ABSTRACT
The objective of this study was to perform an inventory of the extended-spectrum-β-lactamase (ESBL)-producing Enterobacteriaceae isolates responsible for infections in French hospitals and to assess the mechanisms associated with ESBL diffusion. A total of 200 nonredundant ESBL-producing Enterobacteriaceae strains isolated from clinical samples were collected during a multicenter study performed in 18 representative French hospitals. Antibiotic resistance genes were identified by PCR and sequencing experiments. The clonal relatedness between isolates was investigated by the use of the DiversiLab system. ESBL-encoding plasmids were compared by PCR-based replicon typing and plasmid multilocus sequence typing. CTX-M-15, CTX-M-1, CTX-M-14, and SHV-12 were the most prevalent ESBLs (8% to 46.5%). The three CTX-M-type EBSLs were significantly observed in Escherichia coli (37.1%, 24.2%, and 21.8%, respectively), and CTX-M-15 was the predominant ESBL in Klebsiella pneumoniae (81.1%). SHV-12 was associated with ESBL-encoding Enterobacter cloacae strains (37.9%). qnrB, aac(6′)-Ib-cr, and aac(3)-II genes were the main plasmid-mediated resistance genes, with prevalences ranging between 19.5% and 45% according to the ESBL results. Molecular typing did not identify wide clonal diffusion. Plasmid analysis suggested the diffusion of low numbers of ESBL-encoding plasmids, especially in K. pneumoniae and E. cloacae. However, the ESBL-encoding genes were observed in different plasmid replicons according to the bacterial species. The prevalences of ESBL subtypes differ according to the Enterobacteriaceae species. Plasmid spread is a key determinant of this epidemiology, and the link observed between the ESBL-encoding plasmids and the bacterial host explains the differences observed in the Enterobacteriaceae species.
KEYWORDS: ESBL, plasmid-mediated quinolone resistance, AAC(3)-II, AAC(6′)-Ib, plasmid, plasmid-mediated resistance
INTRODUCTION
β-Lactam antibiotics, owing to their bactericidal effect and their low toxicity, are currently the main antibacterial agents used for the treatment of serious infections due to Enterobacteriaceae. However, their increasing use has led to the emergence of extended-spectrum-β-lactamase (ESBL)-producing strains. These enzymes hydrolyze all penicillins, cephalosporins, and oxyimino β-lactams (ceftazidime, cefotaxime, ceftriaxone, cefepime, and aztreonam) but are inactive against carbapenems and cephamycins and are susceptible to clavulanate and tazobactam inhibition activity (1). Their diffusion has compromised the use of extended-spectrum cephalosporins, promoted the use of carbapenems, and, consequently, favored the diffusion of carbapenemase-producing isolates. These strains are also frequently resistant to other major classes of antibiotics such as aminoglycosides and fluoroquinolones because of associated plasmid-encoded resistance mechanisms.
Up to the end of the 1990s, TEM- and SHV-type ESBLs were mainly produced by the Klebsiella pneumoniae and Enterobacter spp. responsible for nosocomial infections (1). Unfortunately, Escherichia coli strains that produce ESBLs belonging to the recent CTX-M group have now diffused worldwide, including in the community. Between their discovery in the middle of the 1980s and now, the prevalence of ESBL-producing Enterobacteriaceae strains has dramatically increased in many countries, especially among E. coli strains, because of the diffusion of CTX-M enzymes (1, 2).
Although ESBLs have been widely investigated, there have been to date no multicenter studies of ESBL-producing Enterobacteriaceae in France. In the present work, we collected 200 ESBL-producing Enterobacteriaceae isolates from clinical samples in 18 French hospitals in order to investigate the diversity of ESBLs in France. We also characterized the plasmid-encoded resistance genes against fluoroquinolones and aminoglycosides. The genetic background of the bacterial isolates and their plasmid content were analyzed to decipher the mechanisms underlying ESBL spread.
(This work was presented in part at the 33rd Réunion Interdisciplinaire de Chimiothérapie Anti-Infectieuse [RICAI], Paris, France, November 2013.)
RESULTS
Epidemiologic context.
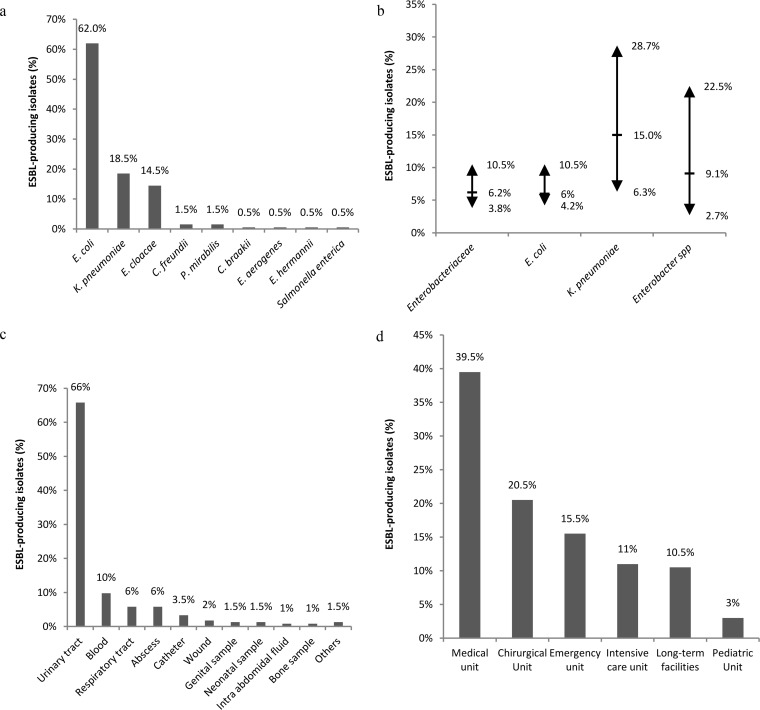
Most of the ESBL-producing Enterobacteriaceae isolates from clinical samples belonged to the E. coli (62%), K. pneumoniae (18.5%), and Enterobacter cloacae (15.5%) species (Fig. 1a). Annual prevalence levels were calculated separately for each hospital; minimum, maximum, and means values across hospitals are presented in Fig. 1b. The annual prevalence of these ESBL-producing Enterobacteriaceae (Fig. 1b) ranged from 3.8% to 10.5% (mean, 6.2%) in the 18 participating hospitals. These annual prevalence measures differed for the three major species of ESBL-producing Enterobacteriaceae as indicated in Fig. 1b. The age range of the subjects was 0 to 101 years and the median age 65 years. The isolates were mainly collected from urinary tract (66%), blood (10%), respiratory tract (5.5%), and sputum (5.5%) samples (Fig. 1c). The patients were hospitalized in medical wards (39.5%), surgical wards (20.5%), emergency units (15.5%), intensive care units (11%), long-term facilities (10.5%), and pediatric wards (3%) (Fig. 1d).
FIG 1.
Epidemiological context of ESBL-producing Enterobacteriaceae in 18 representative French hospitals in 2012 (n = 50,378 Enterobacteriaceae). (a) Distribution of bacterial species in the 200 ESBL-producing Enterobacteriaceae isolates from clinical samples. C. freundii, Citrobacter freundii; P. mirabilis, Proteus mirabilis; C. braakii, Citrobacter braakii; E. aerogenes, Enterobacter aerogenes; E. hermannii, Escherichia hermannii. (b) Annual prevalence of ESBL-producing Enterobacteriaceae (the arrows indicate the extreme values and the horizontal dashes the mean values). (c) The clinical samples from which the 200 ESBL-encoding Enterobacteriaceae were isolated. (d) Hospital units corresponding to patients infected by the 200 ESBL-encoding Enterobacteriaceae strains.
Antimicrobial susceptibility.
Among β-lactam antibiotics, carbapenems, cefoxitin, and the piperacillin-tazobactam combination were the most active, with susceptibility rates of 99.5% (imipenem), 96% (ertapenem), 83% (cefoxitin), and 76.5% (piperacillin-tazobactam). Among extended-spectrum cephalosporins, ceftazidime was the most active, with 24.5% of the isolates susceptible versus only 3% for cefotaxime. The amikacin susceptibility rate was higher than those of gentamicin and tobramycin (79.5% versus 53.5% and 40%, respectively). Only 20.5% of the isolates were susceptible to all quinolones, and ciprofloxacin was active against 31.5% of the strains. Fosfomycin was frequently active, with a resistance rate of 7%.
Characterization of ESBLs.
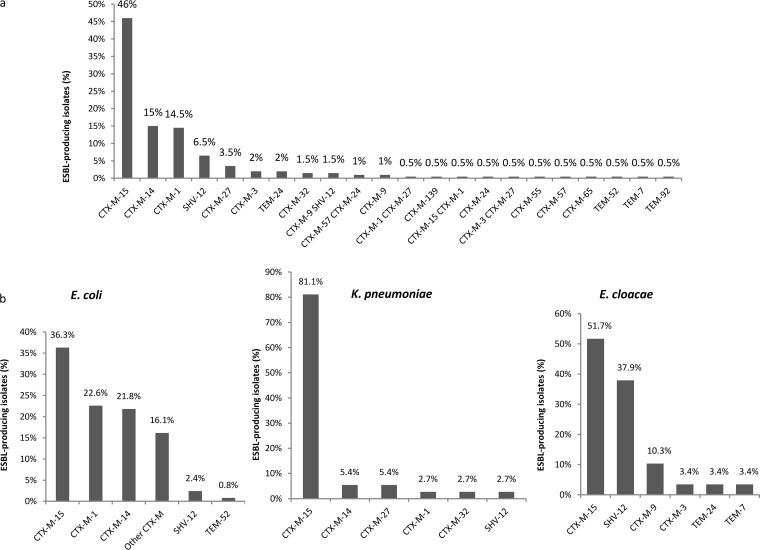
Seventeen ESBL types were identified among the 200 isolates studied (Fig. 2a). The CTX-M type was the most prevalent (90%). The three major CTX-M variants were CTX-M-15 (46.5%), CTX-M-1 (15.5%), and CTX-M-14 (15%). Nine other CTX-M enzymes were observed in 0.5% to 4.5% of the isolates as follows: CTX-M-27 (4.5%), CTX-M-9 (2.5%), CTX-M-3 (2.5%), CTX-M-24 (1.5%), CTX-M-32 (1.5%), CTX-M-57 (1.5%), CTX-M-55 (0.5%), CTX-M-65 (0.5%), and the novel CTX-M-139 (0.5%), which had derived from CTX-M-15 by the Y27F substitution. SHV- and TEM-type ESBLs were observed in 8% (SHV-12, n = 16) and 3.5% (TEM-7, n = 1; TEM-92, n = 1; TEM-52, n = 1; TEM-24, n = 4) of the strains. Eight strains produced two ESBLs.
FIG 2.
The diversity of ESBLs in Enterobacteriaceae isolated from clinical samples in 18 French hospitals (n = 200). (a) Prevalence of ESBLs in Enterobacteriaceae. (b) Distribution of ESBLs in E. coli (n = 124), K. pneumoniae (n = 37), and E. cloacae (n = 29).
The distributions of ESBLs were different in the three major species (Fig. 2b). CTX-M-1 and CTX-M-14 were frequently observed in E. coli strains (24.2% and 21.8% versus 37.1% for CTX-M-15) but were rare in K. pneumoniae, which mainly produced CTX-M-15 (81.1%). CTX-M-15 and SHV-12 were the main ESBLs in E. cloacae (51.7% and 37.9%, respectively). No carbapenemase was observed among the eight isolates intermediate or resistant to ertapenem (five E. cloacae isolates, two K. pneumoniae isolates, and one E. aerogenes isolate).
No plasmid-encoded AmpC was detected among the 200 strains. OXA-1 was detected in 41% of the strains and was especially associated with CTX-M-15 (84%).
Plasmid-borne genes encoding resistance to quinolones and aminoglycosides.
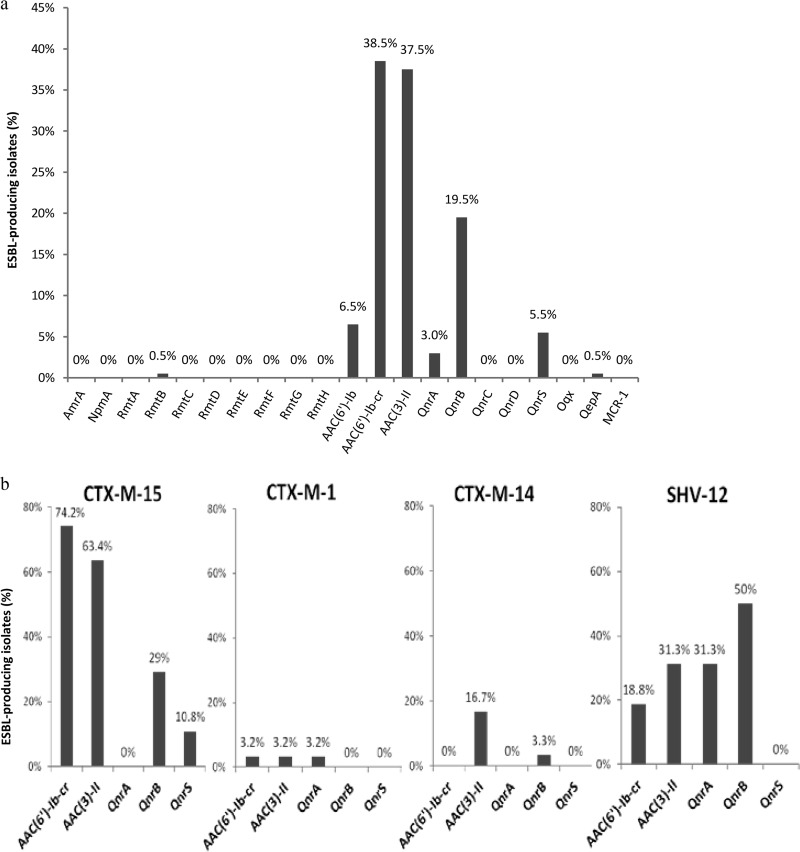
The genes encoding the aminoglycoside acetyltransferases AAC(6′)-Ib and AAC(3)-II were identified in 45% and 37.5% of the isolates, respectively (Fig. 3a). The gene encoding methylase RmtB was detected in only one isolate. The aac(6′)-Ib-cr and qnrB-type genes were the most frequent plasmid-mediated quinolone resistance genes (38.5% and 19.5%). The qnrA1, qnrS1, and qepA genes were more uncommon (3%, 5.5%, and 0.5%, respectively). Among the qnrB variants, the B1 allele was the most frequent (68.4%), with the prevalence of other B alleles ranging from 2.6% to 7.9% (Fig. 3a). AAC(6′)-Ib-cr, AAC(3)-II, and QnrB were mainly associated with ESBLs CTX-M-15 (74.1%, 63.4%, and 29%, respectively) and SHV-12 (18.75%, 31.25%, and 50%, respectively) (Fig. 3b). QnrA1 was observed only in SHV-12-producing strains (31.25%) (Fig. 3b). No qnrC, qnrD, oqx, or mcr-1 genes were detected.
FIG 3.
Percentage of ESBL-encoding isolates harboring plasmid-mediated genes encoding resistance to gentamicin, amikacin, and fluoroquinolones. (a) Distribution of proteins encoded by resistance genes in ESBL-producing Enterobacteriaceae. (b) Distribution of proteins encoded by aac(6′)-Ib-cr, aac(3)-II, qnrB, qnrA1, and qnrS1 in CTX-M-15 (n = 93)-, CTX-M-1 (n = 31)-, CTX-M-14 (n = 30)-, and SHV-12 (n = 16)-producing Enterobacteriaceae.
Analysis of clonal relatedness.
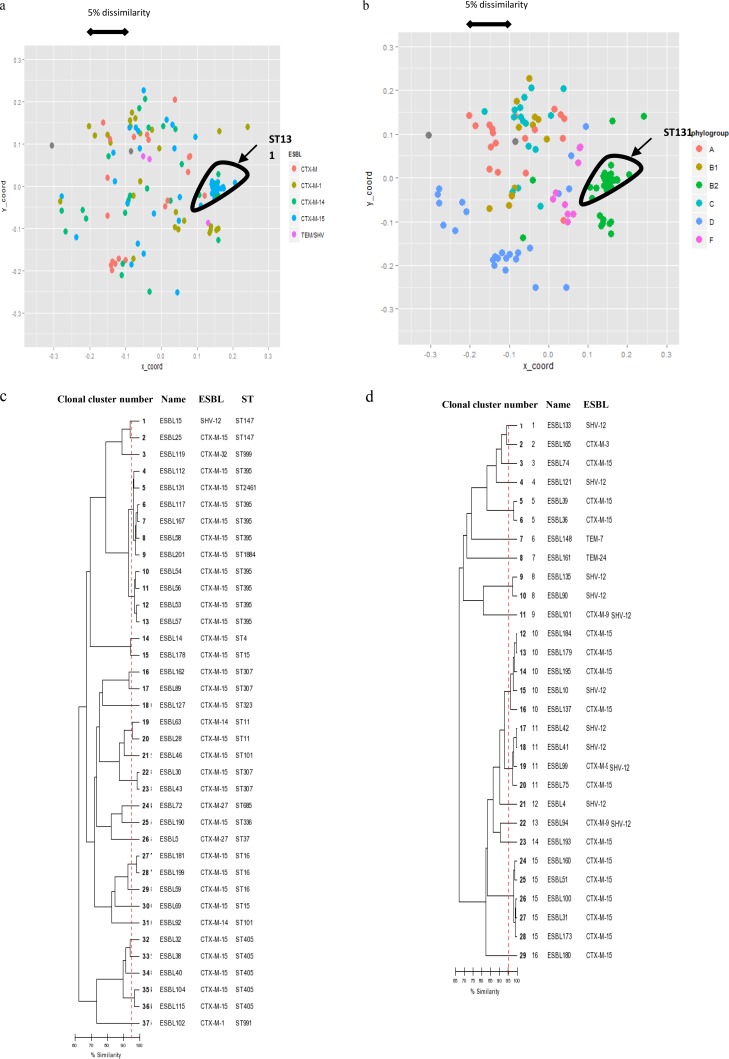
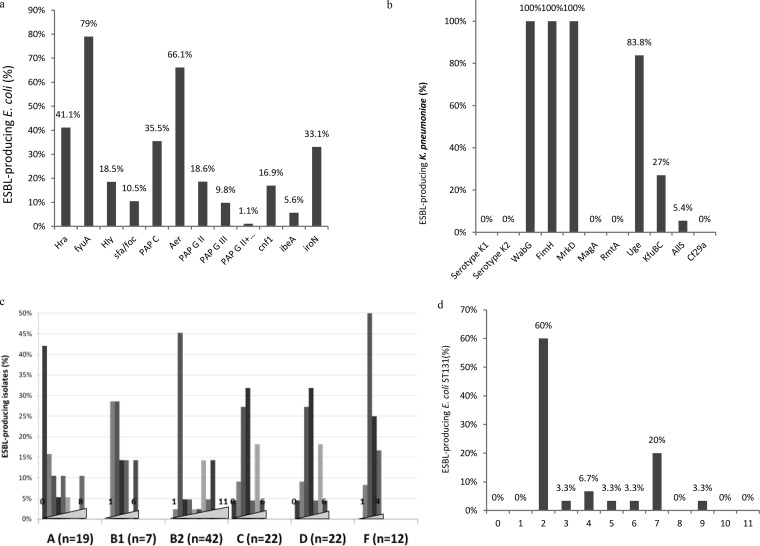
Among the 124 E. coli isolates, 56 clusters were identified by repetitive sequence-based PCR (rep-PCR) (Fig. 4a and b; see also Fig. S1 in the supplemental material). The largest cluster comprised 27 of the 30 isolates belonging to ST131 (90%). They were isolated in 12 different hospitals, with 1 to 4 isolates per hospital: 1 isolate for 3 hospitals, 2 isolates for 4 hospitals, 3 isolates for 4 hospitals, and 4 isolates for 1 hospital. Among the other strains, 62 were classified in 20 clusters comprising 2 to 7 isolates as follows: 2 isolates (n = 10 clusters), 3 isolates (n = 4 clusters), 4 isolates (n = 3 clusters), 5 isolates (n = 1 cluster), 6 isolates (n = 1 cluster), and 7 isolates (n = 1 cluster). Only four were isolated from two different patients in a single hospital.
FIG 4.
(a and b) Multidimensional scaling analysis (MDS) scatter plots derived from DiversiLab data from the ESBL-producing E. coli strain showing ESBL types (a) and the E. coli phylogroup (b). The scale indicates the dissimilarity between strains for the x and y axes. (c and d) Dendrogram of K. pneumoniae (c) and E. cloacae (d) strains. The vertical dashed line indicates the criterion for clonality (≥95% similarity). ST, sequence type.
ST131 isolates were mainly associated with CTX-M-15 (80%). Other ST131 isolates produced CTX-M-14 (13.3%), CTX-M-27 (3.3%), or CTX-M-3 (3%). The major cluster comprising 27 of the 30 isolates isolated from 12 different hospitals produced CTX-M-15 (n = 23), CTX-M-14 (n = 3), or CTX-M-27 (n = 1). The three minor clusters contained only one isolate producing CTX-M-14, CTX-M-15, or CTX-M-3.
Twenty-six clusters were identified among the 37 K. pneumoniae isolates (Fig. 4c). They contained 1 to 6 isolates as follows: 1 isolate (n = 21 clusters), 2 isolates (n = 3 clusters), 4 isolates (n = 1 cluster), and 6 isolates (n = 1 cluster). The five clusters of 2 to 6 isolates comprised only CTX-M-15-producing ones, and all of them were isolated from 2 (n = 4) to 4 (n = 1) patients in a single hospital. The 10 strains of the clusters of 6 and 4 isolates belonged to closely related sequence types ST395, ST1884, and ST2461.
Sixteen clusters were identified among the 29 E. cloacae strains (Fig. 4d). They comprised 1 to 5 isolates as follows: 1 isolate (n = 11), 2 isolates (n = 2), 4 isolates (n = 1), and 5 isolates (n = 2). Four of them were isolated from 2 (n = 3) to 3 (n = 1) patients in a single hospital.
Overall, these data reveal a broad diversity of ESBL-producing Enterobacteriaceae isolates, including the species K. pneumoniae and E. cloacae.
Virulence factor (VF)-encoding genes and E. coli phylogroups.
A total of 33% of E. coli isolates belonged to the B2 phylogroup; 73.1% were ST131isolates. Phylogroups C (17.7%), D (17.7%), and A (15.4%) had similar prevalences. The other phylogroups were more uncommon (F, 9.7%; B1, 5.6%; and B21, 0.8%).
Among the E. coli VF-encoding genes investigated, iron-chelating VFs were the most frequent (fyuA, 79%; iucC, 66%; iroN, 33%), followed by the hra gene (41%) and the papC pilus-encoding gene (35%). The prevalence of the others ranged from 8% to 19% (Fig. 5a). Most E. coli isolates harbored fewer than four VF genes (66.1%). As expected, VFs were more frequent in isolates belonging to B2 phylogroup (Fig. 5c): 43.9% harbored at least five VFs, but few of those belonged to highly prevalent sequence type ST131. ST131 isolates harbored a variable number of VFs ranging from 1 to 9. Of those isolates, 60% (n = 18/30) produced only yersiniabactin and aerobactin, and only 30% harbored at least five VFs and. On the basis of DiversiLab clusters and VFs, two ST131 major groups can be individualized (Fig. 5d). One group comprised 17 isolates that produced only yersiniabactin and aerobactin. The second comprised 6 isolates that produced 7 virulence factors.
FIG 5.
Percentage of ESBL-producing isolates harboring virulence factors for the E. coli (n = 124) (a) and K. pneumoniae (n = 37) (b) species. (c and d) Percentages of ESBL-producing isolates harboring 0 to 11 virulence factors for the different E. coli phylogroups (c) and the ST131 E. coli sequence type (d).
Very few VF genes were detected in the K. pneumoniae strains (Fig. 5b). None of those strains belonged to the K1 or K2 serotype, harbored the rmpA or rmpA2 gene, or accumulated siderophore-encoding genes, which are observed in strains responsible for invasive infections (3). The main VFs were the adhesion-encoding genes fimH and mrkD (100%), wagG (100%), and uge (97.3%). The other genes, kfu and allS, were observed in 27% and 5.4% of K. pneumoniae isolates, respectively.
ESBL-encoding plasmids.
We obtained transformants or transconjuguants harboring only one plasmid for 81 strains. The presence of the ESBL gene on the plasmid was confirmed by Southern blotting.
We studied the plasmid content of the isolates (Table 1) harboring the most prevalent ESBLs in E. coli (CTX-M-15, n = 10; CTX-M-1, n = 10; CTX-M-14, n = 10), K. pneumoniae (CTX-M-15, n = 28), and E. cloacae (CTX-M-15, n = 12; SHV-12, n = 11).
TABLE 1.
Plasmid size and replicon type of ESBL-encoding plasmids in K. pneumoniae, E. cloacae, and E. coli strains
| Species | Plasmid IDa | Plasmid size (kb) | Replicon | ESBL |
|---|---|---|---|---|
| E. coli (n = 30) | ESBL146 | 100 | I1:3 | CTX-M-1 |
| ESBL107 | 120 | I1:3 | CTX-M-1 | |
| ESBL143 | 120 | I1:3 | CTX-M-1 | |
| ESBL86 | 50 | X1 | CTX-M-1 | |
| ESBL172 | 62 | F47: A-: B- | CTX-M-1 | |
| ESBL200 | 90 | I1:3 | CTX-M-1 | |
| ESBL192 | 90 | I1:3 | CTX-M-1 | |
| ESBL134 | 95 | I1:3 | CTX-M-1 | |
| ESBL128 | 120 | I1:3 | CTX-M-1 | |
| ESBL124 | 130 | I1:3 | CTX-M-1 | |
| ESBL37 | 90 | L/M K | CTX-M-14 | |
| ESBL48 | 40 | F-: A6: B- | CTX-M-14 | |
| ESBL130 | 115 | F1: A2: B20 | CTX-M-14 | |
| ESBL67 | 115 | F1: A6: B20 | CTX-M-14 | |
| ESBL191 | 67 | F2: A-: B- | CTX-M-14 | |
| ESBL110 | 62 | F24: A-: B1 | CTX-M-14 | |
| ESBL142 | 100 | F2: A-: B- | CTX-M-14 | |
| ESBL61 | 62 | F2: A-: B- | CTX-M-14 | |
| ESBL85 | 62 | F2: A-: B- | CTX-M-14 | |
| ESBL80 | 90-67 | F47: A-: B- | CTX-M-14 | |
| ESBL145 | 145 | F1: A6: B20 | CTX-M-15 | |
| ESBL194 | 162 | F1: A6: B49 | CTX-M-15 | |
| ESBL22 | 125 | F2: A6: B- | CTX-M-15 | |
| ESBL3 | 90 | F2: A6: B32 | CTX-M-15 | |
| ESBL154 | 120 | F31: A-: B23 | CTX-M-15 | |
| ESBL47 | 80 | F35: A-: B- | CTX-M-15 | |
| ESBL45 | 162 | F48: A6: B49 | CTX-M-15 | |
| ESBL177 | 150 | F16: A-: B1 | CTX-M-15 | |
| ESBL108 | 120 | F2: A1: B- | CTX-M-15 | |
| ESBL29 | 162 | F31: A4: B1 | CTX-M-15 | |
| K. pneumoniae (n = 28) | ESBL104 | 175 | K7: A-: B- | CTX-M-15 |
| ESBL112 | 70 | K1: A-: B- | CTX-M-15 | |
| ESBL115 | 175 | K7: A-: B- | CTX-M-15 | |
| ESBL117 | 165 | K7: A-: B- | CTX-M-15 | |
| ESBL127 | 175 | K7: A-: B- | CTX-M-15 | |
| ESBL131 | 70 | R | CTX-M-15 | |
| ESBL14 | 165 | K8: A13: B- | CTX-M-15 | |
| ESBL162 | 165 | K7: A-: B- | CTX-M-15 | |
| ESBL178 | 165 | K9: A-: B- | CTX-M-15 | |
| ESBL181 | 165 | K5: A-: B- | CTX-M-15 | |
| ESBL190 | 175 | K8: A-: B- | CTX-M-15 | |
| ESBL199 | 175 | K5: A-: B- | CTX-M-15 | |
| ESBL201 | 140 | R | CTX-M-15 | |
| ESBL25 | 165 | K5: A12: B1 | CTX-M-15 | |
| ESBL28 | 70 | R | CTX-M-15 | |
| ESBL30 | 165 | K7: A-: B- | CTX-M-15 | |
| ESBL32 | 175 | K7: A-: B- | CTX-M-15 | |
| ESBL38 | 165 | K7: A-: B- | CTX-M-15 | |
| ESBL40 | 175 | K7: A-: B- | CTX-M-15 | |
| ESBL43 | 175 | K7: A-: B- | CTX-M-15 | |
| ESBL46 | 140 | K-: A-: B- | CTX-M-15 | |
| ESBL54 | 175 | K7: A-: B- | CTX-M-15 | |
| ESBL56 | 70 | R | CTX-M-15 | |
| ESBL57 | 70 | K1: A-: B- | CTX-M-15 | |
| ESBL58 | 70 | R | CTX-M-15 | |
| ESBL59 | 140 | K12: A-: B1 | CTX-M-15 | |
| ESBL69 | 70 | K5: A-: B- | CTX-M-15 | |
| ESBL89 | 175 | K7: A-: B- | CTX-M-15 | |
| E. cloacae (n = 23) | ESBL4 | 170 | HI2 | SHV-12 |
| ESBL10 | 180 | HI2 | SHV-12 | |
| ESBL36 | 170 | HI2 | CTX-M-15 | |
| ESBL39 | 170 | HI2 | CTX-M-15 | |
| ESBL41 | 190 | HI2 | SHV-12 | |
| ESBL42 | 190 | HI2 | SHV-12 | |
| ESBL51 | 180 | HI2 | CTX-M-15 | |
| ESBL74 | 180 | HI2 | CTX-M-15 | |
| ESBL75 | 180 | HI2 | CTX-M-15 | |
| ESBL90 | 170 | HI2 | SHV-12 | |
| ESBL94 | 180 | HI2 | SHV-12 | |
| ESBL99 | 170 | HI2 | SHV-12 | |
| ESBL100 | 180 | HI2 | CTX-M-15 | |
| ESBL101 | 180 | HI2 | SHV-12 | |
| ESBL121 | 180 | HI2 | SHV-12 | |
| ESBL133 | 190 | HI2 | SHV-12 | |
| ESBL135 | 170 | HI2 | SHV-12 | |
| ESBL137 | 180 | HI2 | CTX-M-15 | |
| ESBL160 | 180 | HI2 | CTX-M-15 | |
| ESBL173 | 180 | HI2 | CTX-M-15 | |
| ESBL193 | 180 | HI2 | CTX-M-15 | |
| ESBL31 | 180 | Y | CTX-M-15 | |
| ESBL184 | 136 | Y | CTX-M-15 |
ID, identifier.
Among the K. pneumoniae isolates, CTX-M-15 was mainly encoded by plasmids belonging to the FIIK (n = 23/28; size, 160 to 170 kb) and R (n = 5/28; size, 70 or 140 kb) incompatibility groups. Most FIIK plasmids belonged to the FIIK:7 subtype (n = 12/23). The others belonged to FIIK:5 (n = 4/23), FIIK:1 (n = 3/23), FIIK:8 (n = 2/23), FIIK:9 (n = 1/23), and FIIK12 (n = 1/23). In E. cloacae isolates, CTX-M-15 was mainly encoded by plasmid HI2:1 (n = 10/12; size, 170 to 180 kb) or Y (n = 2/12; size, 130 to 180 kb) and SHV-12 by a HI2:1 plasmid (size, 170 to 190 kb).
In E. coli isolates, blaCTX-M-15 was observed on a wide diversity of 80- to 160-kb IncF plasmids (F1:A6:B20, F1:A6:B49, F2:A1:B-, F2:A6:B-, F2:A6:B32, F16:A-:B1, F31:A-:B23, F31:A4:B1, F35:A-:B-, and F48:A6:B49). blaCTX-M-14 was observed on 40- to 120-kb IncF plasmids F2:A-:B- (n = 4/10), F24:A-:B1 (n = 1/10), F47:A-B- (n = 1/10), F1:A2:B20 (n = 1/10), F1:A6:B20 (n = 1/10), and F-:A6:B- (n = 1/10) and on a 90-kb IncL/M K plasmid (n = 1/10). Less diversity was observed for blaCTX-M-1, which was carried on I1:3 plasmids (n = 8/10; size, 90 to 120 kb), an FII:47 plasmid (n = 1; size, 62 kb), and an X1 plasmid (n = 1; size, 50 kb).
DISCUSSION
The aim of our study was to identify the ESBLs produced by the Enterobacteriaceae isolates responsible for infections and the mechanisms associated with their spread at the national level. The annual prevalence measures of ESBLs in Enterobacteriaceae and E. coli ranged between 3% and 10%. E. coli was the main species isolated (62%), and most of the isolates were collected from urinary tract samples (68%), as described in previous studies (1, 2). Although their frequency was lower than 10% in most of the participating hospitals, the prevalence of ESBL-producing K. pneumoniae and Enterobacter strains was worrying in certain centers that had prevalence values (>20%) similar to those observed during the early 1990s in most French hospitals (1). Most of the isolates were susceptible to fosfomycin (93%) and carbapenems (96% to 99.5%). Cefoxitin and the piperacillin-tazobactam association were inconstantly active (83% and 76.5%).
The main ESBLs characterized during this study belonged, as expected, to the CTX-M family. The three ESBLs CTX-M-15 (46.5%), CTX-M-1 (15.5%), and CTX-M-14 (15%) accounted for more than 75% of the isolates, as observed in other European countries (2). ESBL-encoding genes were frequently associated with the plasmid-encoded mechanisms of resistance to aminoglycosides and quinolones, especially the acc(6′)-Ib-cr (38.5%) and qnr (28.5%) genes. They were especially associated with blaCTX-M-15 [aac(6′)-Ib-cr, 72.6%; aac(3)-II, 62.1%] and blaSHV-12 (qnrB, 43.8%), suggestive of a possible genetic link as previously proposed in K. pneumoniae (4). Studies have suggested an association of qnr and aac(6′)-Ib-cr genes with ESBL genes, but their prevalences differ between countries, with a generally higher diffusion of qnrA and aac(6′)-Ib-cr (5). In contrast to a previous study (6), we observed a higher prevalence of qnrB than qnrA. No carbapenemases were observed, and only one isolate harbored a 16S methytransferase-encoding gene (rmtB). These results give an overview of the diffusion of major plasmid-encoded resistance associated with blaESBL and the wide diffusion of aac(6′)-Ib-cr and qnr in ESBL-producing strains.
Clonal-relatedness analysis and characterization of incompatibility groups of ESBL-encoding plasmids revealed different species-specific rates of ESBL diffusion.
A wider diversity of ESBL was observed among E. coli strains (14 different ESBLs), with 3 major groups producing CTX-M-15 (47/124), CTX-M-1 (28/124), and CTX-M-14 (27/124). DiversiLab analysis identified only 56 clonal clusters for 124 isolates with a major ST131 cluster (30 isolates). However, the analysis of VFs, ESBL content, and other resistance mechanisms revealed only three pairs of identical ST131 isolates. These results suggest that the ST131 clone is endemic in France and has evolved in many different ways (acquisition of different virulence factors, ESBLs, and plasmid-encoded resistance mechanisms), forming different variants that cannot be discriminated by the usual molecular typing methods such as rep-PCR. A whole-genome comparison of these strains would be useful to improve discriminatory ability. Moreover, plasmid analysis of CTX-M-1-, CTX-M-14-, and CTX-M-15-encoding plasmids showed a broadest diversity of plasmids (incompatibility groups I1, FIA, FIA-FIB, FIB, and FII), especially for those harboring blaCTX-M-15 and blaCTX-M-14. These results suggest that the diffusion of CTX-M-15 and CTX-M-14 among E. coli strains in France was not linked to the diffusion of a major epidemic plasmid as suggested by previous studies (7) and suggest very active transfers of CTX-M-15-encoding genes in different plasmids in this species. In contrast, an association was observed between blaCTX-M-1 and I1:3 plasmids of similar sizes. We also observed a strong association of the IncI1 plasmid with the CTX-M-1-encoding gene as previously observed in animal and human strains (7).
Among K. pneumoniae isolates, only 6 different ESBLs, with the great majority being CTX-M-15 (30/37), were identified. DiversiLab analysis revealed a wide diversity of strains of this species (26 clonal clusters for 37 isolates). In contrast to what was observed in E. coli, plasmid analysis identified a strong association of CTX-M-15-encoding plasmids belonging to FIIK and R in K. pneumoniae strains.
ESBL diversity was also low among the E. cloacae isolates, with only 6 different ESBLs representing a great majority of CTX-M-15 (15/29) and SHV-12 (11/27). Clonal diversity was lower in this species, with only 16 clonal clusters identified by DiversiLab analysis among 29 isolates. In this species, blaSHV-12 and blaCTX-M-15 were associated with plasmids belonging to the HI2 and Y incompatibility groups. Moreover, comparisons of plasmid sizes suggested the diffusion of very similar plasmids.
Our findings also indicate that ESBL-encoding plasmids are somewhat species specific. The same blaESBL gene was observed in different plasmids in different species, suggesting that blaESBL has to transfer into a plasmid adapted to the bacterial host to achieve epidemiological success. The relationship between species and plasmid seems to be a strong functional constraint on bacteria evolution and could explain the lack of epidemiological success of certain resistance mechanisms. The epidemiology of ESBLs may therefore not be directly related between species: the relationship between the plasmids and the bacterial species could explain the differences in ESBL prevalence observed according to the Enterobacteriaceae species.
Overall, the ESBL-producing isolates produced a small number of virulence factors. Iron-chelating factors were the most frequently observed factors in these two species, probably because of their major role in bacterial viability in the human body. Most E. coli ST131 isolates harbored fewer than five virulence factors (70%), suggesting a current rare association of ESBL plasmids with highly virulent strains.
In conclusion, our report gives an overview of the main characteristics of French ESBL-producing Enterobacteriaceae strains (main resistance mechanisms, clonal diffusion, plasmid diffusion, and virulence). It confirms the diffusion of the three major CTX-M-enzymes CTX-M-15, CTX-M-1, and CTX-M-14 and identifies the plasmids as major actors in ESBL spread.
MATERIALS AND METHODS
Clinical isolates.
Between June 2012 and October 2012, consecutive, nonrepetitive ESBL-producing Enterobacteriaceae strains isolated from clinical samples of inpatients and outpatients were collected in 18 representative French hospitals (15 teaching hospitals and 3 nonteaching public hospitals) spread over the entire French territory. All species of Enterobacteriaceae were included, and ESBL production was confirmed in each participating center according to the recommendations of the Antimicrobial Committee of the French Society for Microbiology (CA-SFM) (8). Screening samples, such as rectal swabs, were excluded. Between 7 and 17 strains were collected from each participating center according to size. All the strains were sent to the French National Reference Centre for antibiotic resistance for further investigations (Gabriel Montpied Hospital, Clermont-Ferrand, France). Virulence studies of E. coli strains were performed by the French National Reference Centre for E. coli (Robert-Debré Hospital, Paris, France).
Antimicrobial susceptibility testing and ESBL detection.
Identification of strains was confirmed by the Vitek matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) method (bioMérieux, La Balme, France). The production of ESBL was detected according to the recommendations of the Antimicrobial Committee of the French Society for Microbiology (CA-SFM) (8). Antibiotic susceptibilities were determined by the disk diffusion method according to the CA-SFM recommendations (8). ESBL production was confirmed by the double-disk synergy test (8). The following 18 antibiotics were tested: amoxicillin, amoxicillin-clavulanic acid, ticarcillin, piperacillin-tazobactam, cephalothin, cefoxitin, ceftazidime, cefotaxime, ertapenem, imipenem, amikacin, gentamicin, tobramycin, nalidixic acid, ofloxacin, ciprofloxacin, fosfomycin, and co-trimoxazole (Bio-Rad, Marnes-La-Coquette, France).
Characterization of β-lactamase-encoding genes and associated resistance genes.
β-Lactamases were investigated by isoelectric focusing as previously described (9). The ESBL-encoding genes were identified by specific PCR and sequencing experiments using specific primers as described elsewhere (9). Carbapenemase production was investigated in carbapenem-resistant strains by specific PCR as previously described (10). Genes encoding AAC(6′)-Ib, AAC(6′)-Ib-cr, AAC(3)-II, RmtA to RmtH, ArmA, NpmA, QnrA, QnrB, QnrC, QnrD, QnrS, Oqx (for strains not belonging to the Klebsiella genus), and QepA were identified by specific PCR assay and sequencing as previously reported (11–21). Plasmid-mediated colistin resistance mechanism MCR-1 was investigated as previously described (22). Plasmid-encoded AmpC and OXA-1 production was investigated as previously described (23, 24).
Molecular strain typing.
The relatedness between ESBL-producing isolates was assessed by repetitive sequence-based PCR (rep-PCR) using the DiversiLab method according to the manufacturer's instructions (bioMérieux, La Balme, France). The analysis was performed using Pearson's correlation in the dedicated DL software of the manufacturer (version 3.6). Isolates with similarities of <95% were considered different, and isolates with similarities of >98% were considered indistinguishable. All isolates with similarities of >95% and <98% were judged manually using the pattern overlay of the analysis tool in the software (25). A Pearson distance matrix was calculated using DiversiLab, and classical multidimensional scaling was then performed on the distance matrix using R to illustrate the relationship between the isolates (26). The phylogroup of E. coli strains (A, B1, B2, C, D, E, or F) was determined by a PCR method as previously described (27). The highly virulent B21 subgroup was also identified by specific PCR (28). O25b-ST131 E. coli clones were identified according to the PCR method described by Clermont et al. (29). MLST for K. pneumoniae isolates was performed as previously described by Diancourt et al. (30).
Analysis of ESBL-encoding plasmids.
Transferability of the ESBL-encoding genes was studied by mating-out assay. When these experiments failed, plasmid DNA was extracted with NucleoBond Xtra (Macherey-Nagel, Germany) and transferred by electroporation into an E. coli K-12 recipient. Selection was performed on agar plates supplemented with ceftazidime or cefotaxime (4 mg/liter) and rifampin (300 mg/liter) for the mating-out assay and ceftazidime or cefotaxime (4 mg/liter) for electroporation. The size of ESBL-encoding plasmids was determined by Southern blotting with ESBL-specific probes using plasmid DNA extracted by the method of Kado and Liu (31) with plasmids Rsa (39 kb), TP114 (61 kb), pCFF04 (85 kb), and pCFF14 (180 kb) used as standards. The plasmid content of transconjugants or transformants containing one plasmid was further studied by PCR-based replicon typing (PBRT) and plasmid multilocus sequence typing (MLST) (32, 33).
Characterization of virulence factor-encoding genes.
The following 11 virulence factor-encoding genes of extraintestinal pathogenic E. coli (ExPEC) strains were identified by multiplex PCR, as previously described (34): papC (encoding outer membrane usher protein PAPC), papGII and papGIII (P fimbriae), sfa and foc (sfa/foc) (S fimbriae), hlyC (hemolysin), cnf1 (cytotoxic necrotizing factor), iucC (aerobactin), fyuA (yersiniabactin), iroN (salmochelin), hek/hra (hemagglutinin), and ibeA (endothelial invasin). For K. pneumoniae isolates, 9 genes (uge, mrkD, fimH, allS, rmpA, magA, wabG, kfu, and cf29a) were screened by PCR assays (3). Capsule csp-based genotyping was performed as previously described (35). Strains NTUH-K2044, KP52145, and MGH 78578 were used as PCR controls.
Supplementary Material
ACKNOWLEDGMENTS
We thank Laurent Guillouard and Alexis Pontvianne for technical assistance. We thank the team of curators of the Institut Pasteur MLST databases for curating the data and making them publicly available at http://bigsdb.web.pasteur.fr.
We have no commercial affiliations, funding sources, or conflicts of interests to declare.
This research project was supported by the Ministere de l'Education Nationale, de l'Enseignement Superieur et de la Recherche, INRA (USC-2018), Santé Publique, France, and the Centre Hospitalier Regional Universitaire de Clermont-Ferrand, Clermont-Ferrand, France.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01911-16.
REFERENCES
- 1.Paterson DL, Bonomo RA. 2005. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev 18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, Ayala J, Coque TM, Kern-Zdanowicz I, Luzzaro F, Poirel L, Woodford N. 2007. CTX-M: changing the face of ESBLs in Europe. J Antimicrob Chemother 59:165–174. [DOI] [PubMed] [Google Scholar]
- 3.Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, Grimont P. 2009. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 4:e4982. doi: 10.1371/journal.pone.0004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gassama-Sow A, Aïdara-Kane A, Raked N, Denis F, Ploy M-C. 2004. Integrons in Salmonella Keurmassar, Senegal. Emerg Infect Dis 10:1339–1341. doi: 10.3201/eid1007.030666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. 2009. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev 22:664–689. doi: 10.1128/CMR.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lavigne J-P, Marchandin H, Delmas J, Bouziges N, Lecaillon E, Cavalie L, Jean-Pierre H, Bonnet R, Sotto A. 2006. qnrA in CTX-M-producing Escherichia coli isolates from France. Antimicrob Agents Chemother 50:4224–4228. doi: 10.1128/AAC.00904-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comité de l'Antibiogramme de la Société Française de Microbiologie. 2015. Recommandations 2015. Comité de l'Antibiogramme de la Société Française de Microbiologie, Paris, France. [Google Scholar]
- 9.De Champs C, Chanal C, Sirot D, Baraduc R, Romaszko JP, Bonnet R, Plaidy A, Boyer M, Carroy E, Gbadamassi MC, Laluque S, Oules O, Poupart MC, Villemain M, Sirot J. 2004. Frequency and diversity of class A extended-spectrum beta-lactamases in hospitals of the Auvergne, France: a 2 year prospective study. J Antimicrob Chemother 54:634–639. doi: 10.1093/jac/dkh395. [DOI] [PubMed] [Google Scholar]
- 10.Doyle D, Peirano G, Lascols C, Lloyd T, Church DL, Pitout JDD. 2012. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J Clin Microbiol 50:3877–3880. doi: 10.1128/JCM.02117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavaco LM, Hasman H, Xia S, Aarestrup FM. 2009. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovis morbificans strains of human origin. Antimicrob Agents Chemother 53:603–608. doi: 10.1128/AAC.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. 2006. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother 50:3953–3955. doi: 10.1128/AAC.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang M, Guo Q, Xu X, Wang X, Ye X, Wu S, Hooper DC, Wang M. 2009. New plasmid-mediated quinolone resistance gene, qnrC, found in a clinical isolate of Proteus mirabilis. Antimicrob Agents Chemother 53:1892–1897. doi: 10.1128/AAC.01400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, Hooper DC. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother 50:1178–1182. doi: 10.1128/AAC.50.4.1178-1182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirel L, Leviandier C, Nordmann P. 2006. Prevalence and genetic analysis of plasmid-mediated quinolone resistance determinants QnrA and QnrS in Enterobacteriaceae isolates from a French university hospital. Antimicrob Agents Chemother 50:3992–3997. doi: 10.1128/AAC.00597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veldman K, Cavaco LM, Mevius D, Battisti A, Franco A, Botteldoorn N, Bruneau M, Perrin-Guyomard A, Cerny T, De Frutos Escobar C, Guerra B, Schroeter A, Gutierrez M, Hopkins K, Myllyniemi A-L, Sunde M, Wasyl D, Aarestrup FM. 2011. International collaborative study on the occurrence of plasmid-mediated quinolone resistance in Salmonella enterica and Escherichia coli isolated from animals, humans, food and the environment in 13 European countries. J Antimicrob Chemother 66:1278–1286. doi: 10.1093/jac/dkr084. [DOI] [PubMed] [Google Scholar]
- 17.Bueno MFC, Francisco GR, O'Hara JA, de Oliveira Garcia D, Doi Y. 2013. Coproduction of 16S rRNA methyltransferase RmtD or RmtG with KPC-2 and CTX-M group extended-spectrum β-lactamases in Klebsiella pneumoniae. Antimicrob Agents Chemother 57:2397–2400. doi: 10.1128/AAC.02108-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galimand M, Courvalin P, Lambert T. 2012. RmtF, a new member of the aminoglycoside resistance 16S rRNA N7 G1405 methyltransferase family. Antimicrob Agents Chemother 56:3960–3962. doi: 10.1128/AAC.00660-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Hara JA, McGann P, Snesrud EC, Clifford RJ, Waterman PE, Lesho EP, Doi Y. 2013. Novel 16S rRNA methyltransferase RmtH produced by Klebsiella pneumoniae associated with war-related trauma. Antimicrob Agents Chemother 57:2413–2416. doi: 10.1128/AAC.00266-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wachino J, Shibayama K, Kurokawa H, Kimura K, Yamane K, Suzuki S, Shibata N, Ike Y, Arakawa Y. 2007. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob Agents Chemother 51:4401–4409. doi: 10.1128/AAC.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis MA, Baker KNK, Orfe LH, Shah DH, Besser TE, Call DR. 2010. Discovery of a gene conferring multiple-aminoglycoside resistance in Escherichia coli. Antimicrob Agents Chemother 54:2666–2669. doi: 10.1128/AAC.01743-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y-Y, Wang Y, Walsh TR, Yi L- X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 23.Pérez-Pérez FJ, Hanson ND. 2002. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 40:2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beceiro A, Maharjan S, Gaulton T, Doumith M, Soares NC, Dhanji H, Warner M, Doyle M, Hickey M, Downie G, Bou G, Livermore DM, Woodford N. 2011. False extended-spectrum {beta}-lactamase phenotype in clinical isolates of Escherichia coli associated with increased expression of OXA-1 or TEM-1 penicillinases and loss of porins. J Antimicrob Chemother 66:2006–2010. doi: 10.1093/jac/dkr265. [DOI] [PubMed] [Google Scholar]
- 25.Voets GM, Leverstein-van Hall MA, Kolbe-Busch S, van der Zanden A, Church D, Kaase M, Grisold A, Upton M, Cloutman-Green E, Cantón R, Friedrich AW, Fluit AC, DiversiLab Study Group. 2013. International multicenter evaluation of the DiversiLab bacterial typing system for Escherichia coli and Klebsiella spp. J Clin Microbiol 51:3944–3949. doi: 10.1128/JCM.01664-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 27.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 28.Bidet P, Metais A, Mahjoub-Messai F, Durand L, Dehem M, Aujard Y, Bingen E, Nassif X, Bonacorsi S. 2007. Detection and identification by PCR of a highly virulent phylogenetic subgroup among extraintestinal pathogenic Escherichia coli B2 strains. Appl Environ Microbiol 73:2373–2377. doi: 10.1128/AEM.02341-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clermont O, Dhanji H, Upton M, Gibreel T, Fox A, Boyd D, Mulvey MR, Nordmann P, Ruppé E, Sarthou JL, Frank T, Vimont S, Arlet G, Branger C, Woodford N, Denamur E. 2009. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J Antimicrob Chemother 64:274–277. doi: 10.1093/jac/dkp194. [DOI] [PubMed] [Google Scholar]
- 30.Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kado CI, Liu ST. 1981. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol 145:1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villa L, García-Fernández A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 33.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bidet P, Mahjoub-Messai F, Blanco J, Blanco J, Dehem M, Aujard Y, Bingen E, Bonacorsi S. 2007. Combined multilocus sequence typing and O serogrouping distinguishes Escherichia coli subtypes associated with infant urosepsis and/or meningitis. J Infect Dis 196:297–303. doi: 10.1086/518897. [DOI] [PubMed] [Google Scholar]
- 35.Fang C-T, Lai S-Y, Yi W-C, Hsueh P-R, Liu K-L, Chang S-C. 2007. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis 45:284–293. doi: 10.1086/519262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.