ABSTRACT
Sixteen different sequence types (STs) of Escherichia coli isolates from a commercial swine farm in China were confirmed to coharbor the carbapenem resistance gene blaNDM-5 and the colistin resistance gene mcr-1. Whole-genome sequencing revealed that blaNDM-5 and mcr-1 were located on a 46-kb IncX3 plasmid and a 32-kb IncX4 plasmid, respectively. The two plasmids can transfer together with a low fitness cost, which might explain the presence of various STs of E. coli coharboring blaNDM-5 and mcr-1.
KEYWORDS: Escherichia coli, carbapenems, blaNDM-5, colistin, mcr-1, fitness cost
TEXT
Carbapenemase-producing Enterobacteriaceae has become a major public health threat around the world (1). The recently identified carbapenemase New Delhi metallo-β-lactamase confers resistance to all β-lactam antimicrobials except monobactam (2). The NDM-5-encoding gene blaNDM-5 was first identified in an Escherichia coli strain recovered from a patient in the United Kingdom in 2011 (3). Since then, blaNDM-5 was identified in many countries, such as Algeria (4–6), the United States (7), Australia (8), China (9–12), Denmark (13), Japan (14), India (15), and the United Kingdom (3). The widespread occurrence of NDM-5 in recent years should arouse our attention. Colistin is a critically important medication for humans in the treatment of carbapenemase-producing Enterobacteriaceae, and it has been widely used in veterinary medicine in China (16, 17). The first plasmid-mediated colistin resistance gene, mcr-1, was reported in E. coli in 2015 (18). In a short period, colistin-resistant E. coli carrying the mcr-1 gene were reported worldwide (19, 20). Recently, mcr-1 was reported to coexist with blaNDM (21–23) and blaCTX-M (24), which brought great challenges for the treatment of bacterial infection. In the present study, we are the first to report the presence of isolates of various sequence types of E. coli coharboring blaNDM-5 and mcr-1 genes from a commercial pig farm in China.
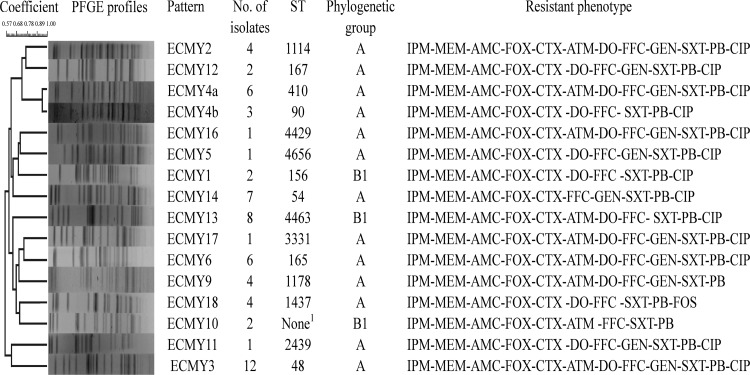
A total of 105 anal swabs samples from swine were collected from a commercial pig farm on 1 October 2015 in Sichuan province. E. coli strains were selected by eosin-methylene blue agar, and only 1 isolate was picked up from each sample. All 105 isolates were identified by BD Phoenix 100 diagnostic systems (Sparks, MD). Sixty-four strains were nonsusceptible to imipenem and polymyxin B, identified by the agar dilution method according to Clinical and Laboratory Standards Institute guidelines (25). Isolates were divided into 16 different clones by pulsed-field gel electrophoresis after XbaI digestion according to the standard PulseNet conditions (26) (Fig. 1). Phylogenetic group (A, B1, B2, and D) typing showed that all 16 different clones belonged to group A (n = 13) and B1 (n = 3) (27). Multilocus sequence typing (MLST) was performed as previously described (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli). All 16 clones belonged to different sequence types. The antimicrobial resistance profile, phylogenetic group, and sequence type of each different clone are shown in Figure 1.
FIG 1.
PFGE profiles, antimicrobial resistance phenotypes, phylogenetic groups, and sequence types (STs) of 16 different E. coli strains. 1Failure to find any corresponding ST type with MLST database via BLAST. Seven housekeeping gene allele types, i.e., adk, fumC, gyrB, icd, mdh, purA, recA, were identified as follows: 10, 11, 4, 8, 274, 8, 42. IPM, imipenem; MEM, meropenem; AMC amoxicillin-clavulanate; FOX, cefoxitin; CTX, cefotaxime; ATM, aztreonam; CIP, ciprofloxacin; DO, doxycycline; FFC, florfenicol; FOS, fosfomycin; GEN, gentamicin; SXT, trimethoprim-sulfamethoxazole; PB, polymyxin B.
The carbapenemase-encoding genes blaGES, blaKPC, blaIMP, blaNDM, blaOXA-48, and blaVIM and the colistin resistance gene mcr-1 were screened in all 16 different clones by PCR as described previously (18, 28, 29). The results showed that they all carried blaNDM-5 and mcr-1. Very recently, mcr-1 and blaNDM-5 coharbored by E. coli ST156 and ST648 were found in a Chinese hospital (23). To the best of our knowledge, this is the first report of the presence of diverse E. coli strains coharboring blaNDM-5 and mcr-1 on a commercial swine farm.
Whole-genome sequencing for 16 different clones was performed on the Illumina MiSeq (Majorbio, Shanghai, China) using a 400-bp paired-end TruSeq library with a 2 × 300 run. The paired-end reads were assembled de novo using the SOAP v2.04 and GapCloser v1.12. The gaps between different contigs were closed by PCR and sequencing. The genetic environments of blaNDM-5 and mcr-1 were analyzed by using the BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequence analysis revealed that blaNDM-5 and mcr-1 were located on two different plasmids, which showed 100% nucleotide identity in all 16 strains. The plasmid carrying blaNDM-5 belonged to the IncX3 group and was designated pECNDM101. It had a length of 46,165 bp with 46.66% G+C content, which had a 9-bp nucleotide substitution, 1-bp insertion, and 3-bp deletion compared with pNDM_MGR194 (GenBank accession number KF220657). An IS5 was inserted with ISAba125 upstream of blaNDM-5, which was also found in isolates in India (15), Japan (14), and China (9, 10).The plasmid carrying mcr-1 belonged to the IncX4 group and was designated pECMCR-1101. It had a length of 32,751 bp with 41.96% G+C content, which had a 559-bp deletion between 1,170 bp and 1,171 bp, an 8-bp nucleotide substitution, and a 6-bp insertion compared with pICBEC72Hmcr (GenBank accession number CP015977), which was recovered from a clinical E. coli strain in Brazil.
Conjugation experiments were carried out among the 16 different clones with rifampin-resistant E. coli EC600 as the recipient. Transconjugants were selected on three different Mueller-Hinton agar (Oxoid) plates that contained 400 μg/ml rifampin with 10 μg/ml imipenem or 2 μg/ml polymyxin B. Positive transconjugants were identified by detection of antimicrobial resistance profiles and screening for the presence of blaNDM-5 and mcr-1. Conjugation frequencies were calculated as the number of transconjugants per recipient cell and are shown in Table 1. When imipenem was used as the selection pressure, transfer frequencies of blaNDM-5 varied from 7.7 × 10−2 to 5.7 × 10−4. When polymyxin B was used as the selection pressure, transfer frequencies of mcr-1 varied from 5.8 × 10−5 to 5.3 × 10−7, and similar transfer frequencies of 2.9 × 10−7 to 2.5 × 10−5 were detected when imipenem and polymyxin B were used as the selection pressure. It was very interesting that the cotransfer of blaNDM-5 and mcr-1 was detected in a few transconjugants when using polymyxin B or imipenem alone. Three different transconjugants were obtained, MYNDM-5 carrying pECNDM101, MYmcr-1 carrying pECMCR-1101, and MYNDM-5+mcr-1 carrying pECNDM101 and pECMCR-1101.
TABLE 1.
Transconjugative frequencies of pECNDM101 and pECmcr-1101 plasmids in 16 different E. coli strains
| E. coli strain (transconjugant) | Transconjugative frequenciesa |
||
|---|---|---|---|
| pECNDM101b | pECmcr-1101c | pECNDM101 and pECmcr-1101d | |
| ECMY1 | 1.1 × 10−3 | 8.7 × 10−6 | 4.0 × 10−6 |
| ECMY2 | 1.1 × 10−2 | 9.2 × 10−7 | 5.5 × 10−7 |
| ECMY3 | 7.7 × 10−2 | 4.6 × 10−6 | 2.8 × 10−6 |
| ECMY4a | 5.3 × 10−2 | 1.6 × 10−5 | 5.7 × 10−6 |
| ECMY4b | 1.3 × 10−2 | 9.4 × 10−7 | 4.3 × 10−7 |
| ECMY5 | 1.7 × 10−2 | 5.3 × 10−6 | 1.2 × 10−6 |
| ECMY6 | 8.0 × 10−3 | 6.3 × 10−6 | 8.9 × 10−7 |
| ECMY9 | 5.7 × 10−4 | 5.8 × 10−5 | 2.5 × 10−5 |
| ECMY10 | 1.1 × 10−3 | 7.1 × 10−7 | 4.3 × 10−7 |
| ECMY11 | 5.3 × 10−3 | 5.6 × 10−5 | 2.3 × 10−5 |
| ECMY12 | 1.8 × 10−3 | 3.0 × 10−5 | 1.7 × 10−5 |
| ECMY13 | 5.6 × 10−3 | 5.3 × 10−7 | 2.9 × 10−7 |
| ECMY14 | 1.4 × 10−3 | 3.0 × 10−6 | 1.3 × 10−6 |
| ECMY16 | 9.1 × 10−4 | 6.6 × 10−6 | 3.9 × 10−6 |
| ECMY17 | 2.2 × 10−3 | 1.7 × 10−5 | 1.2 × 10−5 |
| ECMY18 | 1.8 × 10−3 | 5.0 × 10−5 | 1.6 × 10−5 |
The results shown are presented as the average value of 3 parallel experiments.
Imipenem was used as selection pressure in conjugation experiments.
Polymyxin B was used as selection pressure in conjugation experiments.
Imipenem and polymyxin B were used as selection pressure in conjugation experiments.
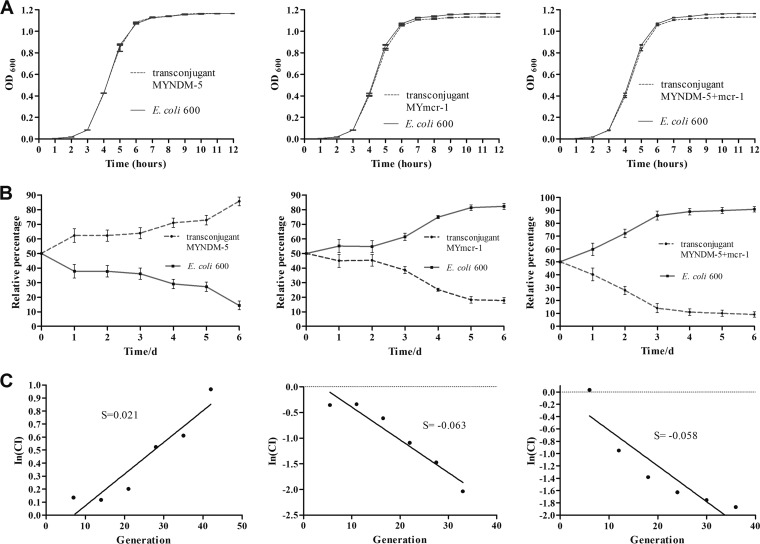
The fitness costs of pECNDM101 and pECMCR-1101 were determined by growth curves and competition experiments as previously described (30, 31). Growth curves of transconjugant MYNDM-5 and recipient E. coli EC600 were similar, and they arrived at the same concentration at the stationary phase (optical density at 600 nm [OD600] = 1.166). However, transconjugants MYmcr-1 and MYNDM-5+mcr-1 had a slight growth disadvantage and a lower concentration at the stationary phase (OD600 = 1.132 and 1.130, respectively) (Fig. 2A). For the competition experiment, a constant increase in the proportion of transconjugant MYNDM-5 was observed from day 3 on. In contrast, constant decreases in the proportion of transconjugants MYmcr-1 and MYNDM-5+mcr-1 were observed from day 2 on and day 1 on, respectively (Fig. 2B). Transconjugants MYmcr-1 and MYNDM-5+mcr-1 presented competitive disadvantages of ∼11.45% and ∼9.67% per 10 generations relative to those of E. coli EC600. Interestingly, transconjugant MYNDM-5 showed a competitive advantage (∼3% per 10 generations) (Fig. 2C), which might contribute to the propagation of blaNDM-5.
FIG 2.
Fitness costs of pECNDM101 and pECmcr-1101 in E. coli EC600. (A) Comparison of the growth kinetics of recipient E. coli EC600 and three transconjugants (MYNDM-5, MYmcr-1, and MYNDM-5+mcr-1) without any antibiotics. Growth curves represent the average results of three independent experiments. (B) Growth competition between recipient E. coli 600 and three transconjugants (MYNDM-5, MYmcr-1, and MYNDM-5+mcr-1). The initial ratio was 1:1. Competition experiments were repeated three times. (C) The selection coefficient (S) used to estimate the difference between the relative fitness of two competitors over the entire competition experiment, and S was calculated as the slope of the linear regression model In(CI)/In(d), where CI is the CFU ratio of the resistant and susceptible strains and d is the dilution factor.
In conclusion, to our knowledge, our study is the first to report that a blaNDM-5-carrying IncX3 plasmid (pECNDM101) and an mcr-1-carrying IncX4 plasmid (pECmcr-1101) coexist in various sequence types of E. coli on a commercial pig farm, and they can transfer together at a low fitness cost. Note that the cotransfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid was detected in a clinical E. coli isolate in China very recently (32). The results highlight that a swine farm is an important reservoir of E. coli carrying blaNDM-5 and mcr-1, which presents a serious challenge for public health via food-chain transmission.
Accession number(s).
The complete nucleotide sequences of the blaNDM-5-carrying IncX3 plasmid (pECNDM101) and the mcr-1-carrying IncX4 plasmid (pECmcr-1101) characterized in this study were submitted to GenBank and assigned accession numbers KX507346 and KX570748, respectively.
ACKNOWLEDGMENTS
This work was supported by the 973 National Basic Research Program of China (project number 2013CB127200), the General Program of the National Natural Science Foundation of China (grant number 31100102), and the Science and Technology Pillar Program in Sichuan Province (grant numbers 2013NZ0025, 13ZC2578, and 2012GZ0001-1).
REFERENCES
- 1.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase- producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hornsey M, Phee L, Wareham DW. 2011. A novel variant, NDM-5, of the New Delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother 55:5952–5954. doi: 10.1128/AAC.05108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sassi A, Loucif L, Gupta SK, Dekhil M, Chettibi H, Rolain JM. 2014. NDM-5 carbapenemase-encoding gene in multidrug-resistant clinical isolates of Escherichia coli from Algeria. Antimicrob Agents Chemother 58:5606–5608. doi: 10.1128/AAC.02818-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yousfi M, Mairi A, Bakour S, Touati A, Hassissen L, Hadjadj L, Rolain JM. 2015. First report of NDM-5-producing Escherichia coli ST1284 isolated from dog in Bejaia, Algeria. New Microbes New Infect 8:17–18. doi: 10.1016/j.nmni.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yaici L, Haenni M, Saras E, Boudehouche W, Touati A, Madec J-Y. 2016. blaNDM-5-carrying IncX3 plasmid in Escherichia coli ST1284 isolated from raw milk collected in a dairy farm in Algeria. J Antimicrob Chemother 71:2671–2672. doi: 10.1093/jac/dkw160. [DOI] [PubMed] [Google Scholar]
- 7.de Man TJ, Perry KA, Avillan JJ, Rasheed JK, Limbago BM. 2015. Draft genome sequence of a New Delhi metallo-β-lactamase-5 (ndm-5)-producing multidrug-resistant Escherichia coli isolate. Genome Announc 3:e00017-15. doi: 10.1128/genomeA.00017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wailan AM, Paterson DL, Kennedy K, Ingram PR, Bursle E, Sidjabat HE. 2016. Genomic characteristics of NDM-producing Enterobacteriaceae isolates in Australia and their blaNDM genetic contexts. Antimicrob Agents Chemother 60:136–141. doi: 10.1128/AAC.01243-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang P, Xie Y, Feng P, Zong Z. 2014. blaNDM-5 carried by an IncX3 plasmid in Escherichia coli sequence type 167. Antimicrob Agents Chemother 58:7548–7552. doi: 10.1128/AAC.03911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen D, Gong L, Walsh TR, Lan R, Wang T, Zhang J, Mai W, Ni N, Lu J, Xu J, Li J. 2016. Infection by and dissemination of NDM-5-producing Escherichia coli in China. J Antimicrob Chemother 71:563–565. doi: 10.1093/jac/dkv352. [DOI] [PubMed] [Google Scholar]
- 11.Zhu YQ, Zhao JY, Xu C, Zhao H, Jia N, Li YN. 2016. Identification of an NDM-5-producing Escherichia coli sequence type 167 in a neonatal patient in China. Sci Rep 6:29934. doi: 10.1038/srep29934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y, Yu X, Xie M, Wang X, Liao K, Xue W, Chan EW, Zhang R, Chen S. 2016. Widespread dissemination of carbapenem-resistant Escherichia coli sequence type 167 strains harboring blaNDM-5 in clinical settings in China. Antimicrob Agents Chemother 60:4364–4368. doi: 10.1128/AAC.00859-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammerum AM, Hansen F, Olesen B, Struve C, Holzknecht BJ, Andersen PS, Thye A-M, Jakobsen L, Røder BL, Stegger M, Hansen DS. 2015. Investigation of a possible outbreak of NDM-5-producing ST16 Klebsiella pneumoniae among patients in Denmark with no history of recent travel using whole-genome sequencing. J Glob Antimicrob Resist 3:219–221. doi: 10.1016/j.jgar.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Nakano R, Nakano A, Hikosaka K, Kawakami S, Matsunaga N, Asahara M, Ishigaki S, Furukawa T, Suzuki M, Shibayama K, Ono Y. 2014. First report of metallo-β-lactamase NDM-5-producing Escherichia coli in Japan. Antimicrob Agents Chemother 58:7611–7612. doi: 10.1128/AAC.04265-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnaraju M, Kamatchi C, Jha AK, Devasena N, Vennila R, Sumathi G, Vaidyanathan R. 2015. Complete sequencing of an IncX3 plasmid carrying blaNDM-5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian J Med Microbiol 33:30–38. doi: 10.4103/0255-0857.148373. [DOI] [PubMed] [Google Scholar]
- 16.Wu S, Zhang F, Huang Z, Liu H, Xie C, Zhang J, Thacker PA, Qiao S. 2012. Effects of the antimicrobial peptide cecropin AD on performance and intestinal health in weaned piglets challenged with Escherichia coli. Peptides 35:225–230. doi: 10.1016/j.peptides.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 17.Rhouma M, Beaudry F, Letellier A. 2016. Resistance to colistin: what is the fate for this antibiotic in pig production? Int J Antimicrob Agents 48:119–126. doi: 10.1016/j.ijantimicag.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 19.Schwarz S, Johnson AP. 2016. Transferable resistance to colistin: a new but old threat: table 1. J Antimicrob Chemother 71:2066–2070. doi: 10.1093/jac/dkw274. [DOI] [PubMed] [Google Scholar]
- 20.Zhi C, Lv L, Yu L-F, Doi Y, Liu J-H. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:292–293. doi: 10.1016/S1473-3099(16)00063-3. [DOI] [PubMed] [Google Scholar]
- 21.Mediavilla JR, Patrawalla A, Chen L, Chavda KD, Mathema B, Vinnard C, Dever LL, Kreiswirth BN. 2016. Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. mBio 7:1191–1116. doi: 10.1128/mBio.01191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao X, Doi Y, Zeng L, Lv L, Liu J-H. 2016. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis 16:288–289. doi: 10.1016/S1473-3099(16)00057-8. [DOI] [PubMed] [Google Scholar]
- 23.Yu H, Qu F, Shan B, Huang B, Jia W, Chen C, Li A, Miao M, Zhang X, Bao C, Xu Y, Chavda KD, Tang YW, Kreiswirth BN, Du H, Chen L. 2016. Detection of mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae (CRE) from different hospitals in China. Antimicrob Agents Chemother 60:5033–5035. doi: 10.1128/AAC.00440-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brauer A, Telling K, Laht M, Kalmus P, Lutsar I, Remm M, Kisand V, Tenson T. 2016. Plasmid with colistin resistance gene mcr-1 in ESBL-producing Escherichia coli strains isolated from pig slurry in Estonia. Antimicrob Agents Chemother. doi: 10.1128/AAC.00443-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement. CLSI document M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 26.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barret JT. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. [DOI] [PubMed] [Google Scholar]
- 27.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Poirel L, Le Thomas I, Naas T, Karlm A, Nordmann P. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother 44:622–632. doi: 10.1128/AAC.44.3.622-632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenski RE, Simpson SC, Nguyen TT. 1994. Genetic analysis of a plasmid-encoded, host genotype-specific enhancement of bacterial fitness. J Bacteriol 176:3140–3147. doi: 10.1128/jb.176.11.3140-3147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foucault ML, Courvalin P, Grillot-Courvalin C. 2009. Fitness cost of VanA-type vancomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 53:2354–2359. doi: 10.1128/AAC.01702-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun J, Yang RS, Zhang Q, Feng Y, Fang LX, Xia J, Li L, Lv XY, Duan JH, Liao XP, Liu YH. 2016. Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nat Microbiol 1:16176. doi: 10.1038/nmicrobiol.2016.176. [DOI] [PubMed] [Google Scholar]