ABSTRACT
We report here a new type of plasmid that carries the mcr-1 gene, the pMCR-1-P3 plasmid, harbored in an Escherichia coli strain isolated from a pig farm in China. pMCR-1-P3 belongs to the IncY incompatibility group and is a phage-like plasmid that contains a large portion of phage-related sequences. The backbone of this plasmid is different from that of other mcr-1-carrying plasmids reported previously.
KEYWORDS: DNA sequencing, antibiotics, plasmid
TEXT
The discovery of the plasmid-mediated colistin resistance gene mcr-1 has captured global attention. Unlike colistin resistance raised by chromosomal mutations, the plasmid carrying the mcr-1 resistance gene can be rapidly disseminated through horizontal gene transfer. Since the first report of plasmid pHNSHP45 carrying the mcr-1 gene (1), a number of plasmids with different backbones from human and animal sources carrying the same mcr-1 gene have been discovered worldwide (2–10). This suggests that the use of colistin in farm animals, particularly in pigs and chickens, is associated with the emergence and spread of colistin resistance (2). Of particular note is evidence of mcr-1 gene transfer from farm animals to humans through the food chain (3, 4). Therefore, analyzing the diversity of mcr-1-carrying plasmids is important to gain an understanding of the pattern of colistin resistance dissemination.
In this study, a total of 262 Escherichia coli strains were isolated from chicken cloacal and pig anal swabs in Shandong Province, China, between July 2015 and June 2016. We screened for the mcr-1 gene by performing PCR, confirmed by Sanger sequencing. Among these E. coli isolates, six mcr-1-positive strains were derived from chicken farms and seven were derived from pig farms. Sequences of the mcr-1 gene from these isolates showed 100% nucleotide identity with the published mcr-1 sequence (1). The results of multilocus sequencing typing (MLST) (11) showed that the 13 mcr-1-carrying isolates belong to 12 different sequence types, indicating wide distribution of the mcr-1 gene (see Table S1 in the supplemental material). To further investigate the genetic background of mcr-1-carrying plasmids, we sequenced all plasmids from each isolate using the Illumina MiSeq platform. The results revealed that one mcr-1-carrying plasmid from E. coli isolate P3 (sequence type 877) displayed a different gene sequence, while the others were mapped to plasmids pHNSHP45 (1), pHNSHP45-2 (GenBank accession number KU341381), and pMCR-1-IncI2 (12). Strain P3 was resistant to ampicillin, colistin, doxycycline, florfenicol, and tetracycline but susceptible to ceftazidime, enrofloxacin, gentamicin, and spectinomycin, among others (see Table S2). Further analysis of plasmid contigs using PlasmidFinder tools (https://cge.cbs.dtu.dk/services/PlasmidFinder/) suggested that isolate P3 contains more than one plasmid. Aiming to isolate the mcr-1-carrying plasmid from this isolate and to determine the host range of the plasmid, conjugation transfer experiments were performed using E. coli J53 Azir (azide resistance) (2 ng/μl colistin and 100 ng/μl NaN3), Pseudomonas aeruginosa ATCC 9027 (2 ng/μl colistin and Pseudomonas isolation agar), and Acinetobacter baumannii ATCC 19606 (2 ng/μl colistin and 8 ng/μl cefepime) as recipient strains. The conjugation frequencies of the mcr-1-positive plasmid were determined to be approximately 10−4, 10−1, and 0 per donor cell, respectively. The conjugation did not succeed with the A. baumannii strain, demonstrating that the mcr-1-positive plasmid is unlikely to be a broad host plasmid. The plasmid DNA was then extracted from positive E. coli J53 Azir conjugants and used for single-molecule real-time (SMRT) sequencing (Pacific Biosciences). The complete sequence of the plasmid, named pMCR-1-P3, was obtained by assembling the PacBio long reads and corrected by assembling Illumina short reads (see Table S3 for sequencing metrics) (10, 11).
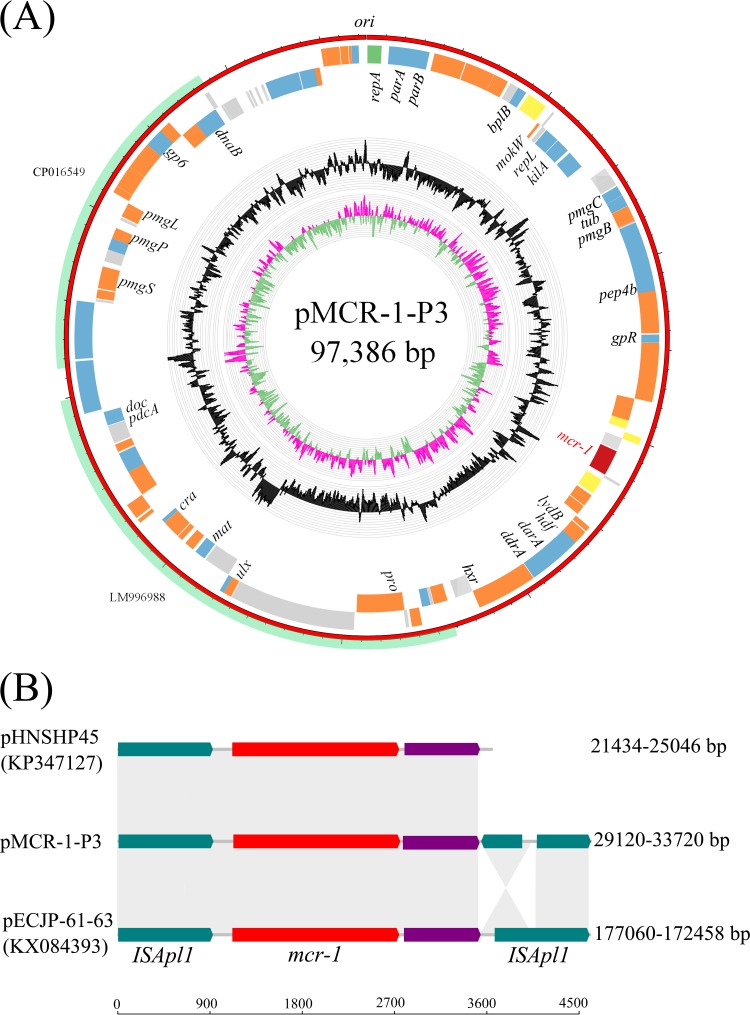
The plasmid sequence was 97,386 bp in length with a GC content of 47.8% (Fig. 1A). It was assigned to the IncY group and encodes a total of 108 open reading frames (ORFs). It contained a fragment of ∼23 kb with 97% nucleotide identity to E. coli genome assembly FHI87 (GenBank accession number LM996988) and a fragment of ∼17 kb with 98% identity to a plasmid sequence from E. coli strain O177:H21 (GenBank accession number CP016549) downstream of the mcr-1 gene (Fig. 1A). It has been found that the mcr-1 gene in some plasmids is surrounded by two ISApl1 elements, one on each end, which may form a composite transposon structure that can potentially move as one complete unit (6, 13–16). In pMCR-1-P3, the sequence of 668 bp on the 5′ end of the second ISApl1 was inverted, and thus the entire 924-bp of the ISApl1 transposase gene was split into two ORFs transcribed in opposite directions (Fig. 1B). This structure was further verified by PCR and sequencing. The fact that the second ISApl1 element is often missing (e.g., in the first reported plasmid pHNSHP45) (Fig. 1B) may indicate that the composite transposon TnApl (16) experiences dynamic changes during the transposition process.
FIG 1.
(A) Structure of plasmid pMCR-1-P3 carrying mcr-1 from Escherichia coli isolate P3. Colored rectangles represent open reading frames, with green, blue, orange, yellow, red, and gray rectangles representing replication genes, ORFs with known function, phage-related genes, mobile elements, the mcr-1 gene, and ORFs with unknown function, respectively. Individual rings range from 1 (outer ring) to 4 (inner ring): ring 1, genes encoded clockwise; ring 2, genes encoded anticlockwise; ring 3, GC content; ring 4, GC skew of the plasmid ([G−C]/[G+C]): magenta, >0; green, <0. Outer ring represents the high similarity of pMCR-1-P3 to LM996988 and CP016549, respectively. (B) Comparison of the immediate genetic environment of representative mcr-1-containing plasmids. Plasmid sequences were retrieved from GenBank; accession numbers are shown in brackets. Gray shading indicates >99% nucleotide identity.
Interestingly, a total of 72 genes encode phage-related proteins in pMCR-1-P3, such as portal protein, tail fiber protein, outer membrane lytic protein, terminase, and others (see Table S4 in the supplemental material). Among them, 47 genes are carried only by phages, while the other 25 genes are carried by both known plasmids and phages. Therefore, this suggests that pMCR-1-P3 is a phage-like plasmid that usually carries many phage-like elements (17–19). No other antibiotic resistance genes were found in pMCR-1-P3.
The IncY plasmids are normally identified from Enterobacteriaceae, such as E. coli, Salmonella, and Klebsiella pneumoniae (1–3). The common antibiotic resistance gene carried by IncY plasmids is blaCTX-M-15, an extended-spectrum beta-lactamase gene. To the best of our knowledge, this is the first report of an IncY plasmid carrying the mcr-1 gene. To date, the mcr-1 gene has been found in seven different plasmid incompatibility groups (IncI2 [20], IncFII [21], IncX4 [22], IncHI1 [15], IncHI2 [23], IncP [22], and IncF [10]), suggesting the wide distribution and high dissemination capacity of mcr-1-carrying plasmids. Given the fact that many phage-related genes are encoded by pMCR-1-P3 and most are homologous to the complete E. coli genome (GenBank accession number CP007392), this plasmid was likely formed via homologous recombination from a prophage that was originally located in the E. coli genome.
Under such heavy selection pressure, the high frequency of antibiotic resistance gene exchange and recombination in animal farms is expected. The identification of new mcr-1-containing phage-like plasmids with an IncY replicon from farm animals suggests that the diversity of mcr-1-carrying plasmids in livestock environments may be higher than expected. Given that phage-like plasmids carrying the mcr-1 gene are not common according to the National Center for Biotechnology Information (57 plasmids carrying the mcr-1 gene currently published), we suggest that pMCR-1-P3 may not have a dissemination advantage. However, results of previous research indicate that exposure to antibiotics induces prophages that can transfer antibiotic resistance genes to susceptible bacterial hosts (24). Therefore, pMCR-1-P3 may have emerged by transducing the phage into the plasmid, transferring the mcr-1 gene into the plasmid at the same time.
Accession number(s).
The nucleotide sequences reported in this work have been deposited in GenBank under accession no. KX880944.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Basic Research Program of China (973 Program, grant 2015CB554200), the National Natural Science Foundation of China (grants 31601081, 31302142, 81401701, and 31471203), the Beijing Municipal Natural Science Foundation (grant 5152019), and the Youth Innovation Promotion Association of Chinese Academy of Sciences (grant 2015069). We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02035-16.
REFERENCES
- 1.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 2.Rhouma M, Beaudry F, Letellier A. 2016. Resistance to colistin: what is the fate for this antibiotic in pig production? Int J Antimicrob Agents 48:119–126. doi: 10.1016/j.ijantimicag.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Hu Y, Liu F, Lin IY, Gao GF, Zhu B. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:146–147. doi: 10.1016/S1473-3099(15)00533-2. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki S, Ohnishi M, Kawanishi M, Akiba M, Kuroda M. 2016. Investigation of a plasmid genome database for colistin-resistance gene mcr-1. Lancet Infect Dis 16:284–285. doi: 10.1016/S1473-3099(16)00008-6. [DOI] [PubMed] [Google Scholar]
- 5.Perrin-Guyomard A, Bruneau M, Houee P, Deleurme K, Legrandois P, Poirier C, Soumet C, Sanders P. 2016. Prevalence of mcr-1 in commensal Escherichia coli from French livestock, 2007 to 2014. Euro Surveill 21:pii=30135 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=21380. [DOI] [PubMed] [Google Scholar]
- 6.Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, Day M, Muller-Pebody B, Ellington MJ, de Pinna E, Johnson AP, Hopkins KL, Woodford N. 2016. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother 71:2300–2305. doi: 10.1093/jac/dkw093. [DOI] [PubMed] [Google Scholar]
- 7.Khalifa HO, Ahmed AM, Oreiby AF, Eid AM, Shimamoto T, Shimamoto T. 2016. Characterisation of the plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli isolated from animals in Egypt. Int J Antimicrob Agents 47:413–414. doi: 10.1016/j.ijantimicag.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 8.Olaitan AO, Chabou S, Okdah L, Morand S, Rolain JM. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:147. doi: 10.1016/S1473-3099(15)00541-1. [DOI] [PubMed] [Google Scholar]
- 9.Fernandes MR, Moura Q, Sartori L, Silva KC, Cunha MP, Esposito F, Lopes R, Otutumi LK, Goncalves DD, Dropa M, Matte MH, Monte DF, Landgraf M, Francisco GR, Bueno MF, de Oliveira Garcia D, Knobl T, Moreno AM, Lincopan N. 2016. Silent dissemination of colistin-resistant Escherichia coli in South America could contribute to the global spread of the mcr-1 gene. Euro Surveill 21:pii=30214 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=22458 [DOI] [PubMed] [Google Scholar]
- 10.McGann P, Snesrud E, Maybank R, Corey B, Ong AC, Clifford R, Hinkle M, Whitman T, Lesho E, Schaecher KE. 2016. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob Agents Chemother 60:4420–4421. doi: 10.1128/AAC.01103-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du H, Chen L, Tang YW, Kreiswirth BN. 2016. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis 16:287–288. doi: 10.1016/S1473-3099(16)00056-6. [DOI] [PubMed] [Google Scholar]
- 13.Stoesser N, Mathers AJ, Moore CE, Day NP, Crook DW. 2016. Colistin resistance gene mcr-1 and pHNSHP45 plasmid in human isolates of Escherichia coli and Klebsiella pneumoniae. Lancet Infect Dis 16:285–286. doi: 10.1016/S1473-3099(16)00010-4. [DOI] [PubMed] [Google Scholar]
- 14.Falgenhauer L, Waezsada SE, Yao Y, Imirzalioglu C, Kasbohrer A, Roesler U, Michael GB, Schwarz S, Werner G, Kreienbrock L, Chakraborty T. 2016. Colistin resistance gene mcr-1 in extended-spectrum beta-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis 16:282–283. doi: 10.1016/S1473-3099(16)00009-8. [DOI] [PubMed] [Google Scholar]
- 15.Zurfluh K, Klumpp J, Nuesch-Inderbinen M, Stephan R. 2016. Full-length nucleotide sequences of mcr-1-harboring plasmids isolated from extended-spectrum-beta-lactamase-producing Escherichia coli isolates of different origins. Antimicrob Agents Chemother 60:5589–5591. doi: 10.1128/AAC.00935-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu H, Qu F, Shan B, Huang B, Jia W, Chen C, Li A, Miao M, Zhang X, Bao C, Xu Y, Chavda KD, Tang YW, Kreiswirth BN, Du H, Chen L. 2016. Detection of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae from different hospitals in China. Antimicrob Agents Chemother 60:5033–5035. doi: 10.1128/AAC.00440-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang H, Dong Y, Yang ZL, Luo H, Zhang X, Gao F. 2014. Complete sequence of pABTJ2, a plasmid from Acinetobacter baumannii MDR-TJ, carrying many phage-like elements. Genomics Proteomics Bioinformatics 12:172–177. doi: 10.1016/j.gpb.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Octavia S, Sara J, Lan R. 2015. Characterization of a large novel phage-like plasmid in Salmonella enterica serovar Typhimurium. FEMS Microbiol Lett 362:fnv044. doi: 10.1093/femsle/fnv044. [DOI] [PubMed] [Google Scholar]
- 19.Falgenhauer L, Yao Y, Fritzenwanker M, Schmiedel J, Imirzalioglu C, Chakraborty T. 2014. Complete genome sequence of phage-like plasmid pECOH89, encoding CTX-M-15. Genome Announc 2:e00356-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang YQ, Zhang AY, Ma SZ, Kong LH, Li YX, Liu JX, Davis MA, Guo XY, Liu BH, Lei CW, Wang HN. 2016. Co-occurrence of mcr-1 and ESBL on a single plasmid in Salmonella enterica. J Antimicrob Chemother 71:2336–2338. doi: 10.1093/jac/dkw243. [DOI] [PubMed] [Google Scholar]
- 21.Xavier BB, Lammens C, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Complete sequence of an IncFII plasmid harbouring the colistin resistance gene mcr-1 isolated from Belgian pig farms. J Antimicrob Chemother 71:2342–2344. doi: 10.1093/jac/dkw191. [DOI] [PubMed] [Google Scholar]
- 22.Li A, Yang Y, Miao M, Chavda KD, Mediavilla JR, Xie X, Feng P, Tang YW, Kreiswirth BN, Chen L, Du H. 2016. Complete sequences of mcr-1-harboring plasmids from extended-spectrum-beta-lactamase- and carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 60:4351–4354. doi: 10.1128/AAC.00550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anjum MF, Duggett NA, AbuOun M, Randall L, Nunez-Garcia J, Ellis RJ, Rogers J, Horton R, Brena C, Williamson S, Martelli F, Davies R, Teale C. 2016. Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J Antimicrob Chemother 71:2306–2313. doi: 10.1093/jac/dkw149. [DOI] [PubMed] [Google Scholar]
- 24.Bearson BL, Allen HK, Brunelle BW, Lee IS, Casjens SR, Stanton TB. 2014. The agricultural antibiotic carbadox induces phage-mediated gene transfer in Salmonella. Front Microbiol 5:52. doi: 10.3389/fmicb.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.