ABSTRACT
IncK plasmids are some of the main carriers of blaCTX-M-14 and blaCMY-2 genes and show high similarity to other plasmids belonging to the I complex, including IncB/O plasmids. Here, we studied the phylogenetic relationship of 37 newly sequenced IncK and IncB/O plasmids. We show that IncK plasmids can be divided into two compatible lineages named IncK1 and IncK2.
KEYWORDS: IncK, plasmid, incompatibility
TEXT
Antimicrobial resistance due to extended-spectrum beta-lactamases (ESBL) and AmpC beta-lactamases is a global problem. Among the most prevalent variants are blaCMY-2 and blaCTX-M-14, both of which are often carried on IncK plasmids from different sources (1–7). IncK plasmids are highly related to IncB/O, IncZ, and IncI plasmids, which all belong to the I complex (8). The compatibility of IncB/O plasmids was extensively studied previously (9). Knowledge on IncZ plasmids was limited until recently, when Moran et al. (10) showed that there are multiple variants of IncZ plasmids. To better understand the complexity of the I complex, the purpose of this study was to further investigate the phylogenetic relationship of IncK plasmids to IncB/O and to determine compatibility within and between potential IncK lineages.
Escherichia coli isolates from The Netherlands carrying blaCMY-2 or blaCTX-M-14 were selected from various ESBL studies performed within our laboratory. These were screened for IncK plasmids using previously described primers (11). Sequencing results were used to confirm the presence of an IncK replicon, as in several cases the PCR-based replicon typing (PBRT) gave an ambiguous result. Additionally, an IncB/O plasmid was added as an outgroup.
For all experiments, IncK or IncB/O plasmids were transferred by either transformation or conjugation. For transformation, plasmids were isolated using the Wizard Plus SV kit (Promega) and transformed to E. coli DH10B ElectroMAX cells (Thermo Fisher Scientific), according to the manufacturer's instructions. Selection of transformants was performed on Luria-Bertani (LB) agar plates (Oxoid/Tritium) supplemented with 2 mg/liter cefotaxime (Sigma). Conjugation was performed as previously described (12) with exconjugants recovered on LB plates supplemented with 2 mg/liter cefotaxime and 75 mg/liter rifampin or 25 mg/liter chloramphenicol. Conjugation was confirmed by PCR on relevant targets. Thirty-six IncK and 1 IncB/O plasmid-carrying E. coli were subjected to whole-genome sequencing (WGS). WGS was performed on an Illumina MiSeq platform using 2 × 250-bp reads and a 300-bp insert size. Assembly was performed using SPAdes (13) with the default settings. Chromosomal contigs were removed by mapping against either DH10B, MG1655, or W3110 genome sequences using BLAST (14). The remaining plasmid contigs were annotated using Prokka (15). Screening for antimicrobial resistance genes was performed using ResFinder (16). Core and pan genome determination and whole-plasmid-based phylogeny were performed with Roary (17), using the nonparalog splitting method. A phylogenetic tree was constructed from both newly sequenced and downloaded sequences using FastTree (18). The phylogenetic tree was visualized using interactive tree of life (iTOL) (19). Plasmid contigs were submitted to the European nucleotide archive (http://www.ebi.ac.uk/ena) with the accession numbers listed in Fig. 1. Additional IncK and IncB/O plasmid sequences were obtained from the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/resources/downloads/plasmids/), GenBank, and the European nucleotide archive.
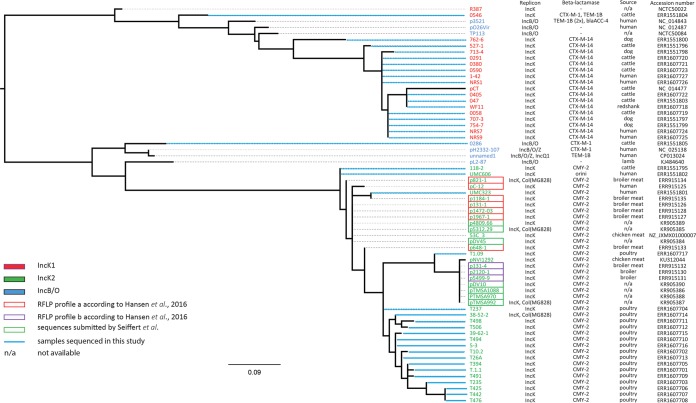
FIG 1.
Phylogenetic tree of IncK and IncB/O plasmids.
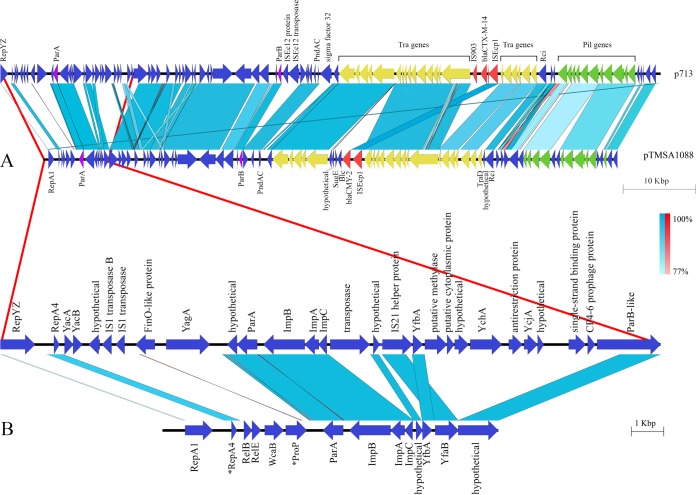
Plasmid sizes ranged from 79,176 to 168,100 bp, with an average GC content of 52.6% and 104 coding sequences. The core genome of all IncK plasmids includes transfer and partition systems and the shufflon recombinase with the pilV gene. blaCTX-M14 was associated with ISEcp1 upstream and IS903 downstream. In contrast, blaCMY-2-carrying plasmids lack IS903, except for one blaCMY-2- and the blaCTX-M-1-carrying plasmid, which both lacked ISEcp1 as well as IS903. IncK1 and IncK2 vary in the presence or absence of several genes (Fig. 2B), which may be caused by the presence of multiple insertion sequences.
FIG 2.
Comparison of the IncK1 and IncK2 plasmid sequences (A) with an additional focus on the highest variable region in the plasmid scaffolds (B). Asterisks indicate partial gene sequences.
Analysis of the phylogenetic tree based on the absence or presence of accessory genes of IncK and IncB/O plasmids (Fig. 1) revealed the presence of two major clusters containing IncK plasmids. The first cluster, designated IncK1, contains plasmids carrying blaCTX-M-14. IncK1 shows high similarity to the previously described pCT (20). The second cluster, IncK2, includes IncK plasmids carrying blaCMY genes. Overall, IncK1 and IncK2 plasmids show high levels of homology (Fig. 2A). Additionally, Hansen et al. (7) presented two IncK plasmid restriction fragment length polymorphism (RFLP) types (SalI digested), which form two distinct clusters, both within the IncK2 lineage (Fig. 1).
Due to accumulation of single nucleotide polymorphisms (SNPs) in the target region of IncK1 plasmids, some typing results were difficult to reproduce using previously designed primers (11). Therefore, a new set of primers was designed (see Table S1 in the supplemental material). New and previously reported primers were mixed in a 1:1 ratio to a final concentration of 2.5 pmol/μl each. PCR was performed using GoTaq green master mix (Promega) with an annealing temperature of 63°C. Additionally, two pairs of primers, targeting RNAI and part of the repY gene, were designed to discriminate between the IncK1 and IncK2 plasmid lineages, using an annealing temperature of 55°C. Selected plasmids were subjected to incompatibility tests, which were performed via conjugation of two E. coli strains carrying different IncK or IncB/O plasmids. To determine the incompatibility of members of the IncK2 lineage, in which all plasmids carry the blaCMY-2 gene, an additional set of primers was used, targeting the ssb gene and ISEcp1 (see Table S1 in the supplemental material), which differed in the two selected IncK2 plasmids. Testing of exconjugants revealed that IncK1 and IncK2 plasmids were compatible (Table 1). In contrast, plasmids belonging to the same group, either IncK1 or IncK2, are incompatible. The compatibility of IncK1 and IncK2 was checked with IncB/O plasmids to confirm that neither of the lineages was a mistyped IncB/O plasmid. Additionally, stability of the plasmids from both lineages in one host was checked (method adapted from Jafar et al. [21]). Stability was determined as a percentage of colonies carrying both plasmids in the absence of selective pressure compared to that of those plated on selective agar. Fifty colonies carrying both IncK1 and IncK2 plasmids were plated on either LB agar or LB agar supplemented with appropriate antibiotics. Subculturing of all colonies was repeated every 24 h for 3 days. The presence of both plasmids was confirmed by PCR, targeting the RNAI and the resistance genes. After 72 h, 98% of the IncK1 plasmids were still present using selective agar and 100% using nonselective agar. The IncK2 plasmid showed 100% stability on both selective and nonselective agar.
TABLE 1.
Incompatibility test results for mating pairs
| Plasmid 1 (plasmid identification) | Plasmid 2 (plasmid identification) | Incompatibility result |
|---|---|---|
| IncK1 (p754) | IncK2 (p118) | Compatible |
| IncK1 (p0291) | IncK2 (p118) | Compatible |
| IncK1 (pWF11) | IncK1 (p527) | Incompatible |
| IncK2 (p39_62_1) | IncK2 (pT1.09) | Incompatible |
| IncK1 (p754) | IncK1 (p0546) | Incompatible |
| IncK2 (pT10.2) | IncK1 (p0546) | Compatible |
| IncK1 (p754) | IncB/O (p0289) | Compatible |
| IncK2 (p118) | IncB/O (p0289) | Compatible |
| IncK2 (p0291) | IncB/O (p0289) | Compatible |
In conclusion, our results show the existence of two IncK plasmid lineages, which confirms the observation of Seiffert et al. (S. N. Seiffert, A. Carattoli, S. Schwendener, A. Endimiani, and V. Perreten, unpublished data), who submitted sequences of seven IncK2 plasmids to GenBank (Fig. 1) (accession no. KR905384 to KR905390). Within one lineage, plasmids are incompatible with each other, but they are compatible between lineages. The phylogenetic analysis could be possibly influenced by the geographical bias of the origin of plasmids included in this study. These findings should therefore be confirmed using an extended collection of plasmids from a more diverse background. The high similarity of IncK, IncB/O, and IncZ RNAI sequences, which are targets in the PBRT classification scheme, causes difficulties with typing. Further analysis is necessary to improve the tools that will allow better detection and discrimination of plasmids of the I complex.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ad C. Fluit from University Medical Center Utrecht (UMCU) for providing E. coli strains carrying IncK plasmids isolated from humans and Nadine Händel and Benno ter Kuile for providing E. coli MG1655 YFP. We also thank Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/resources/downloads/plasmids/) for making the sequences of plasmids R387 and TP113 publicly available. Additionally, we thank Jolien van der Linden for technical assistance.
The research leading to these results has received funding from the European Community's Seventh Framework Program (FP7/2007-2013) under grant agreement number 613754 (EFFORT project).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01954-16.
REFERENCES
- 1.Hordijk J, Wagenaar JA, Kant A, van Essen-Zandbergen A, Dierikx C, Veldman K, Wit B, Mevius D. 2013. Cross-sectional study on prevalence and molecular characteristics of plasmid mediated ESBL/AmpC-producing Escherichia coli isolated from veal calves at slaughter. PLoS One 8:e65681. doi: 10.1371/journal.pone.0065681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hordijk J, Mevius DJ, Kant A, Bos ME, Graveland H, Bosman AB, Hartskeerl CM, Heederik DJ, Wagenaar JA. 2013. Within-farm dynamics of ESBL/AmpC-producing Escherichia coli in veal calves: a longitudinal approach. J Antimicrob Chemother 68:2468–2476. doi: 10.1093/jac/dkt219. [DOI] [PubMed] [Google Scholar]
- 3.Stokes MO, Cottell JL, Piddock LJ, Wu G, Wootton M, Mevius DJ, Randall LP, Teale CJ, Fielder MD, Coldham NG. 2012. Detection and characterization of pCT-like plasmid vectors for blaCTX-M-14 in Escherichia coli isolates from humans, turkeys and cattle in England and Wales. J Antimicrob Chemother 67:1639–1644. doi: 10.1093/jac/dks126. [DOI] [PubMed] [Google Scholar]
- 4.Valverde A, Canton R, Garcillan-Barcia MP, Novais A, Galan JC, Alvarado A, de la Cruz F, Baquero F, Coque TM. 2009. Spread of blaCTX-M-14 is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob Agents Chemother 53:5204–5212. doi: 10.1128/AAC.01706-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhanji H, Khan P, Cottell JL, Piddock LJ, Zhang J, Livermore DM, Woodford N. 2012. Dissemination of pCT-like IncK plasmids harboring CTX-M-14 extended-spectrum beta-lactamase among clinical Escherichia coli isolates in the United Kingdom. Antimicrob Agents Chemother 56:3376–3377. doi: 10.1128/AAC.00313-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baudry PJ, Mataseje L, Zhanel GG, Hoban DJ, Mulvey MR. 2009. Characterization of plasmids encoding CMY-2 AmpC β-lactamases from Escherichia coli in Canadian intensive care units. Diagn Microbiol Infect Dis 65:379–383. doi: 10.1016/j.diagmicrobio.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 7.Hansen KH, Bortolaia V, Nielsen CA, Nielsen JB, Schonning K, Agerso Y, Guardabassi L. 2016. Host-specific patterns of genetic diversity among IncI1-Iγ and IncK plasmids encoding CMY-2 beta-lactamase in Escherichia coli isolates from humans, poultry meat, poultry, and dogs in Denmark. Appl Environ Microbiol 82:4705–4714. doi: 10.1128/AEM.00495-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Praszkier J, Wei T, Siemering K, Pittard J. 1991. Comparative analysis of the replication regions of IncB, IncK, and IncZ plasmids. J Bacteriol 173:2393–2397. doi: 10.1128/jb.173.7.2393-2397.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siemering KR, Praszkier J, Pittard AJ. 1993. Interaction between the antisense and target RNAs involved in the regulation of IncB plasmid replication. J Bacteriol 175:2895–2906. doi: 10.1128/jb.175.10.2895-2906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moran RA, Anantham S, Pinyon JL, Hall RM. 2015. Plasmids in antibiotic susceptible and antibiotic resistant commensal Escherichia coli from healthy Australian adults. Plasmid 80:24–31. doi: 10.1016/j.plasmid.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 12.Dierikx C, van Essen-Zandbergen A, Veldman K, Smith H, Mevius D. 2010. Increased detection of extended spectrum beta-lactamase producing Salmonella enterica and Escherichia coli isolates from poultry. Vet Microbiol 145:273–278. doi: 10.1016/j.vetmic.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 13.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 16.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44(W1):W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cottell JL, Webber MA, Coldham NG, Taylor DL, Cerdeno-Tarraga AM, Hauser H, Thomson NR, Woodward MJ, Piddock LJ. 2011. Complete sequence and molecular epidemiology of IncK epidemic plasmid encoding blaCTX-M-14. Emerg Infect Dis 17:645–652. doi: 10.3201/eid1704.101009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jafar E, Shakibaie MR, Poormasoomi L. 2013. Isolation of a novel antibiotic resistance plasmid DNA from hospital isolates of Pseudomonas aeruginosa. J Clin Exp Pathol 3:140. doi: 10.4172/2161-0681.1000140. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.