ABSTRACT
The substitution N132G in the SDN motif of class A β-lactamases from rapidly growing mycobacteria was previously shown to impair their inhibition by avibactam but to improve the stability of acyl-enzymes formed with clavulanate. The same substitution was introduced in KPC-2 and CTX-M-15 to assess its impact on β-lactamases from Enterobacteriaceae and evaluate whether it may lead to resistance to the ceftazidime-avibactam combination. Kinetic parameters for the inhibition of the β-lactamases by avibactam and clavulanate were determined by spectrophotometry using nitrocefin as the substrate. The substitution N132G impaired (>1,000-fold) the efficacy of carbamylation of KPC-2 and CTX-M-15 by avibactam. The substitution improved the inhibition of KPC-2 by clavulanate due to reduced deacylation, whereas the presence or absence of N132G resulted in the inhibition of CTX-M-15 by clavulanate. The hydrolysis of amoxicillin and nitrocefin by KPC-2 and CTX-M-15 was moderately affected by the substitution N132G, but that of ceftazidime, ceftaroline, and aztreonam was drastically reduced. Isogenic strains producing KPC-2 and CTX-M-15 were constructed to assess the impact of the substitution N132G on the antibacterial activities of β-lactam–inhibitor combinations. For amoxicillin, the substitution resulted in resistance and susceptibility for avibactam and clavulanate, respectively. For ceftazidime, ceftaroline, and aztreonam, the negative impact of the substitution on β-lactamase activity prevented resistance to the β-lactam–avibactam combinations. In conclusion, the N132G substitution has profound effects on the substrate and inhibition profiles of class A β-lactamases, which are largely conserved in distantly related enzymes. Fortunately, the substitution does not lead to resistance to the ceftazidime-avibactam combination.
KEYWORDS: β-lactamase inhibitor, avibactam, CTX-M-15, clavulanate, KPC-2
INTRODUCTION
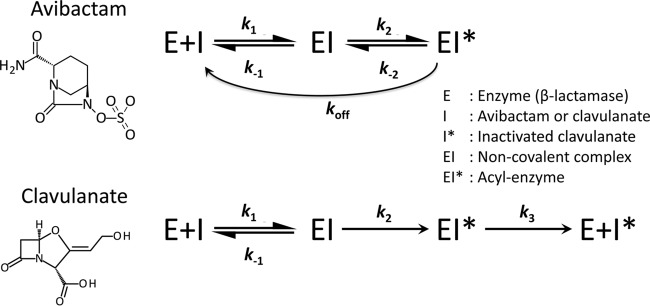
Avibactam is the first representative of a new family of inhibitors active against β-lactamases of classes A and C and certain enzymes of class D (1). Like β-lactam-containing inhibitors of the first generation, such as clavulanate, sulbactam, and tazobactam, avibactam acts as a suicide substrate and forms a covalent adduct with the active-site serine of the enzymes (Fig. 1). However, the carbamylation reaction is reversible in the case of avibactam, whereas the acylation reaction is irreversible in the case of first-generation inhibitors (2). This difference may be accounted for by the presence of a five-membered ring in avibactam, which is sterically less constrained than the four-membered ring of β-lactams. Consequently, the efficacy of the inhibition of the β-lactamases by avibactam depends upon equilibrium between the native (active) and carbamylated (inactive) forms of the enzyme (Fig. 1). This is evaluated by determining the kinetic parameters for the on (k2/Ki) and off (koff) reactions using nitrocefin as a chromogenic substrate. For β-lactam-containing inhibitors, such as clavulanate, the efficacy of the inhibition of β-lactamases depend both on the velocity of the acylation reaction and on the stability of the acyl-enzyme, which may be prone to hydrolysis (3). This was evaluated by determining the extent of β-lactamase acylation by mass spectrometry and the kinetic parameters kcat and Km for the hydrolysis of the inhibitor.
FIG 1.
Reaction schemes for inactivation of β-lactamases by avibactam and clavulanate.
Avibactam in combination with ceftazidime was approved by the U.S. Food and Drug Administration in 2015 to treat complicated intra-abdominal infections, when used in combination with metronidazole, as well as complicated urinary tract infections in patients with limited or no alternative treatment options. Ceftazidime-avibactam was also approved by the European Medicines Agency in June 2016 in patients with additional indications for hospital-acquired pneumonia, including ventilator-associated pneumonia. Although multidrug-resistant Gram-negative bacteria are the main target of the ceftazidime-avibactam combination, avibactam might have an additional application in the treatment of lung infections due to Mycobacterium abscessus in cystic fibrosis patients (4). The β-lactamase BlaMab produced by this rapidly growing mycobacterium is inhibited by avibactam but not by clavulanate, tazobactam, or sulbactam, since those compounds are efficiently hydrolyzed (5). In contrast, the β-lactamase of M. tuberculosis, BlaC, is irreversibly inactivated by clavulanate but slowly inhibited by avibactam (6–8). BlaMab and BlaC differ by the presence of residue N and residue G, respectively, at Ambler position 132, which corresponds to the third position of the conserved motif SDN. Site-directed mutagenesis has shown that this difference fully accounts for the differences in the inhibition profiles of BlaMab and BlaC (5). These observations led to the conclusion that the N132G substitution in BlaMab might result in resistance to β-lactam–avibactam combinations in M. abscessus (5).
In this study, we investigated whether the presence of N or G at the third position of the SDN motif has the same effect on the inhibition of class A β-lactamases from Enterobacteriaceae by avibactam and clavulanate as that previously reported for mycobacterial BlaC and BlaMab. To address this question, we introduced the substitution N132G in the β-lactamases KPC-2 and CTX-M-15 and determined the impact of the substitution on the kinetic parameters for the hydrolysis of β-lactams and the inhibition by avibactam and clavulanate. Isogenic strains were also constructed to determine whether the N132G substitution might be involved in the acquisition of resistance to β-lactam–avibactam combinations.
RESULTS
The N132G substitution impairs the inhibition of KPC-2 and CTX-M-15 by avibactam.
Introduction of the N132G substitution in KPC-2 led to a 4,000-fold reduction in the efficiency of carbamylation (k2/Ki) of the β-lactamase by avibactam (Table 1). The rate of decarbamylation was not affected by the substitution, as shown by koff values of 1.8 × 10−3 s−1 versus 1.0 × 10−3 s−1 for KPC-2 N132G and the parental enzyme, respectively. These results indicate that the substitution shifted the equilibrium between the native and carbamylated forms of KPC-2 toward the native (active) form of the enzyme, resulting in poor inhibition. Similarly, introduction of N132G into CTX-M-15 resulted in a 3,000-fold decrease in the rate of carbamylation. This effect was compensated for to a limited extent by a moderate reduction (20-fold) of the decarbamylation rate. Thus, the N132G substitution decreased the efficacy of inhibition of both KPC-2 and CTX-M-15, as previously shown for class A β-lactamases from mycobacteria (9).
TABLE 1.
Impact of the N132G substitution on kinetic parameters of avibactam and clavulanate and the inhibition of KPC-2 and CTX-M-15
| β-Lactamase | Avibactam |
Clavulanate |
||||
|---|---|---|---|---|---|---|
| k2/Ki (M−1 s−1) | k−2 (s−1) | koff (s−1) | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) | |
| KPC-2 | (2.1 ± 1.0) × 105 | (1.6 ± 0.4) × 10−3 | (1.0 ± 0.6) × 10−3 | 5.2 ± 0.7 | 36 ± 4 | (1.4 ± 0.3) × 105 |
| KPC-2 N132G | 53 ± 3 | (1.1 ± 0.1) × 10−3 | (1.8 ± 1.5) × 10−3 | (1.5 ± 0.2) × 10−4 | 0.35 ± 0.05 | 430 ± 80 |
| CTX-M-15 | (2.1 ± 0.1) × 105 | (1.6 ± 0.3) × 10−3 | (6.7 ± 1.1) × 10−4 | (3.0 ± 0.2) × 10−4 | 0.032 ± 0.005 | (9.4 ± 1.6) × 103 |
| CTX-M-15 N132G | 70 ± 6 | (2.2 ± 1.7) × 10−4 | (3.2 ± 1.7) × 10−5 | (1.2 ± 0.1) × 10−4 | 17 ± 2 | 7.1 ± 1.0 |
Impact of the N132G substitution on the inhibition of KPC-2 and CTX-M-15 by clavulanate.
KPC-2 efficiently hydrolyzed clavulanate with a kcat/Km ratio of 1.4 × 105 M−1 s−1 (Table 1). N132G almost fully abolished the hydrolysis of clavulanate, with an 80,000-fold decrease in kcat. The Km value was reduced 100-fold. These changes in the kinetic parameters kcat and Km are expected for stabilization of an acylated form of the enzyme (10).
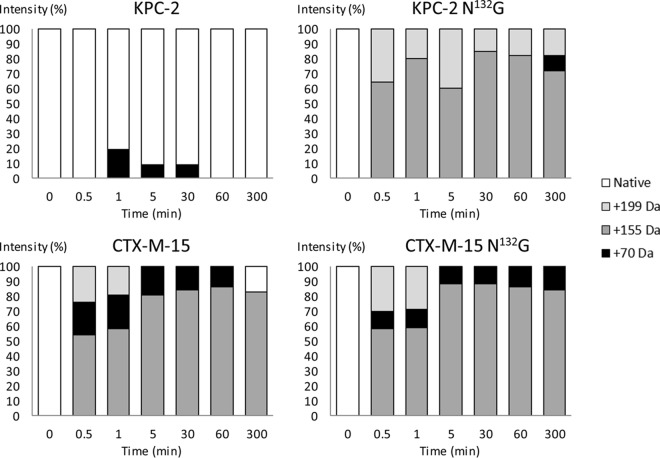
Mass spectrometry analysis revealed that incubation of KPC-2 N132G (10 μM) with clavulanate (500 μM) results in the rapid (<30-s) formation of two acyl-enzymes with mass increments of 199 Da and 155 Da (Fig. 2), which were expected for the acylation of the active serine by clavulanate followed by decarboxylation of the drug within the active site (3). The two acyl-enzymes remained in similar proportions for at least 300 min, in agreement with the low hydrolysis rate (kcat = 1.5 × 10−4 s−1), which implies that only 5.4% of the initial concentration of clavulanate was hydrolyzed in this time period. The native form of the β-lactamase was undetectable, indicating that KPC-2 N132G is efficiently inhibited by clavulanate in vitro. For the parental enzyme, the native from was predominant, in agreement with the high value of kcat. Together, the kinetic and mass spectrometry analyses indicated that introduction of the substitution N132G in KPC-2 almost completely abolished clavulanate hydrolysis due to stabilization of two acyl-enzymes. These modifications are expected to lead to efficient inhibition of the β-lactamase by clavulanate.
FIG 2.
Mass spectrometry of KPC-2 and CTX-M-15 (10 μM) with clavulanate (500 μM) after various times of incubation.
Clavulanate was a poor substrate of CTX-M-15, with a kcat of 3.0 × 10−4 s−1 (Table 1). Mass spectrometry analysis revealed the formation of an acyl-enzyme with a mass increment of 199 Da, which was detected only in the first minute of incubation (Fig. 2). The secondary acyl-enzyme resulting from the decarboxylation of clavulanate (mass increment of 155 Da) was the major form of the enzyme during the whole course of the experiment. This behavior indicates that clavulanate inhibits CTX-M-15. Introduction of the substitution N132G in CTX-M-15 resulted in a marginal decrease in kcat (3.0 × 10−4 s−1 versus 1.2 × 10−4 s−1). The value of Km was increased (from 0.032 μM to 17 μM). This was not associated with any modification of the kinetics of CTX-M-15 inactivation determined by mass spectrometry (Fig. 2). This was expected, since this analysis was performed with a clavulanate concentration of 500 μM, which largely exceeds both Km values. In conclusion, clavulanate is poorly hydrolyzed by CTX-M-15, and the N132G substitution did not further decrease the hydrolysis of the drug. Clavulanate is predicted to inhibit both forms of the enzyme.
Impact of the substitution N132G on the substrate profiles of KPC-2 and CTX-M-15.
Introduction of the substitution N132G in KPC-2 resulted in a moderate reduction in the velocity of hydrolysis of amoxicillin and nitrocefin (Table 2). Fold decreases in the overall catalytic efficacy (kcat/Km) ranged from 3 to 5. The N132G substitution was less well tolerated by CTX-M-15, with a 130-fold reduction in the value of kcat for the hydrolysis of amoxicillin. The kcat/Km ratio could not be estimated since the analysis provided only a lower limit for the value of Km (<50 μM). The kcat/Km ratio for the hydrolysis of nitrocefin by CTX-M-15 was not significantly affected by the N132G substitution (a 1.6-fold decrease in kcat/Km), since the 22-fold decrease in the value of kcat was largely compensated for by a 14-fold decrease in Km.
TABLE 2.
Impact of the N132G substitution in KPC-2 and CTX-M-15 on the kinetic parameters of β-lactams and their hydrolysis
| β-Lactamase | Amoxicillin |
Nitrocefin |
Ceftazidime |
Ceftaroline |
Aztreonam |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) | |
| KPC-2 | 150 ± 10 | 190 ± 50 | (7.9 ± 0.2) × 105 | 42 ± 2 | 11 ± 3 | (3.9 ± 1.0) × 106 | >1.4 | >600 | (3.7 ± 0.1) × 103 | >40 | >600 | (5.1 ± 2.2) × 104 | >350 | >5,000 | (6.9 ± 0.3) ×104 |
| KPC-2 N132G | 7.0 ± 0.4 | 46 ± 9 | (1.5 ± 0.3) × 105 | 35 ± 1 | 27 ± 3 | (1.3 ± 0.1) × 106 | >0.008 | >600 | 14 ± 3 | >2.5 | >600 | (4.5 ± 0.2) × 103 | >0.6 | >5,000 | (9.8 ± 1.9) ×101 |
| CTX-M-15 | 40 ± 4 | 19 ± 1 | (4.8 ± 0.5) × 105 | 190 ± 30 | 47 ± 17 | (4.0 ± 1.5) × 106 | >1.4 | >600 | (2.2 ± 0.2) × 103 | 90 ± 6 | 51 ± 12 | (1.8 ± 0.4) × 106 | 1.2 ± 0.1 | <150 | >8.0 × 103 |
| CTX-M-15 N132G | 0.3 ± 0.01 | <50 | >6.2 × 103 | 8.3 ± 0.3 | 3.3 ± 0.7 | (2.5 ± 0.5) × 106 | >0.01 | >600 | 13 ± 1 | 4.7 ± 0.5 | 240 ± 50 | (1.9 ± 0.4) × 104 | 0.33 ± 0.06 | 3,600 ± 1,100 | (9.0 ± 3.0) × 101 |
The impact of the N132G substitution was also estimated for β-lactams that are partners (ceftazidime) or potential partners (ceftaroline and aztreonam) of avibactam in currently developed combinations (11) (Table 2). These three β-lactams were hydrolyzed by KPC-2 with moderate catalytic efficiencies (kcat/Km ratios ranging from 6.9 × 104 M−1 s−1 to 3.7 × 103 M−1 s−1). The N132G substitution resulted in large decreases in the catalytic efficacy (kcat/Km) of KPC-2 for the hydrolysis of ceftazidime (260-fold) and aztreonam (700-fold) and to a lesser extent for the hydrolysis of ceftaroline (11-fold). Overall, the efficiency of hydrolysis of these β-lactams by KPC-2 was low. Similarly, the N132G substitution resulted in large decreases in the catalytic efficiency of CTX-M-15 (95- to 170-fold).
Impact of the substitution N132G on the antibacterial activity of amoxicillin in combination with avibactam or clavulanate.
MICs of β-lactams in combination with avibactam or clavulanate were determined by the broth microdilution technique against isogenic strains of Escherichia coli producing various β-lactamases (Table 3). The production of KPC-2 conferred high-level resistance to amoxicillin to the E. coli host (MIC > 4,096 μg/ml). Avibactam reduced the MIC of amoxicillin (>16-fold), but the strain remained resistant (MIC = 256 μg/ml) to this antibiotic. This observation suggests that inhibition of the β-lactamase by avibactam cannot restore susceptibility to β-lactams that are hydrolyzed by KPC-2 with a very high catalytic efficacy. Clavulanate was hydrolyzed by KPC-2, and the MIC of amoxicillin remained high in the presence of this inhibitor (2,048 μg/ml).
TABLE 3.
Impact of avibactam on the MICs of β-lactams for E. coli strains producing various β-lactamases
| β-Lactamase | MIC (μg/ml) of indicated β-lactam with or without Avi or Clava |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amoxicillin |
Ceftazidime |
Ceftaroline |
Aztreonam |
|||||||||
| None | Avi | Clav | None | Avi | Clav | None | Avi | Clav | None | Avi | Clav | |
| Noneb | 2 | 2 | 2 | 0.25 | 0.25 | 0.25 | 0.06 | 0.06 | 0.06 | 0.25 | 0.25 | 0.25 |
| KPC-2 | >4,096 | 256 | 2,048 | 128 | 1 | 32 | >1,024 | 1 | 128 | >1,024 | 0.25 | 128 |
| KPC-2 N132G | 2,048 | 2,048 | 8 | 4 | 1 | 0.5 | 32 | 0.5 | 0.06 | 1 | 0.125 | 0.25 |
| CTX-M-15 | 4,096 | 2 | 16 | 16 | 0.5 | 0.5 | >2,048 | 0.06 | 0.06 | 128 | 0.25 | 0.25 |
| CTX-M-15 N132G | 128 | 8 | 4 | 0.5 | 0.5 | 0.25 | 0.5 | 0.06 | 0.06 | 0.25 | 0.25 | 0.125 |
Values are the medians of results from three to five independent determinations. None, absence of avibactam and clavulanate; Avi, presence of avibactam (4 μg/ml); Clav, presence of clavulanate (4 μg/ml).
The control strain, which did not produce any β-lactamase, harbored the vector pTRC99-Km.
Introduction of the substitution N132G in KPC-2 reduced the MIC of amoxicillin in the absence of any inhibitor (from >4,096 μg/ml to 2,048 μg/ml). This difference might be accounted for by the 21-fold reduction in kcat, which was only partially compensated for by a 4-fold reduction in the value of Km. Inhibition of KPC-2 by avibactam was abolished by the substitution N132G, since the MIC of amoxicillin was the same in the presence or absence of this inhibitor (2,048 μg/ml). In contrast, KPC-2 N132G was inhibited by clavulanate, leading to susceptibility to amoxicillin (MIC = 8 μg/ml). Thus, the changes in the inhibition profile of KPC-2 detected in vitro (Table 1) were qualitatively associated with the expected modifications of the profiles of susceptibility to the β-lactam–inhibitor combinations.
For CTX-M-15, the 130-fold reduction in the value of kcat for the hydrolysis of amoxicillin due to the N132G substitution was associated with a 32-fold reduction in the MIC of this drug. CTX-M-15 was fully inhibited by avibactam, leading to a 2,000-fold reduction in the MIC of amoxicillin (from 4,096 μg/ml to 2 μg/ml). In contrast, inhibition of CTX-M-15 N132G was partial, leading to only a 16-fold reduction in the MIC of amoxicillin (from 128 μg/ml to 8 μg/ml). As expected from in vitro data (Table 1), clavulanate inhibited both CTX-M-15 N132G and the parental enzyme, leading to susceptibility to this drug in the former case. As found for KPC-2, the changes in the inhibition profile of CTX-M-15 due to N132G were qualitatively similar in vitro and in the E. coli host.
The main differences between KPC-2 and CTX-M-15 were (i) the higher value of kcat for the hydrolysis of amoxicillin by KPC-2 than by CTX-M-15, which prevented full inhibition of KPC-2 by avibactam in the E. coli host, and (ii) the inhibition of CTX-M-15 by clavulanate, whereas KPC-2 was inhibited only following introduction of the N132G substitution.
Impact of the substitution N132G on the antibacterial activities of drug combinations comprising ceftazidime, ceftaroline, and avibactam.
KPC-2 and CTX-M-15 conferred resistance to ceftazidime, and both β-lactamases were inhibited by avibactam in the E. coli host. The N132G substitution drastically reduced the MICs of ceftazidime, in agreement with the impaired hydrolysis of this drug in vitro. Consequently, the impact of the substitution on the inhibitory activities of avibactam and clavulanate detected in vitro could not result in meaningful changes in the antibiotic susceptibility pattern in the E. coli host. The N132G substitution cannot lead to resistance to the ceftazidime-avibactam combination, since the decrease in the efficacy of avibactam is fully compensated for by the impaired hydrolysis of ceftazidime. As expected, clavulanate restored the activity of ceftazidime in the case of CTX-M-15 but not in the case of KPC-2. Qualitatively, the results obtained for the combinations containing ceftaroline or aztreonam were similar to those obtained with ceftazidime.
DISCUSSION
An extensive analysis of the sequences of class A β-lactamases has shown that N132 at the C-terminal position of the SDN motif is highly conserved (12), but the role of this residue remains poorly understood (13). Crystal structures have shown that N132 forms a hydrogen bond with the side chain carbonyl residues of β-lactams (14) and of avibactam (12). N132 may have an additional role in catalysis, since its side chain oxygen participates in interconnected hydrogen-bonding networks that comprise the catalytic residues E166 and K73 (12, 14).
Here, we show that the substitution N132G has a strong negative impact on the efficacy of KPC-2 and CTX-M-15 carbamylation by avibactam (Table 1). The substitution N132G also drastically reduced the efficacy of clavulanate hydrolysis by KPC-2 (Table 1). Both effects of N132G appear to be conserved in class A β-lactamases, since they have also been detected for BlaMab from M. abscessus (9).
Reduced hydrolysis of clavulanate by KPC-2 was due to impaired deacylation, since mass spectrometry analyses showed that KPC-2 harboring N132G is rapidly acylated by clavulanate (Fig. 2), and kinetic analyses revealed that introduction of this substitution in KPC-2 leads to a reduction in the value of Km (Table 1). N132G has therefore distinct, and in some ways opposite, roles in the interactions of KPC-2 with avibactam and clavulanate in promoting carbamylation and deacylation, respectively. It is worth noting that the N132G substitution (this work) and the S130G substitution (15) in the SDN motif of KPC-2 result in large reductions in the kinetic parameter k2/Ki for the carbamylation reaction by avibactam (4,000-fold and 18,000-fold, respectively) and resistance to β-lactam–avibactam combinations comprising ampicillin or amoxicillin but not ceftazidime. In contrast, the substitution N132A did not reduce the efficacy of carbamylation of CTX-M-15 by avibactam (16). Thus, impaired inhibition of CTX-M-15 by avibactam due to the substitution N132G did not result from the absence of hydrogen bonds involving the carboxamide side chain of N132.
Since avibactam has been very recently introduced in therapeutics, there is little retrospect on the possibility of emergence of resistance to β-lactam–avibactam combinations. Prior to the introduction of the combination in clinical practice, the incidence of resistance to ceftazidime and avibactam was very low. For example, the incidence of resistance to the combination ceftazidime-avibactam (MIC ≥ 16 μg/ml) was 0.3% in a collection of 2,374 multidrug-resistant Klebsiella pneumoniae isolates that produced an Ambler class A enzyme and no class B, C, or D enzymes (17). Likewise, resistance to ceftazidime-avibactam was not detected in a collection of 139 non-carbapenem-susceptible Enterobacteriaceae (18). In agreement, the construction of large panels of isogenic strains of E. coli producing a single β-lactamase revealed that production of none of the TEM, CTX-M, KPC, CMY, ACT, FOX, PDC, GES, PER, VEB, and OXA enzymes that were tested, except PER-4, raised the MIC of ceftazidime above 8 μg/ml when determined in the presence of avibactam (12, 19–21). The first report of ceftazidime-avibactam resistance (32 μg/ml in the presence of 4 μg/ml of avibactam) in a KPC-3-producing Klebsiella pneumoniae isolate was unrelated to prior exposure to the combination (22). Ceftazidime-avibactam resistance following exposure to these drugs was detected in 3 out of 37 consecutive patients receiving this combination (23, 24).
Mutants resistant to the avibactam-ceftazidime combination have been obtained in vitro at a frequency on the order of 10−9 for two strains of Enterobacter cloacae and two strains of Klebsiella pneumoniae producing a KPC-3 β-lactamase (25). The mutations, which were obtained in a single step, produce large (≥16-fold) increases in the MIC of ceftazidime in the presence of avibactam (4 μg/ml). Resistance was due mostly to amino acid substitutions and to short (1- to 6-residue) insertions located within the Ω loop of KPC-3 and immediately adjacent to its C terminus, respectively. None of the mutations affected the SDN motif. The role of residues in the Ω loop of KPC-2 and SHV-1 enzymes was also explored by site-directed mutagenesis (26). For SHV-1, the substitutions that decreased the activity of ceftazidime did not affect the activity of the combination of ceftazidime and avibactam. For KPC-2, certain substitutions affecting R164 and D179, which form a salt bridge in the parental enzyme, led to resistance to the ceftazidime-avibactam combination. Resistance involved improved kinetic parameters for the hydrolysis of ceftazidime rather than decreased inhibition by avibactam. In contrast, we show here that the substitution N132G in KPC-2 and CTX-M-15 decreased the efficacies of both the hydrolysis of ceftazidime by the β-lactamases and the carbamylation of their active-site Ser by avibactam (Tables 1 and 2).
In conclusion, our analyses demonstrate that the substitution N132G severely impairs the carbamylation of class A β-lactamases by avibactam. However, two factors are expected to limit the emergence of this substitution under the selective pressure of drug combinations comprising avibactam. First, resistance to avibactam–β-lactam combinations was observed only for amoxicillin, since the substitution N132G impaired the hydrolysis of ceftazidime. Second, N (asparagine) and G (glycine) are specified by codons AA(U or C) and GG(A, U, G, or C). This implies that the substitution N132G requires replacement of two A's by two G's, a mutagenic event expected to be very rare.
MATERIALS AND METHODS
Plasmid construction.
Genes encoding KPC-2 and CTX-M-15 with the N132G substitution were obtained by site-directed mutagenesis using primers listed in Table 4. Briefly, two overlapping fragments of the gene encoding KPC-2 were amplified with primers T7 plus KPCa and T7-term plus KPCb. The purified PCR products were denatured and annealed via the complementary sequences present in the primers KPCa and KPCb, and the resulting heteroduplex was used as a template for amplification with primers T7 and T7-term. The same approach was used to amplify the gene encoding CTX-M-15 N132G with primers T7, CTXa, T7-term, and CTXb. The amplified genes were digested with the restriction endonucleases BamHI and NdeI and ligated into the vector pET-29a, digested by the same enzymes. For subcloning into the vector pTRC-99k, conferring resistance to kanamycin, the mutated genes were amplified with KPCfor plus KPCrev and CTXfor plus CTXrev, digested with EcoRI plus SalI, and cloned into pTRC-99k digested by the same enzymes. The sequence of the genes cloned into pTRC-99k was confirmed. Derivatives of pET29a and pTRC-99k were used for the production of the β-lactamases and antibiotic susceptibility testing, respectively. The vector pTRC-99k is a derivative of pTRC99a (Pharmacia), obtained by replacing the β-lactamase resistance gene with a kanamycin resistance gene (Km lacI, pTRC promoter, oriV colEI; D. Mengin-Lecreulx, unpublished data).
TABLE 4.
Primers used for site-directed mutagenesis
| Primer | Sequencea |
|---|---|
| KPCa | 5′-GTGCAATACAGTGATGGCGCCGCCGCCAATTTGTTG |
| KPCb | 5′-GCCATCACTGTATTGCACGGC |
| CTXa | 5′-CTACAGTACAGCGATGGCGTGGCGATGAATAAGCTG |
| CTXb | 5′-GCCATCGCTGTACTGTAGCGCGGCCGC |
| T7 | 5′-TAATACGACTCACTATAGGG |
| T7-term | 5′-CTAGTTATTGCTCAGCGGT |
| KPCfor | 5′-ATTTGAATTCATGTCACTGTATCGCC |
| KPCrev | 5′-ATTTGTCGACTTAGCAGCCGGATCCTTC |
| CTXfor | 5′-ATTTGAATTCATGGCGACGGCAACCGT |
| CTXrev | 5′-ATTTGTCGACTTACAAACCGTCGGTGAC |
The bases introduced in the primers to generate the N132G substitutions in KPC-2 and CTX-M-15 are underlined. The portions of the primers that display complementary sequences are italicized. The recognition sites for EcoRI and SalI are indicated in bold.
Production and purification of β-lactamases.
E. coli BL21(DE3) harboring pTRC-99k derivatives that encoded the β-lactamases were grown in brain heart infusion broth (Difco) containing kanamycin (50 μg/ml) and cultured at 37°C until the optical density at 600 nm reached 0.8. Isopropyl-β-d-1-thiogalactopyranoside (IPTG; 500 μM) was added, and the incubation was continued for 18 h at 16°C. Bacteria were collected by centrifugation (5,200 × g; 4°C), resuspended in buffer A (Tris-HCl, 50 mM, pH 8.8), and lysed by sonication. Cell debris was removed by centrifugation (17,400 × g; 4°C). The clarified lysate was filtered and loaded onto an anion-exchange chromatography column (Q Sepharose FF, 1 ml; GE Healthcare) equilibrated in buffer A. Elution was performed with a NaCl gradient in Tris-HCl (25 mM, pH 7.5), NaCl (300 mM, buffer B). The β-lactamases were recovered in the flowthrough and loaded onto a size exclusion chromatography column (Superdex 200 HL26/60 column; Amersham Pharmacia Biotech) equilibrated in buffer B. The β-lactamases, which eluted as monomers, were concentrated by ultrafiltration to ca. 5 mg/ml (cutoff, 10 kDa; Amicon Ultra-15; Millipore) and stored at −20°C in buffer B. Protein concentration was determined by the Bio-Rad protein assay using bovine serum albumin as a standard. The purified enzymes were checked by mass spectrometry and SDS-PAGE analyses.
Determination of kinetic parameters.
The kinetic parameters kcat, Km, and kcat/Km were determined at 20°C in 2-(N-morpholino)ethanesulfonic acid (MES; 100 mM, pH 6.4) by spectrophotometry, as previously described (7). Briefly, the initial velocity (vi) was determined by spectrophotometry for various concentrations of β-lactams [S] and a fixed concentration of β-lactamase [E]. The values of vi were plotted as a function of [S]. The kinetic constants Km and kcat were determined by fitting the equation vi = kcat [E]/Km + [S] to the resulting curve. The molecular extinction coefficients were 14,600 M−1 cm−1 at 486 nm for nitrocefin, −1,200 M−1 cm−1 at 244 nm for amoxicillin, −9,800 M−1 cm−1 at 256 nm for ceftazidime, −6,300 M−1 cm−1 at 306 nm for ceftaroline, −400 M−1 cm−1 at 318 nm for aztreonam, and −2,500 M−1 cm−1 at 227 nm for clavulanate. For the last compound, the concentration of MES was reduced to 30 mM in order to limit the absorbance of the buffer. For the hydrolysis of clavulanate, the value of Km, which could not be evaluated by the method described above (7), was determined by competition with nitrocefin (100 μM) at 20°C in 100 mM MES (pH 6.4) (9, 10). For avibactam, kinetic parameters for the carbamylation (k2/Ki and k−2) and decarbamylation (koff) reactions (2, 27) were determined at 37°C using nitrocefin (100 μM) in MES (100 mM, pH 6.4), as previously described (4) (9). Kinetics constants were deduced from a minimum of 12 progress curves obtained in a minimum of two independent experiments.
Mass spectrometry analyses.
Formation of clavulanate-enzyme adducts was assessed by incubating β-lactamases (10 μM) with clavulanate (500 μM) at 20°C in water. After various times of incubation, the samples were injected into the mass spectrometer (QStar Pulsar I; Applied Biosystems) with a mixture of acetonitrile (50%), water (49.5%), and formic acid (0.5%) at a flow rate of 0.05 ml/min. Spectra were acquired in positive mode as previously described (6).
Antibiotic susceptibility testing.
MICs were determined in Mueller-Hinton broth using the microdilution method in 96-well plates as described by the Clinical and Laboratory Standards Institute (28). Expression of the β-lactamase genes was induced with IPTG (500 μM) both in the preculture and in the 96-well plates. Kanamycin (50 μg/ml) was present in the preculture used to prepare the inoculum but not in the 96-well plates used for MIC determination.
ACKNOWLEDGMENTS
We thank L. Dubost and A. Marie for technical assistance in the collection of mass spectra and J. D. Docquier for the gift of plasmids encoding KPC-2 and CTX-M-15.
Avibactam was provided by AstraZeneca. C.O. was supported by the Année Recherche program from the Assistance Publique-Hôpitaux de Paris.
We have no competing interests to declare.
REFERENCES
- 1.Falcone M, Paterson D. 2016. Spotlight on ceftazidime/avibactam: a new option for MDR Gram-negative infections. J Antimicrob Chemother 71:2713–2722. doi: 10.1093/jac/dkw239. [DOI] [PubMed] [Google Scholar]
- 2.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Kern G, Walkup GK, Fisher SL. 2012. Avibactam is a covalent, reversible, non-beta-lactam beta-lactamase inhibitor. Proc Natl Acad Sci U S A 109:11663–11668. doi: 10.1073/pnas.1205073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drawz SM, Bonomo RA. 2010. Three decades of beta-lactamase inhibitors. Clin Microbiol Rev 23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubee V, Bernut A, Cortes M, Lesne T, Dorchene D, Lefebvre A-L, Hugonnet J-E, Gutmann L, Mainardi J-L, Herrmann J-L, Gaillard J-L, Kremer L, Arthur M. 2015. β-Lactamase inhibition by avibactam in Mycobacterium abscessus. J Antimicrob Chemother 70:1051–1058. doi: 10.1093/jac/dku510. [DOI] [PubMed] [Google Scholar]
- 5.Soroka D, Li de la Sierra-Gallay I, Dubee V, Triboulet S, van Tilbeurgh H, Compain F, Ballell L, Barros D, Mainardi JL, Hugonnet JE, Arthur M. 2015. Hydrolysis of clavulanate by Mycobacterium tuberculosis beta-lactamase BlaC harboring a canonical SDN motif. Antimicrob Agents Chemother 59:5714–5720. doi: 10.1128/AAC.00598-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhar N, Dubée V, Ballell L, Cuinet G, Hugonnet J-L, Signorino-Gelo F, Barros D, Arthur M, McKinney J. 2015. Rapid cytolysis of Mycobacterium tuberculosis by faropenem, an orally bioavailable beta-lactam antibiotic. Antimicrob Agents Chemother 59:1308–1319. doi: 10.1128/AAC.03461-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hugonnet JE, Blanchard JS. 2007. Irreversible inhibition of the Mycobacterium tuberculosis beta-lactamase by clavulanate. Biochemistry 46:11998–12004. doi: 10.1021/bi701506h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hugonnet JE, Tremblay LW, Boshoff HI, Barry CE III, Blanchard JS. 2009. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science 323:1215–1218. doi: 10.1126/science.1167498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soroka D, Ourghanlian C, Compain F, Fichini M, Dubée V, Mainardi J, Hugonnet J, Arthur M. 30 December 2016. Inhibition of β-lactamases of mycobacteria by avibactam and clavulanate. J Antimicrob Chemother doi: 10.1093/jac/dkw546. [DOI] [PubMed] [Google Scholar]
- 10.Frere JM, Dormans C, Duyckaerts C, De Graeve J. 1982. Interaction of beta-iodopenicillanate with the beta-lactamases of Streptomyces albus G and Actinomadura R39. Biochem J 207:437–444. doi: 10.1042/bj2070437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bush K. 2015. A resurgence of beta-lactamase inhibitor combinations effective against multidrug-resistant Gram-negative pathogens. Int J Antimicrob Agents 46:483–493. doi: 10.1016/j.ijantimicag.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Lahiri SD, Bradford PA, Nichols WW, Alm RA. 2016. Structural and sequence analysis of class A beta-lactamases with respect to avibactam inhibition: impact of Omega-loop variations. J Antimicrob Chemother 71:2848–2855. doi: 10.1093/jac/dkw248. [DOI] [PubMed] [Google Scholar]
- 13.Pratt RF, Krishnaraj R, Xu H. 1992. Effect of side-chain amide thionation on turnover of beta-lactam substrates by beta-lactamases. Further evidence on the question of side-chain hydrogen-bonding in catalysis. Biochem J 286:857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matagne A, Frere JM. 1995. Contribution of mutant analysis to the understanding of enzyme catalysis: the case of class A beta-lactamases. Biochim Biophys Acta 1246:109–127. doi: 10.1016/0167-4838(94)00177-I. [DOI] [PubMed] [Google Scholar]
- 15.Papp-Wallace KM, Winkler ML, Taracila MA, Bonomo RA. 2015. Variants of beta-lactamase KPC-2 that are resistant to inhibition by avibactam. Antimicrob Agents Chemother 59:3710–3717. doi: 10.1128/AAC.04406-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King DT, King AM, Lal SM, Wright GD, Strynadka NC. 2015. Molecular mechanism of avibactam-mediated beta-lactamase inhibition. ACS Infect Dis 1:175–184. doi: 10.1021/acsinfecdis.5b00007. [DOI] [PubMed] [Google Scholar]
- 17.Hackel M, Kazmierczak KM, Hoban DJ, Biedenbach DJ, Bouchillon SK, de Jonge BL, Stone GG. 2016. Assessment of the in vitro activity of ceftazidime-avibactam against multidrug-resistant Klebsiella spp. collected in the INFORM Global Surveillance Study, 2012 to 2014. Antimicrob Agents Chemother 60:4677–4683. doi: 10.1128/AAC.02841-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dupont H, Gaillot O, Goetgheluck AS, Plassart C, Emond JP, Lecuru M, Gaillard N, Derdouri S, Lemaire B, Girard de Courtilles M, Cattoir V, Mammeri H. 2015. Molecular characterization of carbapenem-nonsusceptible enterobacterial isolates collected during a prospective interregional survey in France and susceptibility to the novel ceftazidime-avibactam and aztreonam-avibactam combinations. Antimicrob Agents Chemother 60:215–221. doi: 10.1128/AAC.01559-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giani T, Cannatelli A, Di Pilato V, Testa R, Nichols WW, Rossolini GM. 2016. Inhibitory activity of avibactam against selected beta-lactamases expressed in an isogenic Escherichia coli strain. Diagn Microbiol Infect Dis 86:83–85. doi: 10.1016/j.diagmicrobio.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Papp-Wallace KM, Bajaksouzian S, Abdelhamed AM, Foster AN, Winkler ML, Gatta JA, Nichols WW, Testa R, Bonomo RA, Jacobs MR. 2015. Activities of ceftazidime, ceftaroline, and aztreonam alone and combined with avibactam against isogenic Escherichia coli strains expressing selected single beta-lactamases. Diagn Microbiol Infect Dis 82:65–69. doi: 10.1016/j.diagmicrobio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lahiri SD, Alm RA. 2016. Identification of novel VEB beta-lactamase enzymes and their impact on avibactam inhibition. Antimicrob Agents Chemother 60:3183–3186. doi: 10.1128/AAC.00047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humphries RM, Yang S, Hemarajata P, Ward KW, Hindler JA, Miller SA, Gregson A. 2015. First report of ceftazidime-avibactam resistance in a KPC-3-expressing Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 59:6605–6607. doi: 10.1128/AAC.01165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shields RK, Potoski BA, Haidar G, Hao B, Doi Y, Chen L, Press EG, Kreiswirth BN, Clancy CJ, Nguyen MH. 13 September 2016. Clinical outcomes, drug toxicity and emergence of ceftazidime-avibactam resistance among patients treated for carbapenem-resistant Enterobacteriaceae infections. Clin Infect Dis doi: 10.1093/cid/ciw636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spellberg B, Bonomo RA. 2016. Ceftazidime-avibactam and carbapenem-resistant Enterobacteriaceae: “we're gonna need a bigger boat.” Clin Infect Dis 63:1619–1621. doi: 10.1093/cid/ciw639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livermore DM, Warner M, Jamrozy D, Mushtaq S, Nichols WW, Mustafa N, Woodford N. 2015. In vitro selection of ceftazidime-avibactam resistance in Enterobacteriaceae with KPC-3 carbapenemase. Antimicrob Agents Chemother 59:5324–5330. doi: 10.1128/AAC.00678-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winkler ML, Papp-Wallace KM, Bonomo RA. 2015. Activity of ceftazidime/avibactam against isogenic strains of Escherichia coli containing KPC and SHV beta-lactamases with single amino acid substitutions in the Omega-loop. J Antimicrob Chemother 70:2279–2286. doi: 10.1093/jac/dkv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Durand-Reville TF, Lahiri S, Thresher J, Livchak S, Gao N, Palmer T, Walkup GK, Fisher SL. 2013. Kinetics of avibactam inhibition against class A, C, and D beta-lactamases. J Biol Chem 288:27960–27971. doi: 10.1074/jbc.M113.485979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clinical and Laboratory Standards Institute. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 10th ed. Document M07-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]