genotoxic agents initiate the SOS response, during which RecA induces the autoproteolysis of PrtR and the upregulation of pyocin biosynthetic genes (31). Since the release of pyocins is through bacterial cell lysis, the production of the pyocins is under tight regulation. For an individual bacterium, pyocin production and release are detrimental to its survival in response to DNA damage. As a proof, mutations in the pyocin biosynthesis genes increase bacterial resistance to genotoxic agents, such as ciprofloxacin and UV light (2, 3).
However, pyocins are also believed to play a role in bacterial competition by targeting other P. aeruginosa strains (35, 36). Therefore, the production and release of pyocins in an unfavorable environment might secure the niche by killing other bacterial strains (37–40). In addition, Oliveira et al. demonstrated that pyocins released by one strain stimulate biofilm formation by a susceptible strain (41). Thus, foreign pyocins might be regarded as a danger signal that induces defense mechanisms for protection and competition.
Besides stimulating biofilm formation in heterologous strains, pyocins have been shown to play important roles in biofilm formation in the isogenic population (42). Turnbull et al. demonstrated that PA0629, which encodes a peptidoglycan-degrading endolysin, is required for the release of extracellular DNA (eDNA), cytosolic contents, and membrane vesicles (MVs) (42). MVs and eDNA play important roles in biofilm formation and integrity (43–47). Interestingly, mutation of orn resulted in the upregulation of pyocin biosynthesis genes as well as increased levels of the biofilm-promoting molecule c-di-GMP (22). Consistent with this, the orn mutant displays a hyperbiofilm phenotype (23). Overexpression of PA2133 (an EAL domain-containing protein) reduced the intracellular c-di-GMP level and the level of biofilm formation in the orn mutant (22). Given the role of pyocin in biofilm formation, it will be interesting to examine whether the upregulated pyocin biosynthesis genes contribute to the hyperbiofilm phenotype of the orn mutant.
In this study, we found that the upregulation of pyocin biosynthesis genes in the orn mutant was due to the RecA-mediated autoproteolysis of PrtR. Overexpression of PA2133 in the orn mutant did not restore the expression levels of those genes or bacterial resistance to ciprofloxacin (data not shown). In addition, we observed higher expression levels of SOS genes in the orn mutant. These results indicate that mutation of orn might impair the integrity of the genome, which activates RecA. One of the major effects of orn mutation is the accumulation of nanoRNAs, which function as primers in transcription initiation, thus altering global gene expression (26). It might be possible that genes involved in DNA replication or repair are aberrantly regulated in the orn mutant, and this aberrant regulation leads to activation of the SOS response. Another possibility is that the accumulated nanoRNAs might anneal to single-stranded DNA during DNA replication, interfering with normal DNA synthesis, which induces the SOS response. Further studies are needed to fully elucidate the biological role of Orn.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 4. Bacteria were grown in L broth (5 g of NaCl, 5 g of yeast extract, 10 g of tryptone per liter, pH 7.0) or L agar (L broth with 1.5% [wt/vol] agar) at 37°C. The concentrations of the antibiotics used were as follows: for Escherichia coli, ampicillin at 100 μg/ml, gentamicin at 10 μg/ml, and tetracycline at 10 μg/ml; for P. aeruginosa, carbenicillin at 150 μg/ml, tetracycline at 100 μg/ml, and gentamicin at 150 μg/ml.
TABLE 4.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics or function | Source |
|---|---|---|
| P. aeruginosa strains | ||
| PA14 | Wild-type P. aeruginosa strain | David Bradley |
| PA14 Δorn | orn deletion mutant of PA14 | This study |
| PA14 Δorn/orn | PA14 Δorn mutant complemented with the orn gene inserted into the chromosome | This study |
| PA14 ΔprtN | prtN deletion mutant of PA14 | This study |
| PA14 ΔPA0629 | PA0629 deletion mutant of PA14 | This study |
| PA14 Δorn ΔprtN | orn and prtN double deletion mutant of PA14 | This study |
| PA14 Δorn ΔPA0629 | orn and PA0629 double deletion mutant of PA14 | This study |
| PA14 ΔrecA::Gm | PA14 with recA disrupted by insertion of a Gm resistance cassette; Gmr | This study |
| PA14 Δorn ΔrecA::Gm | PA14 Δorn with recA disrupted by insertion of a Gm resistance cassette; Gmr | This study |
| Plasmids | ||
| pMMB67EH | Shuttle vector between E. coli and P. aeruginosa; Ampr | S. Jin lab |
| pUC18T-Mini-Tn7T-Tc | For gene insertion in chromosome; Tcr | Herbert Schweizer |
| pEX18Tc | Gene replacement vector; Tcr oriT+ sacB+ | Herbert Schweizer |
| pEX18Ap | Gene replacement vector; Apr oriT+ sacB+ | Herbert Schweizer |
| pE1923 | pEX18Tc with prtN deleted; Tcr | This study |
| pE1797 | pEX18Ap with recA deleted; Ampr | This study |
| pE1962 | pEX18Tc with PA0629 deleted; Tcr | This study |
| pE1777 | prtR gene of PA14 on pUC18T-Mini-Tn7T-Tc with 500-bp upstream regions of prtR ORFa; Tcr | This study |
| pE1990 | pMMB67EH-prtR-His | This study |
| pE1986 | pMMB67EH-prtRS162A-His | This study |
ORF, open reading frame.
Plasmid construction.
For DNA manipulations, standard protocols or the instructions of the manufacturers of commercial products were followed. Plasmid pE1923 was constructed by cloning a 1,069-bp upstream fragment and a 941-bp downstream fragment of the prtN coding region into the EcoRI-HindIII sites of plasmid pEX18Tc. pE1962 was constructed by cloning a 1,245-bp upstream fragment and a 1,208-bp downstream fragment of the PA0629 coding region into the EcoRI-HindIII sites of plasmid pEX18Tc. A 500-bp upstream region of the prtR open reading frame was amplified from the chromosomal DNA of wild-type P. aeruginosa strain PA14 by PCR and cloned into the KpnI-HindIII sites of pUC18T-Mini-Tn7T-Tc, resulting in pE1777. The prtR coding region was amplified from PA14 chromosomal DNA and cloned into the KpnI-HindIII sites of pMMB67EH, resulting in pE1990, where the prtR gene is under the control of a tac promoter and fused with a His tag at the C terminus. A plasmid carrying the prtRS162A mutation was constructed by replacing the serine at codon 162 with an alanine codon by overlapping PCR.
RNA sequencing and data analysis.
Wild-type strain PA14 and the orn mutant were cultured in LB broth at 37°C and grown to log phage (optical density at 600 nm [OD600], 0.8 to 1.0). Total RNA was isolated with an RNeasy Protect Bacteria minikit with on-column DNase I digestion (Qiagen, Shanghai, China). A second round of DNase treatment was performed following a vigorous protocol with a Turbo DNA-free kit (Ambion). 16S, 23S, and 5S rRNAs were removed by using a Ribo-Zero magnetic kit (Bacteria; Epicentre).
Gene expression profiling was carried out by use of the Illumina RNA sequencing (RNA-Seq) technology. Three biological replicates of each sample were performed for RNA-Seq. The rRNA-depleted RNA was fragmented to 150 to 200 bp, and then first- and second-strand cDNAs were synthesized, followed by end repair and adapter ligation. After 12 cycles of PCR enrichment, the libraries were sequenced using an Illumina HiSeq 2500 platform with a paired-end protocol and read lengths of 100 nt.
The sequence data were analyzed using a method described previously (48). Briefly, sequence reads were mapped onto the reference genome of PA14 (GenBank accession number NC_008463) using the CLC Genomics Workbench (version 8.0) program (CLC Bio-Qiagen, Aarhus, Denmark). Then the count data for the expression values were analyzed using the DESeq package of R/Bioconductor software. Genes with different expression levels were identified by performing a negative binomial test using the DESeq package, a cutoff fold change of greater than 2, and a Benjamini-Hochberg (BH)-adjusted P value of less than 0.05. The raw sequence reads were normalized by dividing by size factors and then transformed to the log2(N + 1) format, where N is the count number.
RNA extraction and quantitative RT-PCR.
Overnight cultures of bacteria were diluted 50-fold into fresh LB medium and grown to an OD600 of 2.0. Total RNA was isolated with an RNeasy minikit (Tiangen Biotech, Beijing, China). cDNA was synthesized from total RNA using PrimeScript reverse transcriptase (TaKaRa, Dalian, China) and random primers. For quantitative real-time PCR (RT-PCR), cDNA was mixed with 4 pmol of reverse and forward primers and SYBR Premix Ex TaqTM II (TaKaRa) in a 20-μl reaction system. The primers used in this study are described in Table 5. Quantitative RT-PCR was performed with a CFX Connect real-time system (Bio-Rad, USA). The 16S ribosomal gene PA0668.1 was used as an internal control (49).
TABLE 5.
Primers used in this study
| Primer | Sequence (5′ → 3′) | Function |
|---|---|---|
| KpnI-prtR1-S | CAGGAGGGTACCGAGGCGAGCCAGGACCAGTT | prtR cloning (500-bp upstream region of prtR ORFa) |
| HindIII-prtR-His-AS | ATTATAAAGCTTTCAGTGGTGGTGGTGGTGGTG ACCTCCCCGCACCAGGGACGGGCCGC | prtR cloning |
| KpnI-prtR2-S | ATCGTCGGTACCTAGGCTCTTTACAGAAAATCCATCG | prtR cloning (43-bp upstream region of prtR ORF) |
| prtR-PM-S | GGCAACGCGATGGAACCGCTGATCAT | prtRS162A mutant cloning |
| prtR-PM-AS | GTTCCATCGCGTTGCCGGTGAGTTGGG | prtRS162A mutant cloning |
| EcoRI-PA0629up-S | CCGGAATTCCAGCCTCTGCTACCACGTCTATGGC | prtR deletion |
| BamHI-PA0629up-AS | CGCGGATCCCGATCCTCCTGCACTCCGATGGGTT | prtR deletion |
| BamH I-PA0629down-S | CGCGGATCCATGAGCCGGCTCGCTCTGCTCCTGC | prtR deletion |
| HindIII-PA0629down-AS | CCCAAGCTTCGTCTCGCCATCTTTCTCGGACAGC | prtR deletion |
| prtN-RT-S | CGACGATAGCCACAAG | RT-PCR |
| prtN-RT-AS | GGATGCGATGCTGTC | RT-PCR |
| prtR-RT-S | GATGCGCAACCTGAAGCA | RT-PCR |
| prtR-RT-AS | TGAATGGTGTTCTGCGAAACC | RT-PCR |
| PA0613-RT-S | GTGGTGGTGGAGCACTATCTCA | RT-PCR |
| PA0613-RT-AS | CCGCAGTGGCGGTACTTC | RT-PCR |
| PA0614-RT-S | CGCTGCCTGCCAAGGA | RT-PCR |
| PA0614-RT-AS | ATCAGTACCCAGAGCGGCATT | RT-PCR |
| PA0629-RT-S | GTGGAGAACCTCAATTACAG | RT-PCR |
| PA0629-RT-AS | TAGGTGTTGTCGGCAATC | RT-PCR |
| PA0636-RT-S | TGGAAGACCCGGCAGAAG | RT-PCR |
| PA0636-RT-AS | CGTTGAGCTTGGACAGATCCT | RT-PCR |
| PA0985-RT-S | TCAAGCCTCTCCATCTAT | RT-PCR |
| PA0985-RT-AS | TCCAGTTCATCTCTAACAAG | RT-PCR |
| recA-RT-S | ATATCAAGAACGCCAACT | RT-PCR |
| recA-RT-AS | TAGAACTTCAGTGCGTTA | RT-PCR |
| recN-RT-S | CCACCAGTTGCTCAG | RT-PCR |
| recN-RT-AS | GTTGCTCAGGGTCTTC | RT-PCR |
| lexA-RT-S | AATCCCGCCTTCTTCAAT | RT-PCR |
| lexA-RT-AS | AATGCCGATGTCCTTCAT | RT-PCR |
| uvrA-RT-S | TCCATCGAACAGAAGTCC | RT-PCR |
| uvrA-RT-AS | CGCAGGTAGTCGTAGATC | RT-PCR |
| uvrB-RT-S | TACTTCGTTTCCTACTAC | RT-PCR |
| uvrB-RT-AS | GAGTCCTTCTCGATATAG | RT-PCR |
| uvrD-RT-S | GAGCTGATCGAGAATCTT | RT-PCR |
| uvrD-RT-AS | TTTCTTCCTTGTGGTAGC | RT-PCR |
| PA0668.1-RT-S | AAGGTCTTCGGATTGTAA | RT-PCR |
| PA0668.1-RT-AS | GTGCTTATTCTGTTGGTAA | RT-PCR |
ORF, open reading frame.
Antibiotic susceptibility assay.
The MICs of various antibiotics for P. aeruginosa were determined by serial 2-fold dilution in LB medium, as described previously (50, 51). The MIC was recorded as the lowest concentration of antibiotic which inhibited visible growth after incubation at 37°C for 24 h.
PrtR stability assay.
To measure the stability of PrtR, overnight bacterial cultures of various P. aeruginosa strains were diluted 50-fold in fresh LB with carbenicillin and cultured for 2 h. When the OD600 reached 0.4 to 0.6, IPTG was added to the medium to reach 0.01 mM. After 1 h, 500 μg/ml chloramphenicol was added to the medium to block protein synthesis (52). Bacterial cells were collected at the time points indicated above after the addition of chloramphenicol. Samples from equivalent numbers of bacterial cells were separated on SDS-polyacrylamide gels (15% polyacrylamide). The proteins were then transferred onto a polyvinylidene difluoride (PVDF) membrane and incubated with rabbit anti-His antibody (1:1,000; Cell Signaling Technology, USA) at room temperature for 1 h. After washing with 1× phosphate-buffered saline (1× PBS; 274 mM NaCl, 5.4 mM KCl, 20 mM Na2HPO4, 4 mM KH2PO4, pH 7.4) containing 2% Tween 20 four times, the membrane was incubated with the secondary antibody, horseradish peroxidase-conjugated goat anti-rabbit IgG (1:2,000; Promega, USA), at room temperature for 1 h. Signals were detected with an ECL Plus kit (Millipore), and bands were visualized with a Bio-Rad molecular imager (ChemiDoc XRS+).
Accession number(s).
The raw RNA sequencing data has been deposited in the NCBI Short Read Archive (SRA) database under accession no. SRP063080.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation of China (31670130, 31370168, and 31370167), the Program of International S&T Cooperation (2015DFG32500), and the Science and Technology Committee of Tianjin (15JCYBJC53900 and 15JCZDJC33000).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02256-16.
REFERENCES
- 1.Driscoll JA, Brody SL, Kollef MH. 2007. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67:351–368. doi: 10.2165/00003495-200767030-00003. [DOI] [PubMed] [Google Scholar]
- 2.Brazas MD, Hancock RE. 2005. Ciprofloxacin induction of a susceptibility determinant in Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:3222–3227. doi: 10.1128/AAC.49.8.3222-3227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breidenstein EB, Khaira BK, Wiegand I, Overhage J, Hancock RE. 2008. Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob Agents Chemother 52:4486–4491. doi: 10.1128/AAC.00222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dotsch A, Becker T, Pommerenke C, Magnowska Z, Jansch L, Haussler S. 2009. Genomewide identification of genetic determinants of antimicrobial drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:2522–2531. doi: 10.1128/AAC.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghequire MG, De Mot R. 2015. The tailocin tale: peeling off phage tails. Trends Microbiol 23:587–590. doi: 10.1016/j.tim.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol 38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- 7.Ghequire MG, De Mot R. 2014. Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol Rev 38:523–568. doi: 10.1111/1574-6976.12079. [DOI] [PubMed] [Google Scholar]
- 8.Matsui H, Sano Y, Ishihara H, Shinomiya T. 1993. Regulation of pyocin genes in Pseudomonas aeruginosa by positive (prtN) and negative (prtR) regulatory genes. J Bacteriol 175:1257–1263. doi: 10.1128/jb.175.5.1257-1263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Penterman J, Singh PK, Walker GC. 2014. Biological cost of pyocin production during the SOS response in Pseudomonas aeruginosa. J Bacteriol 196:3351–3359. doi: 10.1128/JB.01889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hengzhuang W, Wu H, Ciofu O, Song Z, Hoiby N. 2011. Pharmacokinetics/pharmacodynamics of colistin and imipenem on mucoid and nonmucoid Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 55:4469–4474. doi: 10.1128/AAC.00126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Tseng BS, Zhang W, Harrison JJ, Quach TP, Song JL, Penterman J, Singh PK, Chopp DL, Packman AI, Parsek MR. 2013. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol 15:2865–2878. doi: 10.1111/1462-2920.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flemming HC, Neu TR, Wozniak DJ. 2007. The EPS matrix: the “house of biofilm cells.” J Bacteriol 189:7945–7947. doi: 10.1128/JB.00858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 15.Nadell CD, Xavier JB, Foster KR. 2009. The sociobiology of biofilms. FEMS Microbiol Rev 33:206–224. doi: 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- 16.Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. 2008. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol Microbiol 68:223–240. doi: 10.1111/j.1365-2958.2008.06152.x. [DOI] [PubMed] [Google Scholar]
- 17.Brooun A, Liu S, Lewis K. 2000. A Dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 44:640–646. doi: 10.1128/AAC.44.3.640-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol 75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fazli M, Almblad H, Rybtke ML, Givskov M, Eberl L, Tolker-Nielsen T. 2014. Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ Microbiol 16:1961–1981. doi: 10.1111/1462-2920.12448. [DOI] [PubMed] [Google Scholar]
- 20.Römling U, Amikam D. 2006. Cyclic di-GMP as a second messenger. Curr Opin Microbiol 9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem 280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 22.Cohen D, Mechold U, Nevenzal H, Yarmiyhu Y, Randall TE, Bay DC, Rich JD, Parsek MR, Kaever V, Harrison JJ, Banin E. 2015. Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:11359–11364. doi: 10.1073/pnas.1421450112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orr MW, Donaldson GP, Severin GB, Wang J, Sintim HO, Waters CM, Lee VT. 2015. Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover. Proc Natl Acad Sci U S A 112:E5048–E5057. doi: 10.1073/pnas.1507245112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Zhu L, Deutscher MP. 1998. Oligoribonuclease is encoded by a highly conserved gene in the 3′-5′ exonuclease superfamily. J Bacteriol 180:2779–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh S, Deutscher MP. 1999. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc Natl Acad Sci U S A 96:4372–4377. doi: 10.1073/pnas.96.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldman SR, Sharp JS, Vvedenskaya IO, Livny J, Dove SL, Nickels BE. 2011. NanoRNAs prime transcription initiation in vivo. Mol Cell 42:817–825. doi: 10.1016/j.molcel.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen G, Zhao Q, Zhu F, Chen R, Jin Y, Liu C, Pan X, Jin S, Wu W, Cheng Z. 2016. Oligoribonuclease is required for the type III secretion system and pathogenesis of Pseudomonas aeruginosa. Microbiol Res 188-189:90–96. doi: 10.1016/j.micres.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Gupta K, Liao J, Petrova OE, Cherny KE, Sauer K. 2014. Elevated levels of the second messenger c-di-GMP contribute to antimicrobial resistance of Pseudomonas aeruginosa. Mol Microbiol 92:488–506. doi: 10.1111/mmi.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicastro GG, Kaihami GH, Pereira TO, Meireles DA, Groleau MC, Déziel E, Baldini RL. 2014. Cyclic-di-GMP levels affect Pseudomonas aeruginosa fitness in the presence of imipenem. Environ Microbiol 16:1321–1333. doi: 10.1111/1462-2920.12422. [DOI] [PubMed] [Google Scholar]
- 30.Wu W, Jin S. 2005. PtrB of Pseudomonas aeruginosa suppresses the type III secretion system under the stress of DNA damage. J Bacteriol 187:6058–6068. doi: 10.1128/JB.187.17.6058-6068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Z, Shi J, Liu C, Jin Y, Li K, Chen R, Jin S, Wu W. 2014. PrtR homeostasis contributes to Pseudomonas aeruginosa pathogenesis and resistance against ciprofloxacin. Infect Immun 82:1638–1647. doi: 10.1128/IAI.01388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cirz RT, O'Neill BM, Hammond JA, Head SR, Romesberg FE. 2006. Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J Bacteriol 188:7101–7110. doi: 10.1128/JB.00807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slilaty SN, Little JW. 1987. Lysine-156 and serine-119 are required for LexA repressor cleavage: a possible mechanism. Proc Natl Acad Sci U S A 84:3987–3991. doi: 10.1073/pnas.84.12.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedberg EC, Walker GC, Siede W. 2005. DNA repair and mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- 35.Michel-Briand Y, Baysse C. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499–510. doi: 10.1016/S0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 36.Ghequire MG, Dillen Y, Lambrichts I, Proost P, Wattiez R, De Mot R. 2015. Different ancestries of R tailocins in rhizospheric Pseudomonas isolates. Genome Biol Evol 7:2810–2828. doi: 10.1093/gbe/evv184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waite RD, Curtis MA. 2009. Pseudomonas aeruginosa PAO1 pyocin production affects population dynamics within mixed-culture biofilms. J Bacteriol 191:1349–1354. doi: 10.1128/JB.01458-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cornforth DM, Foster KR. 2013. Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol 11:285–293. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- 39.Riley MA, Gordon DM. 1999. The ecological role of bacteriocins in bacterial competition. Trends Microbiol 7:129–133. doi: 10.1016/S0966-842X(99)01459-6. [DOI] [PubMed] [Google Scholar]
- 40.Dingemans J, Ghequire MG, Craggs M, De Mot R, Cornelis P. 2016. Identification and functional analysis of a bacteriocin, pyocin S6, with ribonuclease activity from a Pseudomonas aeruginosa cystic fibrosis clinical isolate. Microbiologyopen 5:413–423. doi: 10.1002/mbo3.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliveira NM, Martinez-Garcia E, Xavier J, Durham WM, Kolter R, Kim W, Foster KR. 2015. Biofilm formation as a response to ecological competition. PLoS Biol 13:e1002191. doi: 10.1371/journal.pbio.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turnbull L, Toyofuku M, Hynen AL, Kurosawa M, Pessi G, Petty NK, Osvath SR, Carcamo-Oyarce G, Gloag ES, Shimoni R, Omasits U, Ito S, Yap X, Monahan LG, Cavaliere R, Ahrens CH, Charles IG, Nomura N, Eberl L, Whitchurch CB. 2016. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun 7:11220. doi: 10.1038/ncomms11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaparakis-Liaskos M, Ferrero RL. 2015. Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol 15:375–387. doi: 10.1038/nri3837. [DOI] [PubMed] [Google Scholar]
- 44.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schooling SR, Hubley A, Beveridge TJ. 2009. Interactions of DNA with biofilm-derived membrane vesicles. J Bacteriol 191:4097–4102. doi: 10.1128/JB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toyofuku M, Roschitzki B, Riedel K, Eberl L. 2012. Identification of proteins associated with the Pseudomonas aeruginosa biofilm extracellular matrix. J Proteome Res 11:4906–4915. doi: 10.1021/pr300395j. [DOI] [PubMed] [Google Scholar]
- 47.Toyofuku M, Tashiro Y, Hasegawa Y, Kurosawa M, Nomura N. 2015. Bacterial membrane vesicles, an overlooked environmental colloid: biology, environmental perspectives and applications. Adv Colloid Interface Sci 226:65–77. doi: 10.1016/j.cis.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 48.Chua SL, Liu Y, Yam JKH, Chen Y, Vejborg RM, Tan BGC, Kjelleberg S, Tolker-Nielsen T, Givskov M, Yang L. 2014. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat Commun 5:4462. doi: 10.1038/ncomms5462. [DOI] [PubMed] [Google Scholar]
- 49.Li M, Long Y, Liu Y, Liu Y, Chen R, Shi J, Zhang L, Jin Y, Yang L, Bai F, Jin S, Cheng Z, Wu W. 2016. HigB of Pseudomonas aeruginosa enhances killing of phagocytes by up-regulating the type III secretion system in ciprofloxacin induced persister cells. Front Cell Infect Microbiol 6:125. doi: 10.3389/fcimb.2016.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jo JT, Brinkman FS, Hancock RE. 2003. Aminoglycoside efflux in Pseudomonas aeruginosa: involvement of novel outer membrane proteins. Antimicrob Agents Chemother 47:1101–1111. doi: 10.1128/AAC.47.3.1101-1111.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alvarez-Ortega C, Wiegand I, Olivares J, Hancock RE, Martinez JL. 2010. Genetic determinants involved in the susceptibility of Pseudomonas aeruginosa to beta-lactam antibiotics. Antimicrob Agents Chemother 54:4159–4167. doi: 10.1128/AAC.00257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leslie DJ, Heinen C, Schramm FD, Thüring M, Aakre CD, Murray SM, Laub MT, Jonas K. 2015. Nutritional control of DNA replication initiation through the proteolysis and regulated translation of DnaA. PLoS Genet 11:e1005342. doi: 10.1371/journal.pgen.1005342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.