ABSTRACT
Reversal of antimicrobial resistance is an appealing and largely unexplored strategy in drug discovery. The objective of this study was to identify potential targets for “helper” drugs reversing cephem resistance in Escherichia coli strains producing β-lactamases. A CMY-2-encoding plasmid was transferred by conjugation to seven isogenic deletion mutants exhibiting cephem hypersusceptibility. The effect of each mutation was evaluated by comparing the MICs in the wild type and the mutant harboring the same plasmid. Mutation of two genes encoding proteins involved in cell wall biosynthesis, dapF and mrcB, restored susceptibility to cefoxitin (FOX) and reduced the MICs of cefotaxime and ceftazidime, respectively, from the resistant to the intermediate category according to clinical breakpoints. The same mutants harboring a CTX-M-1-encoding plasmid fell into the intermediate or susceptible category for all three drugs. Individual deletion of dapF and mrcB in a clinical isolate of CTX-M-15-producing E. coli sequence type 131 (ST131) resulted in partial reversal of ceftazidime and cefepime resistance but did not reduce MICs below susceptibility breakpoints. Growth curve analysis indicated no fitness cost in a ΔmrcB mutant, whereas a ΔdapF mutant had a 3-fold longer lag phase than the wild type, suggesting that drugs targeting DapF may display antimicrobial activity, in addition to synergizing with selected cephems. DapF appeared to be a potential FOX helper drug target candidate, since dapF inactivation resulted in synergistic potentiation of FOX in the genetic backgrounds tested. The study showed that individual inactivation of two nonessential genes involved in cell wall biogenesis potentiates cephem activity according to drug- and strain-specific patterns.
KEYWORDS: extended-spectrum β-lactamase (ESBL), cephem resistance, helper drug
INTRODUCTION
Resistance to cephems, a group of β-lactam antibiotics including cephalosporins and cephamycins, is clinically important, as these antimicrobial agents, especially oxyimino-cephalosporins, are commonly used in clinical practice, are easy to administer, and have low toxicity (1). With treatment options limited to “last-resort” antimicrobial agents, mainly carbapenems (2), no adequate therapeutic options exist to face the worldwide increase of infections caused by cephem-resistant Enterobacteriaceae producing extended-spectrum β-lactamase (ESBL) or AmpC β-lactamase, which have a considerable clinical and economic impact (3). There is an urgent need for new therapeutic strategies to manage both community- and hospital-acquired infections caused by these bacteria. One of the possible strategies is to suppress resistant subpopulations by combination therapy (4). This approach includes combinations with “helper” drugs designed to enhance antimicrobial activity (5). β-Lactamase inhibitors are a prominent example of helper drugs used in clinical practice. Numerous inhibitors with novel mechanisms of inhibition are currently being developed against β-lactamase-producing strains that have resistance to traditional inhibitors, such as clavulanic acid, tazobactam, and sulbactam (6). A new β-lactam–β-lactamase inhibitor combination, ceftazidime-avibactam, has recently been approved for clinical use (7). Efflux pump inhibitors (EPIs) are another example of helper drugs suppressing resistance mechanisms, but despite the numerous EPIs described in the scientific literature (8), none of them has reached clinical development, mainly due to toxicity. In addition to suppression of resistance, helper drugs can also reduce the likelihood of resistance development during antimicrobial therapy (9).
The antimicrobial susceptibility phenotype of a given bacterial species depends on the concerted activities of several elements, which as a whole are known as the intrinsic resistome (10, 11). The various components of the Escherichia coli intrinsic resistome have been inferred by previous studies measuring the susceptibility and fitness of in-frame gene deletion mutants in the presence of antimicrobial agents (12, 13). Despite the potential of the intrinsic resistome to serve as a source for helper drug targets, no studies have investigated whether inactivation of these genes restores susceptibility in resistant strains by reducing the MIC below the clinical breakpoint. In this study, we evaluated the helper drug target potential of 16 selected genes important for intrinsic resistance to cephems in E. coli (12, 13). The effect of each gene deletion on cephem activity was studied by comparing the MICs of the deletion mutant and the wild type in different E. coli genetic backgrounds harboring plasmids encoding clinically important ESBLs (CTX-M-1 and CTX-M-15) or AmpC β-lactamase (CMY-2). We identified two gene deletions, ΔdapF and ΔmrcB, that potentiate cephem activity against β-lactamase-producing strains in a drug-specific fashion.
RESULTS
Effects of gene deletions on acquired cephem resistance.
The MICs of cefoxitin (FOX) and cefotaxime (CTX) in the E. coli wild-type strain BW25113 and in the 16 derivative isogenic Keio mutants selected for this study are shown in Table 1. Based upon the magnitude of the MIC change determined by each gene deletion relative to the wild type, six and two gene deletions exhibited synergy with FOX and CTX, respectively. One gene deletion (ΔsurA) had an antagonistic effect with FOX. Seven gene deletions resulted in MIC reductions greater than 2-fold (Table 1), and their effects were studied in the same genetic background (BW25113) containing a CMY-2-encoding plasmid. As expected, the plasmid conferred resistance to ceftazidime (CAZ), CTX, and FOX in BW25113 (Table 2). However, in the presence of the same plasmid, two individual gene deletions (ΔmrcB and ΔdapF) reduced the MICs of FOX and CTX by more than 2-fold compared to wild-type strain BW25113. Both deletions exhibited synergistic interactions with FOX, while CAZ and CTX synergistic interactions were limited to ΔmrcB and ΔdapF, respectively (Table 2). According to the CLSI breakpoints, these two mutants were susceptible to FOX and fell into the intermediate category for CAZ and CTX, respectively (Table 2). The effects of these two gene deletions were further evaluated in the presence of a CTX-M-1-encoding plasmid to check whether the effect of each gene deletion is restricted to the specific β-lactamase type. The MICs in both ΔdapF and ΔmrcB mutants harboring the CTX-M-1-encoding plasmid were 8- to 16-fold lower than in the wild-type strain containing the same plasmid (Table 3). CTX resistance was completely reversed by deletion of dapF, while the mrcB deletion resulted in only partial reversal of resistance to the cephalosporin. Both deletions exhibited synergistic interactions with CAZ and FOX, although both the wild type and the mutants producing CTX-M-1 fell into the susceptible category, according to the CLSI breakpoints for the drugs (Table 3).
TABLE 1.
MICs of FOX and CTX in E. coli wild-type BW25113 and in 16 derivative isogenic Keio mutants
| Drug | MICa (μg/ml) |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | ΔmrcB | ΔdacA | ΔycfM | ΔybgF | ΔdapF | ΔycjU | Δfis | ΔrimK | ΔrplA | ΔdksA | ΔbamB | ΔsurA | ΔtolC | ΔacrA | ΔrecA | ΔyciM | |
| FOX | 4 | 0.25 | 1 | 0.5 | 1 | 0.5 | 2 | 4 | 4 | 4 | 2 | 4 | 16b | 1 | 2 | 2 | 8 |
| CTX | 0.031 | 0.016 | 0.016 | 0.031 | 0.031 | 0.008 | 0.016 | 0.031 | 0.031 | 0.031 | 0.008 | 0.016 | 0.031 | 0.016 | 0.016 | 0.031 | 0.031 |
The MICs of drugs for which individual gene deletion resulted in a synergistic effect (interaction index ≤ 0.25) are underlined.
Deletion of surA had an antagonistic effect on FOX (interaction index ≥ 4).
TABLE 2.
MICs of FOX, CTX, and CAZ in wild-type BW25113 E. coli and derivative isogenic Keio mutants carrying a CMY-2-encoding plasmid
| Drug | MICa (μg/ml) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | ΔmrcB | ΔdacA | ΔycfM | ΔybgF | ΔdapF | ΔdksA | ΔtolC | Susceptibility breakpoint | Resistance breakpoint | |
| FOX | 32 | 8 | 32 | 16 | 16 | 2 | 16 | 32 | ≤8 | ≥32 |
| CTX | 8 | 4 | 8 | 16 | 8 | 2 | 4 | 8 | ≤1 | ≥4 |
| CAZ | 32 | 8 | 32 | 32 | 32 | 32 | 16 | 32 | ≤4 | ≥16 |
MICs equal to or below the CLSI susceptibility breakpoints for Enterobacteriaceae (37) are indicated in boldface. The MICs of drugs for which individual gene deletion resulted in a synergistic effect (interaction index ≤ 0.25) are underlined.
TABLE 3.
MICs of FOX, CTX, and CAZ in E. coli wild-type BW25113 and in two derivative isogenic Keio mutants carrying a CTX-M-1-encoding plasmid (ΔdapF and ΔmrcB)
| Cephem | MICa (μg/ml) |
||||
|---|---|---|---|---|---|
| Wild type | ΔmrcB | ΔdapF | Susceptibility breakpoint | Resistance breakpoint | |
| FOX | 4 | 0.5 | 0.5 | ≤8 | ≥ 32 |
| CTX | 16 | 2 | 1 | ≤1 | ≥ 4 |
| CAZ | 2 | 0.25 | 0.25 | ≤4 | ≥ 16 |
MICs below the CLSI susceptibility breakpoints for Enterobacteriaceae (37) are indicated in boldface. The MICs of drugs for which individual gene deletion resulted in a synergistic effect (interaction index ≤ 0.25) are underlined.
Based on the susceptibility results, dapF and mrcB were deleted in a clinical isolate of CTX-M-15-producing E. coli sequence type 131 (ST131) (UR40) to determine if the two individual gene deletions could potentiate cephems in this epidemic clone. Both the wild type and the mutants producing CTX-M-15 were susceptible to FOX, according to the CLSI breakpoints (Table 4). Complete and partial reversal of CAZ resistance was observed in ΔdapF and ΔmrcB mutants, respectively. The two mutants fell into the intermediate category for cefepime (FEP) but retained resistance to ceftriaxone (CRO) and CTX, most likely because the wild-type strain displays very high MICs of these two oxyimino-cephalosporins (MICs, >64 and >128 μg/ml, respectively). Deletion of dapF had a synergistic effect with all five tested cephems, whereas only synergy with FOX was detected following mrcB deletion (Table 4).
TABLE 4.
MICs of CRO, CTX, CAZ, FOX, and FEP in a clinical isolate of E. coli ST131 (UR40) harboring a CTX-M-15-encoding plasmid and in two derivative isogenic mutants (ΔdapF and ΔmrcB)
| Drug | MICa (μg/ml) |
||||
|---|---|---|---|---|---|
| Wild-type UR40 | ΔmrcB | ΔdapF | Susceptibility breakpoint | Resistance breakpoint | |
| CRO | >128 | 128 | 64 | ≤1 | ≥ 4 |
| CTX | >64 | 64 | 32 | ≤1 | ≥ 4 |
| CAZ | 16 | 8 | 4 | ≤4 | ≥ 16 |
| FOX | 8 | <2 | <2 | ≤8 | ≥ 32 |
| FEP | 16 | 8 | 4 | ≤2 | ≥ 16 |
MICs equal to or below the CLSI susceptibility breakpoints for Enterobacteriaceae (37) are indicated in boldface. The MICs of drugs for which individual gene deletion resulted in a synergistic effect (interaction index ≤ 0.25) are underlined.
Real-time growth analysis of E. coli ST131 wild type and mutants.
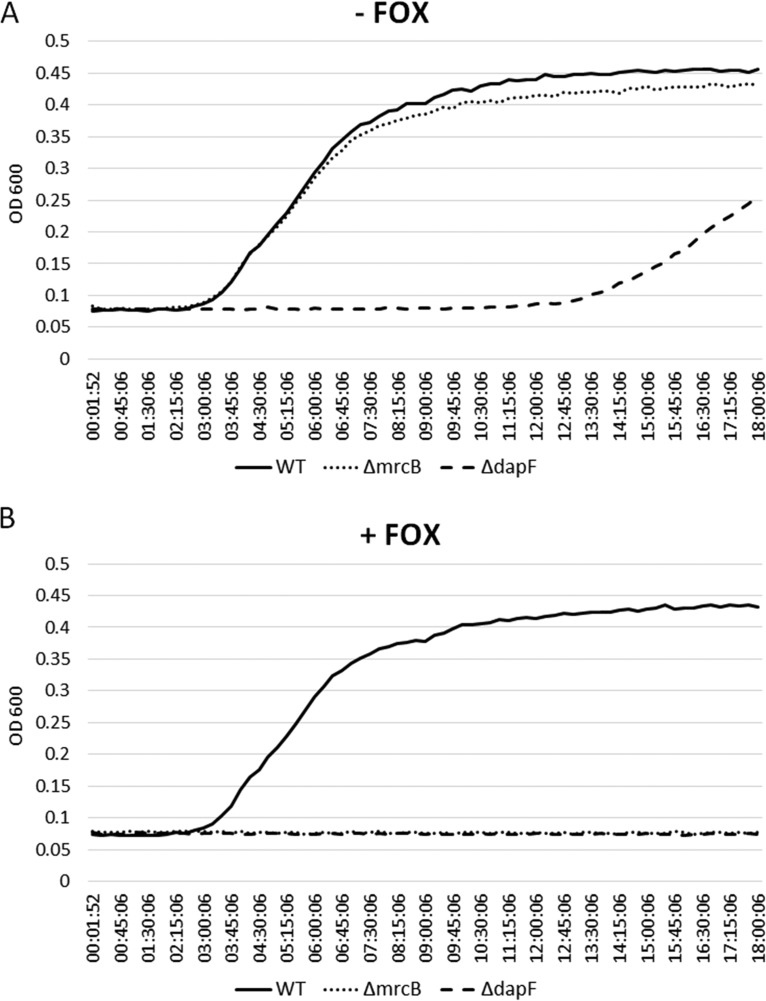
Real-time growth curve analysis was performed to generate information regarding the fitness of ΔdapF and ΔmrcB mutants compared to the wild type and to assess how these gene deletions affected growth in the presence of FOX. This cephamycin was selected because it was the only cephem that exhibited synergy with both deletions (Table 4). Analysis of the lag phase of the two mutants and the wild type in FOX-free medium showed no effect of mrcB deletion on bacterial growth. However, ΔdapF displayed a 3-fold-longer lag phase than the wild type and the ΔmrcB mutant (Fig. 1A). As expected, supplementation of the medium with 2 μg/ml of FOX had no effect on the growth of the wild type but inhibited growth of both mutants (Fig. 1B).
FIG 1.
Growth curves of wild-type (WT) E. coli ST131 producing CTX-M-15 and derivative deletion mutants (ΔdapF and ΔmrcB) in the absence (A) and presence (B) of 2 μg/ml FOX.
MrcB and DapF homology between Enterobacteriaceae and humans.
Considerable homology in amino acid sequence (100% coverage and 87 to 99% identity relative to the sequence in E. coli) was detected for both proteins between Enterobacteriaceae species: Klebsiella pneumoniae (87% for MrcB and 95% for DapF), Salmonella enterica (91% for MrcB and 96% for DapF), Shigella flexneri (99% for MrcB and 99% for DapF), and Citrobacter freundii (93% for MrcB and 98% for DapF). In contrast, no human homologues were identified for MrcB and DapF.
DISCUSSION
We employed a novel, straightforward approach to identify potential antimicrobial helper drug targets in ESBL/AmpC-producing E. coli. This approach applied knowledge generated from previous high-throughput drug-gene interaction profiling studies showing that individual gene deletions can cause hypersusceptibility in wild-type E. coli (12, 13). Here, the effects of such gene deletions on cephem susceptibility were studied in resistant E. coli producing different types of clinically important β-lactamases. Interestingly, deletion of mrcB or dapF enhanced the activity of oxyimino-cephalosporins (CAZ, CTX, and FEP) and cephamycins (FOX) according to drug- and strain-specific patterns. Complete reversal of resistance, defined as a MIC reduction from the resistant to the susceptible category according to clinical breakpoints, was observed only for FOX, by inactivation of either gene in the CMY-2-producing strain (Table 2), and for CAZ, by dapF inactivation in the CTX-M-15-producing strain (Table 4). Both gene products are involved in cell wall biogenesis, i.e., mrcB encodes a penicillin-binding protein (PBP1b) and dapF encodes an epimerase involved in the synthesis of peptidoglycan precursors.
DapF appears to be a promising FOX helper drug target, since deletion mutants in dapF displayed FOX potentiation and were consistently susceptible to the cephamycin regardless of the strain genetic background and β-lactamase type (Tables 2, 3, and 4). Although the CTX-M-1 and CTX-M-15 β-lactamases did not confer resistance in the wild-type strain, dapF deletion mutants consistently showed MICs ≥6-fold lower than that of the wild type, which is a desirable feature for a helper drug target. Deletion of the gene also restored full susceptibility to CAZ in the CTX-M-15-producing strain (Table 4) but did not reduce the MIC of CAZ in the strain that produced CMY-2 (Table 2). We do not know why the MIC of CAZ was not lowered in the presence of CMY-2, but this observation suggests that combining CAZ with DapF inhibitors would not enhance CAZ efficacy against strains expressing this AmpC type β-lactamase. DapF, a cytoplasmic diaminopimelate (DAP) epimerase, is part of the DAP pathway that converts diaminopimelic acid (LL-DAP) to a better peptidoglycan precursor (meso-DAP), which incorporates into the cell wall 1,000 times more efficiently than LL-DAP (14, 15). Deletion of dapF increases the LL-DAP pool compared to meso-DAP, likely reducing the peptidoglycan synthesis rate, which in turn may explain the reduced fitness of the ΔdapF ST131 mutant in the growth assay (Fig. 1). It should be noted that the increased cephem susceptibility observed in this mutant may well be influenced by its reduced fitness. The poor fitness of ΔdapF ST131 suggests that compounds interfering with DapF function may exhibit a growth-inhibitory effect in vivo. The absence of DapF homologues in mammals (15, 16) is another characteristic favorable for developing a FOX helper drug targeting this protein.
Similar to ΔdapF, deletion of mrcB reversed FOX resistance in the CMY-2-producing strain and increased susceptibility to both FOX and oxyimino-cephalosporins in the CTX-M-producing strains (Tables 3 and 4). However, unlike ΔdapF, mrcB deletion decreased the MIC of CAZ in the CMY-2-producing strain (Table 2) and did not affect the strain's fitness (Fig. 1). Previous studies have shown reduced fitness of ΔmrcB in stationary-phase competition assays using model E. coli strains and decreased tolerance for bile in Salmonella enterica serovar Typhi (17, 18). The mrcB gene product, PBP1b, is one of the two major bifunctional penicillin-binding proteins in E. coli, with both transglycosylase and transpeptidase enzymatic activities. PBP1b is uniquely important for biofilm formation and motility and is implicated in outer membrane constriction during cell division (19–21). While the enhanced cephem activity observed in ΔmrcB could be due to reduced integrity of the peptidoglycan layer (22), the known synthetic lethality between mrcB and mrcA (PBP1a) (20, 23–25) suggests a more specific mechanism. In particular, the potentiation of FOX in the ΔmrcB mutant implies that PBP1a may be a primary target of the cephamycin. Interestingly, mrcA deletion did not cause increased cephem sensitivity, suggesting a more critical role of PBP1b in cephem resistance (12, 13, 23). PBP1b function requires interaction with the outer membrane lipoprotein YcfM (20, 24, 26). In the absence of its interaction partner, aberrant YcfM interactions with other proteins may introduce additional envelope stress in the presence of cephems (20). These data suggest that lack of PBP1b-YcfM interaction not only reduces peptidoglycan synthesis, but may also induce collateral damage in the presence of cephems. It can be hypothesized that compounds interfering with PBP1b-YcfM interaction may inhibit PBP1b functions and produce secondary deleterious effects.
This study identified an antagonistic effect of surA deletion on FOX antimicrobial activity but no interaction between the gene deletion and CTX in the CMY-2-producing strain (Table 1). This observation suggests that the antimicrobial activity of FOX partially depends upon SurA. This protein is a periplasmic chaperone that assists outer membrane porin maturation and outer membrane biogenesis (27). Inactivation of surA results in fewer porins in the outer membrane and defects in this membrane barrier (28, 29). The observed antagonism between surA deletion and FOX may not be solely due to the reduced outer membrane porin-dependent diffusion of FOX into the periplasmic space, since the activities of other cephems were not affected in ΔsurA. Other, unknown factors, possibly related to SurA chaperone functions, could be responsible for the increased MIC of FOX in this gene deletion mutant.
In conclusion, this study identified two potential cephem helper drug target candidates, DapF and PBP1b. Cephem activity was potentiated by inactivation of the genes encoding these proteins, although full reversal of resistance was obtained only for selected drugs and was generally influenced by the strain's genetic background and the β-lactamase type, which may be regarded as undesirable features for a novel helper drug. The only exception was dapF inactivation, which synergistically potentiated FOX antimicrobial activity regardless of the strain's genetic background and β-lactamase type. The reduced fitness of the ΔdapF mutant can be regarded as a desirable feature, since drugs targeting DapF are likely to display their own antimicrobial activity, in addition to synergizing with cephems. Moreover, the high homology among Enterobacteriaceae and low similarity with human orthologues are also favorable features for the helper drug target potential of DapF. On the basis of these in vitro findings, more research is needed to assess the possible use of the protein as a target for the discovery of helper drugs that enhance FOX activity when used in combination.
MATERIALS AND METHODS
Antimicrobial agents and media.
All the antimicrobial agents and growth media used in this study were purchased from Sigma-Aldrich (Denmark) and Becton Dickinson (USA), respectively. Bacteria were routinely grown on Luria-Bertani agar (LA) and in Luria-Bertani broth (LB). Susceptibility testing and growth curve analysis were performed in cation-adjusted Mueller-Hinton Broth II (MHB II). SOC tryptone-based medium (30) was used to enhance the recovery of transformants after electroporation. PCR primers were synthesized by TAG (Copenhagen, Denmark).
Bacterial isolates and plasmids.
Sixteen Keio mutants (31) and the wild-type E. coli K-12 (BW25113) were purchased from the Coli Genetic Stock Center, Yale University (http://cgsc.biology.yale.edu/). These mutants were selected based on the findings of two previous studies using the Keio collection (12, 13). Our selection included (i) mutants that had reduced fitness (3-fold or greater) in the presence of different cephems, such as cefaclor, cefoxitin, cefsulodin, and ceftazidime (ΔmrcB, ΔdacA, ΔycfM, ΔybgF, ΔdapF, Δfis, ΔrplA, and ΔbamB) (13), and (ii) mutants that had increased susceptibility to 8 or more antimicrobial agents, including at least one cephem (ΔybgF, ΔdapF, ΔycjU, ΔrimK, ΔrplA, ΔdksA, ΔsurA, ΔtolC, ΔacrA, ΔrecA, and ΔyciM) (12). Donor strains carrying IncI1 plasmids encoding different β-lactamases (CMY-2 and CTX-M-1) were used for the conjugation experiments (32, 33) (Table 5). A representative of the epidemic E. coli ST131 clone harboring blaCTX-M-15 on an IncF plasmid (strain UR40) (34) was used for genetic engineering.
TABLE 5.
Bacterial strains and plasmids used in this study
| Strain | Plasmid | β-Lactamase enzyme | Origin | Relevant genotype or featuresa | Reference |
|---|---|---|---|---|---|
| BW25113 | Laboratory strain, K-12 derivative | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− rph-1 Δ(rhaDrhaB)568 hsdR514 | 31 | ||
| R7AC | IncI1 | CMY-2 | Dog feces | ST297 | 32 |
| UR40 | IncF | CTX-M-15 | Human urine | ST131 | 34 |
| 2007-10-2348-1 | IncI1 | CTX-M-1 | Cattle feces | ST49 | 33 |
| JW0145-1 | BW25113 ΔmrcB::Kan | 31 | |||
| JW0627-1 | BW25113 ΔdacA::Kan | 31 | |||
| JW5157-1 | BW25113 ΔycfM::Kan | 31 | |||
| JW0732-1 | BW25113 ΔybgF::Kan | 31 | |||
| JW5592-1 | BW25113 ΔdapF::Kan | 31 | |||
| JW1310-1 | BW25113 ΔycjU::Kan | 31 | |||
| JW0141-1 | BW25113 ΔdksA::Kan | 31 | |||
| JW2496-3 | BW25113 ΔbamB::Kan | 31 | |||
| JW0052-1 | BW25113 ΔsurA::Kan | 31 | |||
| JW5503-1 | BW25113 ΔtolC::Kan | 31 | |||
| JW0452-3 | BW25113 ΔacrA::Kan | 31 | |||
| JW2669-1 | BW25113 ΔrecA::Kan | 31 | |||
| JW0836-1 | BW25113 ΔrimK::Kan | 31 | |||
| JW3947-1 | BW25113 ΔrplA::Kan | 31 | |||
| JW1272-3 | BW25113 ΔyciM::Kan | 31 | |||
| JW3229-1 | BW25113 Δfis::Kan | 31 | |||
| pKD46-Ampr | Synthetic plasmid | λ-red genes (γ, β, and exo) and araC-ParaB; Ampr | 36 | ||
| pKD46-Tetr | Synthetic plasmid | pKD46-Ampr derivative; Tetr | This study | ||
| UR40 ΔdapF | IncF | CTX-M-15 | UR40 (ST131) dapF::Cam | This study | |
| UR40 ΔmrcB | IncF | CTX-M-15 | UR40 (ST131) mrcB::Cam | This study |
Amp, ampicillin; Kan, kanamycin; Cam, chloramphenicol.
Transfer of β-lactamase-encoding plasmids into Keio mutants.
The well-characterized IncI1 CMY-2-encoding plasmid from E. coli R7AC (32, 35) was transferred individually into each selected Keio mutant and the wild-type strain by conjugation and transformation, respectively. A second IncI1 plasmid encoding CTX-M-1 (Table 5) was transferred by conjugation into two selected mutants, ΔmrcB and ΔdapF. Briefly, 500 μl of overnight donor and recipient cultures was centrifuged, resuspended in 50 μl LB, and incubated at 37°C. After 90 min of mating, the suspension was streaked on a selective LA plate containing 25 μg/ml kanamycin (Kan) and 1 μg/ml CTX. Due to the lack of a marker for counterselection, the same plasmids could not be introduced into the Keio wild-type strain BW25113 by conjugation and thus were transformed by electroporation according to a standard protocol (30). The wild-type transformants obtained were used as a reference to assess the effects of each gene deletion in the Keio mutant transconjugants containing the same plasmid.
Targeted gene deletions in CTX-M-15-producing ST131.
Two genes, mrcB and dapF, were deleted individually in UR40 (ST131) using the λ-red recombination system (36). Briefly, a FLP recombination target (FRT)-flanked chloramphenicol resistance cassette was amplified from a template plasmid, pKD3, using primer pairs tagged with sequences containing 39 bases up- or downstream of the target gene. As the UR40 strain was resistant to ampicillin (Amp) and Kan, a tetracycline resistance (Tetr) marker was cloned into the original Amp resistance (Ampr) marker of the temperature-sensitive helper plasmid, pKD46, carrying λ-red recombinase genes γ, β, and exo under control of an arabinose inducible promoter, araC-ParaB. The Tetr marker was PCR amplified from pBR322 using primers containing 5′ XmnI digestion sites and digested with XmnI (Thermo, USA). In parallel, pKD46 was digested with XmnI and subsequently dephosphorylated with alkaline phosphatase (Thermo, USA). Finally, the two fractions were ligated using T4 DNA ligase (Thermo, USA), resulting in the construction of pKD46 with the Tetr resistance marker. The newly constructed pKD46-Tetr plasmid was transformed into the UR40 strain, and recombinase proteins were induced by adding arabinose to the culture. Subsequently, the induced culture was used to prepare electrocompetent cells following a standard protocol (30), and the amplified FRT-flanked resistance cassette was electroporated into the competent cells, followed by recovery in SOC medium and selection on LA plates supplemented with 24 μg/ml chloramphenicol. Finally, gene deletions were confirmed by PCR amplification and sequencing (Macrogen Inc.). Primer sequences are available upon request.
Antimicrobial susceptibility testing.
MICs of FOX, CTX, CAZ, CRO, and FEP were determined by broth microdilution using two different types of commercial 96-well plates (ESB1F and COMPAN1F; Trek Diagnostic Systems Inc.) according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (37). Each MIC value was reported as the average of at least two biological replicates. Complete reversal of antimicrobial resistance was defined as a reduction of the MIC to equal to or below the CLSI susceptibility breakpoint. Reduction of the MIC to the intermediate category was categorized as partial reversal of resistance. To enable quantitative assessment of antibiotic–gene-deletion interaction, the fractional inhibitory concentration index calculation (38) was modified to calculate the interaction index, defined as follows: antibiotic MICmutant/antibiotic MICwild type. A stringent interpretation of interaction index values of ≤0.25 and ≥4 was used to indicate synergistic and antagonistic interaction, respectively. Values from >0.5 to <4 were considered “no interaction,” similar to the standard designations of fractional inhibitory concentration index values (39). In addition, the synergistic and antagonistic designations also correspond to a 4-fold minimum MIC decrease or increase, respectively, consistent with interpretations in standard checkerboard assays (38).
Growth curve analyses.
Growth curve analyses were performed using BioScreen (Oy Growth Curves Ab Ltd., Finland). Broth microcultures of the mutants and the wild type in MHB II were distributed into a 100-well plate with or without 2 μg/ml FOX and incubated in a BioScreen chamber for 18 to 24 h at 37°C under continuous shaking. The FOX concentration, 4-fold below the MIC of the wild-type UR40 strain, was chosen based on the interaction index for synergy, 0.25. The optical density at 600 nm (OD600) of each microculture was measured and recorded every 15 min, after a standstill of 5 s in automatic mode. The recorded ODs were transferred to an Excel file and subsequently used to plot growth curves.
In silico study of protein homology.
The amino acid sequences of E. coli MrcB and DapF were downloaded in FASTA format from Uniprot Web resources (http://www.uniprot.org) (uniprot entries P02919 [MrcB] and P0A6K1 [DapF]) and were aligned with those of other Enterobacteriaceae (taxonomy identification number [taxid], 91347) and human (taxid, 9606) homologues using protein BLAST on the website of the National Center for Biotechnology Information (NCBI). Except for expansion of the maximum target sequences, default BLAST parameters were used, with an expected cutoff value of 10. The accession numbers used for the protein sequences were WP_016529988.1 (MrcB) and ACI11814.1 (DapF) for K. pneumoniae, WP_000918132.1 (MrcB) and WP_001160672.1 (DapF) for S. enterica, EIQ27511.1 (MrcB) and EIQ17403.1 (DapF) for S. flexneri, and CAA90232.1 (MrcB) and WP_038634512.1 (DapF) for C. freundii.
ACKNOWLEDGMENTS
We acknowledge Heidi Gumpert for assistance with bioinformatics tools.
The study was supported by the University of Copenhagen Research Centre for Control of Antibiotic Resistance (UC-Care) and by a grant from Zoetis.
REFERENCES
- 1.Bassetti M, Ginocchio F, Mikulska M. 2011. New treatment options against Gram-negative organisms. Crit Care 15:215. doi: 10.1186/cc9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. 2011. Carbapenems: past, present, and future. Antimicrob Agents Chemother 55:4943–4960. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giske CG, Monnet DL, Cars O, Carmeli Y. 2008. Clinical and economic impact of common multidrug-resistant Gram-negative bacilli. Antimicrob Agents Chemother 52:813–821. doi: 10.1128/AAC.01169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drusano GL, Louie A, MacGowan A, Hope W. 2015. Suppression of emergence of resistance in pathogenic bacteria: keeping our powder dry, part 1. Antimicrob Agents Chemother 60:1183–1193. doi: 10.1128/AAC.02177-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalan L, Wright GD. 2011. Antibiotic adjuvants: multicomponent anti-infective strategies. Expert Rev Mol Med 13:e5. doi: 10.1017/S1462399410001766. [DOI] [PubMed] [Google Scholar]
- 6.Drawz SM, Papp-Wallace KM, Bonomo RA. 2014. New β-lactamase inhibitors: a therapeutic renaissance in an MDR world. Antimicrob Agents Chemother 58:1835–1846. doi: 10.1128/AAC.00826-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zasowski EJ, Rybak JM, Rybak MJ. 2015. The β-lactams strike back: ceftazidime-avibactam. Pharmacother J Hum Pharmacol Drug Ther 35:755–770. doi: 10.1002/phar.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kourtesi C, Ball AR, Huang Y-Y, Jachak SM, Vera DM, Khondkar P, Gibbons S, Hamblin MR, Tegos GP. 2013. Microbial efflux systems and inhibitors: approaches to drug discovery and the challenge of clinical implementation. Open Microbiol J 7:34–52. doi: 10.2174/1874285801307010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright GD. 2016. Antibiotic adjuvants: rescuing antibiotics from resistance. Trends Microbiol 24:862–871. doi: 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Olivares J, Bernardini A, Garcia-Leon G, Corona F, Sanchez MB, Martinez JL. 2013. The intrinsic resistome of bacterial pathogens. Front Microbiol 4:103. doi: 10.3389/fmicb.2013.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernal P, Molina-Santiago C, Daddaoua A, Llamas MA. 2013. Antibiotic adjuvants: identification and clinical use. Microb Biotechnol 6:445–449. doi: 10.1111/1751-7915.12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu A, Tran L, Becket E, Lee K, Chinn L, Park E, Tran K, Miller JH. 2010. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic bar code. Antimicrob Agents Chemother 54:1393–1403. doi: 10.1128/AAC.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, Shales M, Lovett S, Winkler ME, Krogan NJ, Typas A, Gross CA. 2011. Phenotypic landscape of a bacterial cell. Cell 144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mengin-Lecreulx D, Michaud C, Richaud C, Blanot D, Van Heijenoort J. 1988. Incorporation of LL-diaminopimelic acid into peptidoglycan of Escherichia coli mutants lacking diamino pimelate epimerase encoded by dapF. J Bacteriol 170:2031–2039. doi: 10.1128/jb.170.5.2031-2039.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutton CA, Perugini MA, Gerrard JA. 2007. Inhibition of lysine biosynthesis: an evolving antibiotic strategy. Mol Biosyst 3:458–465. doi: 10.1039/b705624a. [DOI] [PubMed] [Google Scholar]
- 16.Hor L, Dobson RCJ, Downton MT, Wagner J, Hutton CA, Perugini MA. 2013. Dimerization of bacterial diaminopimelate epimerase is essential for catalysis. J Biol Chem 288:9238–9248. doi: 10.1074/jbc.M113.450148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pepper ED, Farrell MJ, Finkel SE. 2006. Role of penicillin-binding protein 1b in competitive stationary-phase survival of Escherichia coli. FEMS Microbiol Lett 263:61–67. doi: 10.1111/j.1574-6968.2006.00418.x. [DOI] [PubMed] [Google Scholar]
- 18.Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, Wain J, Parkhill J, Turner AK. 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res 19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar A, Sarkar SK, Ghosh D, Ghosh AS. 2012. Deletion of penicillin-binding protein 1b impairs biofilm formation and motility in Escherichia coli. Res Microbiol 163:254–257. doi: 10.1016/j.resmic.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Typas A, Banzhaf M, Van Den Berg Van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, Von Rechenberg M, Breukink E, Den Blaauwen T, Gross CA, Vollmer W. 2010. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray AN, Egan AJF, van't Veer IL, Verheul J, Colavin A, Koumoutsi A, Biboy J, Altelaar MAF, Damen MJ, Huang KC, Simorre JP, Breukink E, den Blaauwen T, Typas A, Gross CA, Vollmer W. 2015. Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. eLife 4:1–29. doi: 10.7554/eLife.07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Typas A, Banzhaf M, Gross CA, Vollmer W. 2011. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yousif SY, Broome-Smith JK, Spratt BG. 1985. Lysis of Escherichia coli by beta-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J Gen Microbiol 131:2839–2845. [DOI] [PubMed] [Google Scholar]
- 24.Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. 2010. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato J, Suzuki H, Hirota Y. 1985. Dispensability of either penicillin-binding protein-1a or -1b involved in the essential process for cell elongation in Escherichia coli. Mol Gen Genet 200:272–277. doi: 10.1007/BF00425435. [DOI] [PubMed] [Google Scholar]
- 26.Egan AJF, Jean NL, Koumoutsi A, Bougault CM, Biboy J, Sassine J, Solovyova AS, Breukink E, Typas A, Vollmer W, Simorre J-P. 2014. Outer-membrane lipoprotein LpoB spans the periplasm to stimulate the peptidoglycan synthase PBP1B. Proc Natl Acad Sci U S A 111:8197–8202. doi: 10.1073/pnas.1400376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong M, Ferrell B, Lu W, Chai Q, Wei Y. 2013. Insights into the function and structural flexibility of the periplasmic molecular chaperone surA. J Bacteriol 195:1061–1067. doi: 10.1128/JB.01143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricci DP, Schwalm J, Gonzales-Cope M, Silhavy TJ. 2013. The activity and specificity of the outer membrane protein chaperone SurA are modulated by a proline isomerase domain. mBio 4:e00540-13. doi: 10.1128/mBio.00540-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chai Q, Ferrell B, Zhong M, Zhang X, Ye C, Wei Y. 2014. Diverse sequences are functional at the C-terminus of the E. coli periplasmic chaperone SurA. Protein Eng Des Sel 27:111–116. doi: 10.1093/protein/gzu003. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Russel DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 31.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Damborg P, Gaustad IB, Olsen JE, Guardabassi L. 2011. Selection of CMY-2 producing Escherichia coli in the faecal flora of dogs treated with cephalexin. Vet Microbiol 151:404–408. doi: 10.1016/j.vetmic.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 33.Jakobsen L, Bortolaia V, Bielak E, Moodley A, Olsen SS, Hansen DS, Frimodt-Møller N, Guardabassi L, Hasman H. 2015. Limited similarity between plasmids encoding CTX-M-1 β-lactamase in Escherichia coli from humans, pigs, cattle, organic poultry layers and horses in Denmark. J Glob Antimicrob Resist 3:132–136. doi: 10.1016/j.jgar.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 34.Cerquetti M, Giufrè M, García-Fernández A, Accogli M, Fortini D, Luzzi I, Carattoli A. 2010. Ciprofloxacin-resistant, CTX-M-15-producing Escherichia coli ST131 clone in extraintestinal infections in Italy. Clin Microbiol Infect 16:1555–1558. doi: 10.1111/j.1469-0691.2010.03162.x. [DOI] [PubMed] [Google Scholar]
- 35.Hansen KH, Bortolaia V, Nielsen CA, Nielsen JB, Schønning K, Agersø Y, Guardabassi L. 2016. Host-specific patterns of genetic diversity among IncI1-Iγ and IncK plasmids encoding CMY-2 β-lactamase in Escherichia coli isolates from humans, poultry meat, poultry and dogs in Denmark. Appl Environ Microbiol 82:4705–4714. doi: 10.1128/AEM.00495-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.CLSI. 2013. Performance standards for antimicrobial testing. M100-S23. CLSI, Wayne, PA. [Google Scholar]
- 38.Johnson MD, MacDougall C, Ostrosky-Zeichner L, Perfect JR, Rex JH. 2004. Combination antifungal therapy. Antimicrob Agents Chemother 48:693–715. doi: 10.1128/AAC.48.3.693-715.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Odds FC. 2003. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother 52:1. doi: 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]