Abstract
Background
More and more evidence indicates that microRNAs are present and involved in many tumor-related diseases. The function of microRNA-622 (miR-622) in colorectal cancer (CRC) remains controversial. Dual specificity tyrosine phosphorylation-regulated kinase 2 (DYRK2) has been reported as a tumor suppressor gene in different cancers. The detailed regulation mechanism of DYRK2 in CRC remains unclear.
Methods
miR-622 and DYRK2 expression levels were detected at tissue and cellular level respectively by using real time polymerase chain reaction (PCR), Western blot, and immunohistochemical staining. Pearson’s correlation analysis was used to evaluate the correlation between miR-622 and DYRK2. Transwell assay was applied to measure the effect of miR-622 on migration and invasion of SW1116 and SW480. We used dual luciferase reporter assay to confirm the targeted binding effect of miR-622 and DYRK2 3′-untranslated region (3′UTR). An antisense experiment was executed to further confirm the role miR-622 had played with regard to migration and invasion by targeting regulation of DYRK2 pathway in CRC cells.
Results
In our research, we found that the expression of miR-622 was elevated in CRC tissues and cell lines compared to that of nonCRC tissues and the normal human colon epithelial cell line NCM460. Correspondingly, the expression of DYRK2 in CRC tissues and cell lines showed a contrary tendency. The different expression level of DYRK2 was closely correlated with clinicopathological characteristics of CRC patients. We demonstrated that down-regulation of miR-622 could inhibit the ability of migration and invasion of CRC cell lines SW1116 and SW480. Also, we confirmed that DYRK2 was negatively regulated by miR-622 via a specific targeted binding site within the 3′UTR. We finally verified that the migration and invasion ability of CRC cells in the conducted DYRK2 3′UTR defect plasmid transfection group were lower compared to miR-622 and cotransfection group.
Conclusion
The findings of this study indicate that a decrease of miR-622 expression could suppress migration and invasion by targeting regulation of DYRK2 and miR-622/DYRK2 could be a potential molecular treating target of CRC.
Keywords: miR-622, DYRK2, migration, invasion, colorectal cancer
Introduction
Colorectal cancer (CRC) is the most common malignant tumor associated with the digestive system and is regarded as one of the main causes of cancer-related deaths worldwide.1,2 Continuous accumulation of genetic and epigenetic events may cause the occurrence and development of CRC. Though a great deal of progress has been made, including surgery and combined radio and chemotherapy, CRC remains a problematic disease, with invariable manifestations of tumor recurrence, and the 5-year survival rate for colon cancer is not high.3 Besides local recurrence, another reason for unfavorable prognosis of CRC is distant metastasis. Therefore, it is urgent to seek metastasis-related molecules and identify its regulatory mechanism in CRC. Recently, microRNAs (miRNAs) have become the primary starting point in both trials and associated therapies at the molecular level to treat cancer-related diseases.4,5
miRNAs are a group of evolutionarily conserved small non-coding RNAs. miRNAs were certified as an oncogene or cancer suppressor gene in multiple cancers via regulating its target genes by interfering with transcription or inhibiting translation.6 Ghanbari et al reported that plasma miRNA-142-3p and miRNA-26a-5p were down-regulated and could be a novel biomarker for the diagnosis of CRC.7 miRNA-622 lies in the human genomic region of chromosome 13q31.3 and was reported as a tumor suppressor gene in hepatocellular carcinoma, gastric cancer, and glioma cells, but the function of miR-622 in CRC remains controversial.8–11 Fang et al reported miR-622 was down-regulated in colorectal tumor tissues and cell lines, and works as a tumor suppressor in CRC metastasis.12 Through an miRNA profiles study, Balaguer et al revealed a marked over-expression of miR-622 in the CRC samples compared to the normal colorectal mucosa; a similar result was found in the recent study of Ma et al.13,14 Up to now, whether miR-622 could affect metastasis and its specific regulatory mechanism in CRC remains unclear.
Like most malignant tumors, abnormal cell division caused by dysregulation of the cell cycle may lead to the development of CRC. As one of the most important cell cycle-related proteins, dual specificity tyrosine phosphorylation-regulated kinase 2 (DYRK2) and its downstream Snail, which is initially phosphorylated by DYRK2, plays a key role during cell division. For example, DYRK2 can regulate G1/S transition as a priming kinase for GSK3 and promote G2/M transition with an EDVP E3 ligase complex. Another function of DYRK2 is involved with cancer metastasis.15 Yan et al reported that DYRK2 could inhibit cell invasion and migration in CRC cell lines.16 Enomoto et al showed DYRK2 was closely correlated with lymph node involvement and its recurrence in breast cancers.17
In this study, we verified that miR-622 was highly expressed in CRC tissues and two CRC cell lines SW1116/SW480. We confirmed that miR-622 works as a tumor initiator during migration and invasion of CRC cells. Furthermore, we confirmed that miR-622 could negatively target DYRK2 and promote the aggressive phenotypes in CRC cells.
Materials and methods
Patients and tissue samples
One hundred and sixty-nine samples of tumor tissues and adjacent tissues were obtained from patients undergoing tumor resections in the Central Hospital affiliated to Shenyang Medical College. All patients had a clear histologic diagnosis of CRC based on the American Joint Committee on Cancer. All cases were diagnosed with CRC and treated between January 2014 and January 2016 at Central Hospital affiliated to Shenyang Medical College. Written informed consent was provided by the patients whose tissues were used in this research. The Institute Research Medical Ethics Committee of Central Hospital affiliated to Shenyang Medical College granted approval for this study.
Cell culture
Human colon cancer cell lines HT29, SW1116, SW480 and normal human colon epithelial cell line NCM460 were purchased from Institute of Biochemistry and Cell Biology (Chinese Academy of Sciences, Shanghai, People’s Republic of China) and maintained in a Roswell Park Memorial Institute 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% (v/v) fetal bovine serum (FBS; Sigma-Aldrich Co, St Louis, MO, USA), 100 IU/mL penicillin and 100 mg/mL streptomycin (Shanghai Baoman Biotechnology Co., Ltd., People’s Republic of China), at 37°C in a humidified atmosphere containing 5% CO2.
Immunohistochemical staining of DYRK2 in CRC sections
All the procedures were carried out as described.18 In brief, all CRC tissues were treated using the following procedures: 4% paraformaldehyde fixation, paraffin-embedding, section (4 μm thickness), deparaffinization, rehydration, hydrogen peroxide incubation, antigen retrieval, 10% goat serum (BioWorld, Visalia, CA, USA) blocking, rabbit anti-DYRK2 antibody incubation (1:1,000; Cell Signaling Technology, Danvers, MA, USA) 4°C overnight, biotinylated anti-rabbit secondary antibody incubation (1:100; BioWorld, 37°C 20 minutes), streptavidin-horseradish peroxidase complex incubation, diaminobenzidine tetrahydrochloride (DAB; MedChemExpress, Monmouth Junction, NJ, USA) stain, hematoxylin (Amresco, Solon, OH, USA) counterstain. All sections were independently assessed by two experienced pathologists who were ignorant of patients’ clinical pathology and other information. DYRK2 expression level was evaluated as described elsewhere.16 Score for the percent of positive cells was set as: ≤10%=0; >10% to ≤25%=1; >25% to ≤50%=2; >50% to ≤75%=3; and >75%=4. Another score for the intensity of cytoplasm staining was set as negative =0, weak =1, moderate =2, or strong =3.
Reverse transcription and quantitative real time polymerase chain reaction
Total RNA was extracted by TRIzol (Thermo Fisher Scientific) according to the manufacturer’s instructions.19–21 For CRC tissues detection, total RNAs were extracted from 20 randomly selected cases of metastatic (m)CRC, 20 cases of non-metastatic (nm)CRC and 40 nonCRC. The expression of miR-622 was detected with Stem-Loop RT-PCR assay as reported. The U6 small nuclear RNA and GAPDH mRNA were used as internal references. Primer sequences were synthesized as follows:
DYRK2 primer: forward, 5′-CCTGAACAAGCAATGAAGCA-3′; reverse, 5′-GGTCATCATCATAG CCACCA-3′
Snail primer: forward, 5′-GTATCCAGAGCTGTTTGGA-3′; reverse, 5′-AACATTTTCCTCCCAGGCC-3′
GAPDH primer: forward, 5′-CTCCTCCACCTT TGACGCTG-3′; reverse, 5′-TCCTCTTGTGCTCTT GCTGG-3′.
RT-PCR for miR-622 was performed with real time PCR Universal Reagent (GenePharma, Shanghai, People’s Republic of China). All the reactions were performed three times.
Oligonucleotide transfection
SW1116 and SW480 cells were seeded in 6-well plates with a density of 1×105/L cells per well, for further incubation. Twenty-four hours later, 50 nmol of the miR-622 mimics or mimic-negative control, miR-622 inhibitor or inhibitor-negative control were transfected into the SW1116 and SW480 cells with Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. The antisense oligonucleotides used in these studies were the miR-622 inhibitor, 5′-GCUCCAACCUCAGCAGACUGU-3′ and the miRNA inhibitor-negative control (inhibitor control), 5′-UCACAACCUCCUAGAAAGAGUAGA-3′. The mimics used in these studies were the miR-622 mimic: 5′-ACAGUCUGCUGAGGUUGGAGC-3′ a n d 5 ′-UCCAACCUCAGCAGACUGUUU-3 ′ and the miRNA mimic-negative control (mimic control): 5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′. All miRNA oligonucleotides were purchased from GenePharma.
Western blot analysis
Total cellular and tissue protein were extracted by radioimmunoprecipitation lysis buffer (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Samples were subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene difluoride (PVDF) membrane. Target proteins were probed with specific antibodies of DYRK2, Snail, p-Snail and GAPDH. The details of the antibodies mentioned are the following: rabbit anti-DYRK2 antibody (1:1,000; Cell Signaling Technology); rabbit anti-snail antibody (1:1,000; Cell Signaling Technology); rabbit anti-p-snail antibody (1:500; Novus, USA); rabbit anti- GAPDH antibody (0.5 μg/mL; Abcam, Cambridge, UK).
Dual luciferase reporter assay
The DYRK2 3′ untranslated region (3′UTR) full length was PCR amplified and cloned at the SacI and XhoI sites into pmirGLO vector (Promega Corporation, Fitchburg, WI, USA). The mutant construct of DYRK2 3′UTR was generated by a Quick Change mutagenesis kit (Life Sciences-Germany, Bremen, Germany). SW1116 and SW480 cells were seeded on a 24-well plate and cotransfected with reporter vectors and miR-622 mimics or negative control using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. After 36 hours, luciferase activity was measured by a dual luciferase reporter assay system according to the manufacturer’s instructions (Promega Corporation).
Transwell assay
Transwell assays were used to evaluate CRC cell aggressive phenotypes. As described before, SW1116 and SW480 cells were seeded onto the upper part of a Transwell chamber (BD Biosciences, San Jose, CA, USA) containing a gelatin-coated polycarbonate membrane filter (pore size: 8 mm), for the migration and invasion assays respectively.22 For invasion assays, the chambers were preconditioned using Matrigel (1:8, BD Biosciences). Medium without serum was supplemented into the upper wells, and medium containing 20% FBS was supplemented into the lower wells. After 24 hours incubation at 37°C with 5% CO2, the non-migrating or non-invading cells were wiped out carefully. Then the filters were fixed in 90% alcohol and stained with crystal violet (Amresco). Five random fields were counted per chamber by using an inverted microscope (Olympus, Tokyo, Japan), and each experiment was repeated three times.
Statistical analysis
All data were analyzed with SPSS17.0. All experiments were repeated three times and all data from three independent experiments were expressed with a mean ± standard deviation (SD). The relationship between miR-622 and DYRK2 expressions was tested with a two-tailed Pearson’s correlation analysis. Correlation between DYRK2 expression and clinicopathological features was analyzed by using the Pearson’s chi-squared test. Difference of miR-622 and DYRK2 expression in different groups of tissues and cell lines was analyzed by the Wilcoxon signed rank test. A two-sided P-value of less than 0.05 was considered to be statistically significant.
Results
Elevated expression of miR-622 but depressed DYRK2 in CRC tissue and cell lines
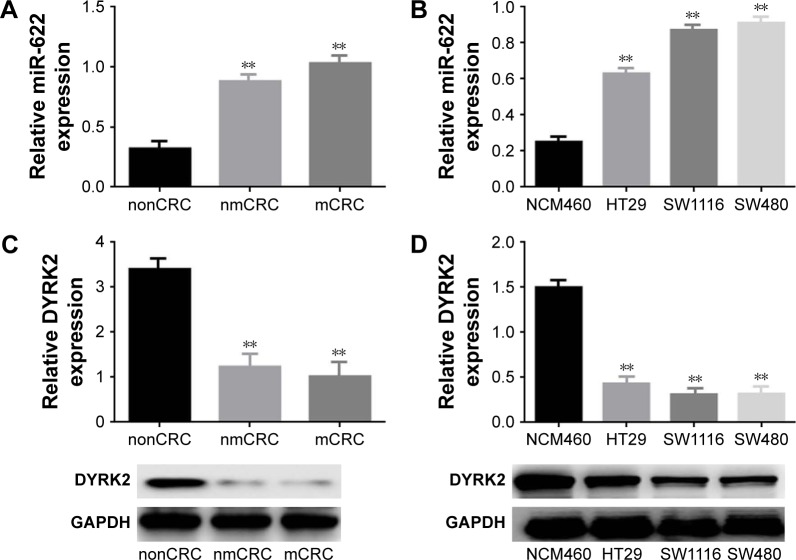
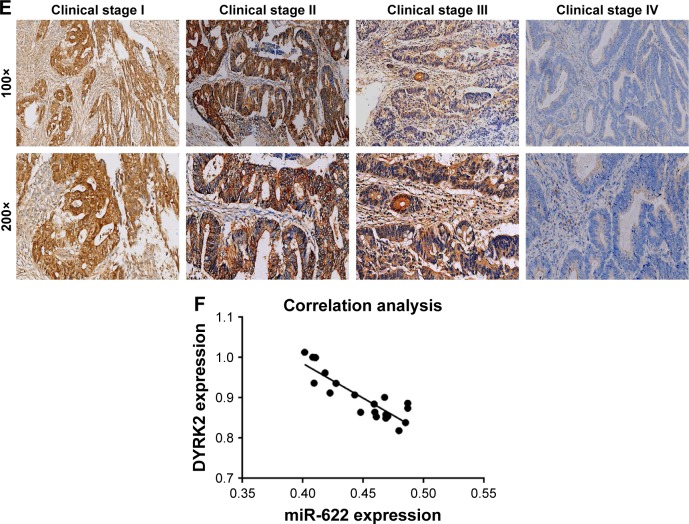
Firstly, we detected the expression of miR-622 in 20 cases of mCRC tissues, 20 cases of nmCRC tissues, and matched non-colorectal cancer (nonCRC) tissues by RT-PCR (all cases were randomly selected from the 169 patients). As shown in Figure 1A, the relative expression of miR-622 mRNA was obviously elevated in CRC tissues (mCRC and nmCRC) compared to nonCRC (miR-622 mRNA was 0.321±0.012/mCRC and 0.882±0.034/nmCRC vs 1.032±0.104/nonCRC, P<0.01). Secondly, we detected the expression of miR-622 in three CRC cell lines, HT29, SW1116 and SW480, compared to normal human colon epithelial cell line NCM460. The results of RT-PCR showed the same tendency as the tissue level (miR-622 mRNA was 0.253±0.014/NCM460 vs 0.632±0.052/HT29, 0.872±0.032/SW1116 and 0.914±0.093/SW480, P<0.01) (Figure 1B). Thirdly, we measured the expression of DYRK2 both at tissue and cell level by RT-PCR and Western blot. The following test showed obvious depression of DYRK2 in CRC tissues and cell lines compared to non-cancerous tissues and normal human colon epithelial cell line NCM460 (DYRK2 mRNA was 3.412±0.122/nonCRC vs 1.232±0.226/nmCRC and 1.021±0.114/mCRC, P<0.01; DYRK2 mRNA was 1.524±0.132/NCM460 vs 0.433±0.023/HT29, 0.315±0.037/SW1116 and 0.324±0.042/SW480, P<0.01) (Figure 1C and D). Furthermore, the results of immunohistology (IHC) staining showed the expression of DYRK2 was gradually decreased in different clinical stages of CRC (Figure 1E). A further correlation analysis confirmed that DYRK2 expression was closely correlated with clinicopathological features, especially with clinical stage (P=0.026), nodal (N) classification (P=0.026) and metastasis (M) classification (P=0.011) (determined by using IHC staining) (Table 1). Finally, the expression of miR-622 and DYRK2 mRNA in the same CRC specimens was detected by RT-PCR and a further correlation analysis was executed. As shown in Figure 1F, we found a significant inverse correlation between miR-622 and DYRK2 (two-tailed Pearson’s correlation analysis, r=0.7703, P<0.01). In short, our findings showed elevated miR-622 but depressed DYRK2 expression in CRC tissues and cell lines.
Figure 1.
Elevated expression of miR-622 but depressed DYRK2 in colorectal cancer tissue and cell lines. (A) miR-622 expression in 20 cases of CRC tissues with metastasis (mCRC) and 20 cases of CRC tissues without metastasis (nmCRC) was higher than in non-cancerous tissues (nonCRC) as measured by using real time PCR. **P<0.01 vs non-cancerous tissues. (B) The expression of miR-622 was increased in three CRC cell lines (HT29, SW1116, and SW480) compared to normal human colon epithelial cell line NCM460, detected by using real time PCR. **P<0.01 vs NCM460. (C, D) Depressed expression of DYRK2 was shown in CRC tissues (mCRC and nmCRC) compared to non-cancerous tissues (nonCRC) (**P<0.01 vs non-cancerous tissues) and in three CRC cell lines (HT29, SW1116, and SW480) compared to normal human colon epithelial cell line NCM460 (**P<0.01 vs NCM460), detected by using real time PCR and Western blot. All the data are shown as mean ± standard deviation. (E) A gradual decline of DYRK2 expression at four different clinical stages was shown by IHC staining, at two magnifications (100× and 200×). (F) Analysis of correlation between the expression of miR-622 and DYRK2 in CRC tissues (two tailed Pearson’s correlation analysis, r=0.7703, **P<0.01) by detecting the mRNA expression.
Abbreviations: miR-622, microRNA-622; DYRK2, dual specificity tyrosine phosphorylation-regulated kinase 2; CRC, colorectal cancer; mCRC, metastatic colorectal cancer; nonCRC, non-colorectal cancer; PCR, polymerase chain reaction; nmCRC, non-metastatic colorectal cancer; IHC, immunohistology.
Table 1.
Correlation of DYRK2 expression and clinicopathological features in CRC
| Features | Number of cases | DYRK2
|
P-value | |
|---|---|---|---|---|
| High | Low | |||
| Age at diagnosis (years) | 0.920 | |||
| ≤50 | 57 | 26 | 31 | |
| >50 | 112 | 52 | 60 | |
| Gender | 0.786 | |||
| Female | 108 | 49 | 59 | |
| Male | 60 | 29 | 31 | |
| Location | 0.814 | |||
| Colon | 82 | 39 | 43 | |
| Rectal | 87 | 39 | 48 | |
| Clinical stage | 0.026 | |||
| I | 21 | 14 | 7 | |
| II | 26 | 16 | 10 | |
| III | 50 | 21 | 29 | |
| IV | 72 | 27 | 45 | |
| T classification | 0.486 | |||
| T1 + T2 | 32 | 13 | 19 | |
| T3 + T4 | 137 | 65 | 72 | |
| N classification | 0.026 | |||
| No | 48 | 29 | 19 | |
| Yes | 121 | 49 | 72 | |
| M classification | 0.011 | |||
| M0 | 92 | 50 | 42 | |
| M1 | 77 | 28 | 49 | |
| Pathologic differentiation | 0.535 | |||
| Poor | 30 | 16 | 14 | |
| Moderate | 125 | 57 | 68 | |
| Well | 14 | 5 | 9 | |
Notes: Correlation between DYRK2 expression and clinicopathological characteristics of CRC patients. Through the chi-square test we found DYRK2 was closely correlated with clinical stage (P=0.026), nodal (N) classification (P=0.026) and metastasis (M) classification (P=0.011) (determined by IHC staining).
Abbreviations: CRC, colorectal cancer; DYRK2, dual specificity tyrosine phosphorylation-regulated kinase 2; IHC, immunohistology.
Down-regulation of miR-622 inhibited migration and invasion of CRC cells
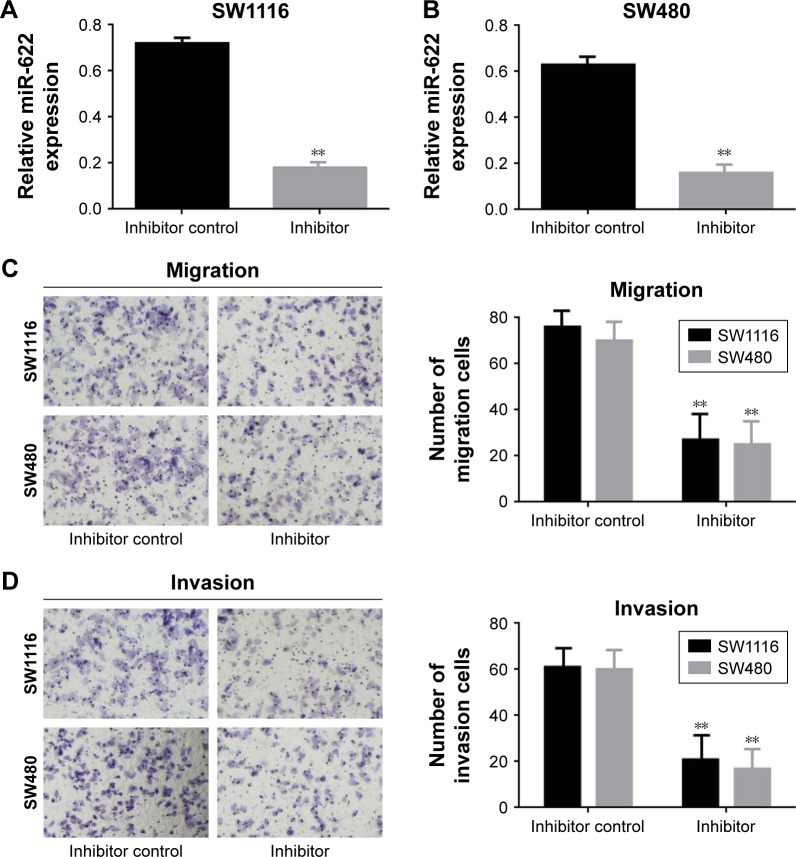
In the first part of the Results section we detected the expression of miR-622 in CRC tissues and cell lines, then we tried to clarify the potential function of miR-622 and how it affects migration and invasion of CRC cell lines. We transfected the miR-622 inhibitor or inhibitor control into CRC cell lines SW1116 and SW480 respectively; we then observed the changes of their migration and invasion ability through Transwell assay. As we predicted, the expression of miR-622 in CRC cells had declined by transfection of miR-622 inhibitor as compared to inhibitor control groups (miR-622 mRNA was 0.722±0.052/inhibitor control vs 0.181±0.023/inhibitor in SW1116 and 0.633±0.047/inhibitor control vs 0.163±0.031/inhibitor in SW480, P<0.01) (Figure 2A and B). We then detected the changes of migration and invasion ability in miR-622 intervened CRC cells. After 48 hours for transfection, Transwell assays were executed and we found that compared to the inhibitor control groups the number of migratory and invading cells in the miR-622 inhibitor group were remarkably decreased (migration cells were 76.523±3.823/inhibitor control vs 27.621±3.125/inhibitor in SW1116 and 71.576±3.612/inhibitor control vs 23.942±3.132/inhibitor in SW480, P<0.01; invasion cells were 61.231±2.943/inhibitor control vs 20.154±2.452/inhibitor in SW1116 and 60.633±3.126/inhibitor control vs 19.637±2.531/inhibitor in SW480, P<0.01) (Figure 2C and D). These findings revealed that miR-622 works as a tumor initiator in the aggressive phenotypes of CRC cells.
Figure 2.
Down-regulation of miR-622 could inhibit migration and invasion of CRC cells. (A, B) miR-622 mRNA was remarkably decreased by transfection of miR-622 inhibitor compared to inhibitor control in SW1116 and SW480 cells detected by real time PCR. (C, D) Down-regulation of miR-622 could inhibit the migration and invasion of SW1116 and SW480 cells. Data are presented as mean ± standard deviation. Two tailed Student’s t-test was used to analyze the significant differences, **P<0.01. Scale bars 20 μm.
Abbreviations: miR-622, microRNA-622; DYRK2, dual specificity tyrosine phosphorylation-regulated kinase 2; CRC, colorectal cancer; PCR, polymerase chain reaction.
miR-622 targeting regulated DYRK2 in CRC cells
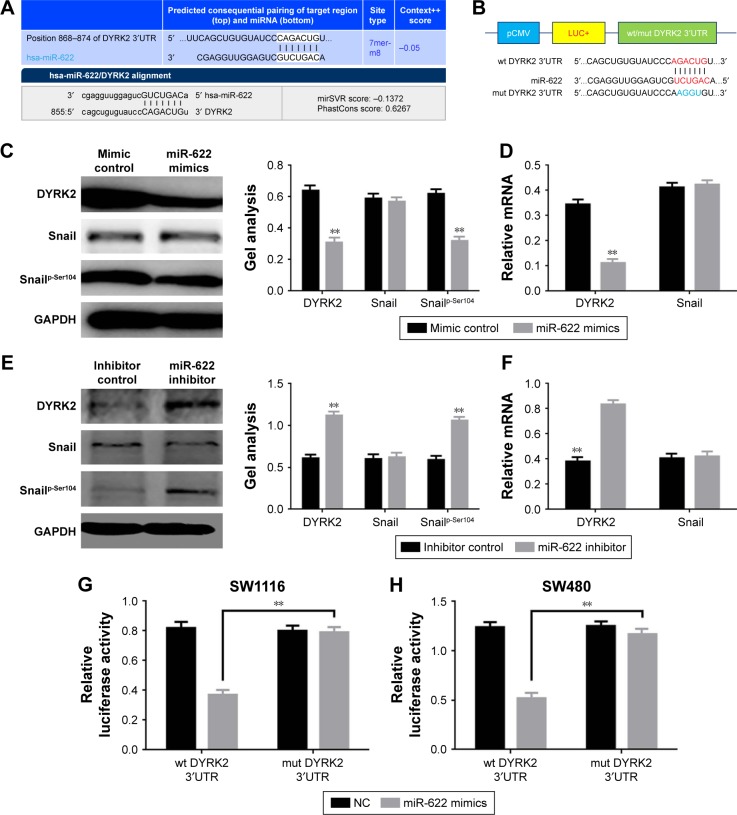
The above results showed miR-622 could affect the aggressive phenotypes of CRC cells. Combined with the detection of DYRK2 at tissue and cell level, we wondered whether the regulation of miR-622 was implemented through a DYRK2 pathway in CRC cells.23,24 miRNAs are known to be able to regulate hundreds of mRNA targets, resulting in changes to cellular phenotypes, so we first tried to explore the potential target of miR-622 by predictive software. As we expected, DYRK2 3′UTR has a theoretical binding site of miR-622 by prediction of miRanda, miRDB, and targetscan (Figure 3A). Secondly, we confirmed that up- and down-regulation of miR-622 expression could apparently affect the expression of DYRK2 and the phosphorylation level of Snail (snailp-Ser104), which is the downstream of DYRK2 in CRC cells, through RT-PCR and Western blot (Figure 3C–F). Thirdly, we conducted the reporter plasmids containing wild type and mutant DYRK2 3′UTR (Figure 3B). And finally, a further luciferase reporter assay proved miR-622 could directly target DYRK2. As shown in Figure 3G, when we cotransfected the reporter vectors containing wild type DYRK2 3′UTR (pmirGLO-DYRK2-3′UTR-wt) and miR-622 mimics into SW1116 cells, the activity of luciferase was significantly decreased. To the contrary, when we co-transfected the reporter vectors containing mutant DYRK2 3′UTR (pmirGLO-DYRK2-3′UTR-mut) and miR-622 mimics, the activity of luciferase was not changed. The same results were also found in SW480 cells (Figure 3H).
Figure 3.
miR-622 targeting regulated DYRK2 in CRC cells. (A) DYRK2 was a target gene of miR-622 predicted by miRanda and targetscan. (B) Diagram of the luciferase reporter plasmids with the wild-type or mutant DYRK2 3′UTR. (C, D) The expression of DYRK2 and the activity of its downstream snailp-Ser104 was obviously decreased when transfection of miR-622 mimics, elucidated by Western blot and real time PCR. (E, F) When transfection of miR-622 inhibitor, DYRK2 and snailp-Ser104 was increased. (G, H) The relative luciferase activity in SW1116 and SW480 cells was determined by fluorescence changes after co-transfection of miR-622 mimics and the plasmid with wild-type or mutant DYRK2 3′UTR. Three independent experiments were performed in duplicate. All the data mentioned are shown as mean ± standard deviation. **P<0.01 vs control group.
Abbreviations: miR-622, microRNA-622; DYRK2, dual specificity tyrosine phosphorylation-regulated kinase 2; 3′UTR, 3′-untranslated region; CRC, colorectal cancer; PCR, polymerase chain reaction; mut, mutant; wt, wild-type; NC, negative control.
DYRK2 suppressed the activity of miR-622 on migration and invasion of CRC cells
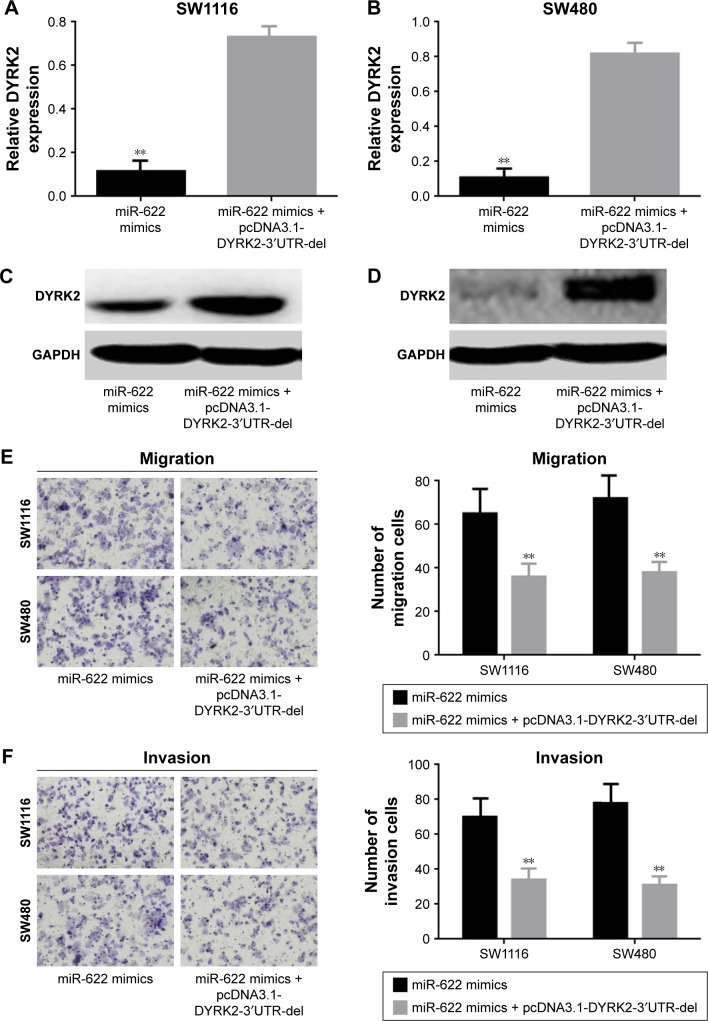
In the Results section, we found miR-622 could target the regulation of DYRK2 and work as an initiator in migration and invasion of CRC cells. To further confirm the role miR-622 had played with regard to migration and invasion by targeting regulation of DYRK2 pathway in CRC cells, we designed an antisense experiment by cotransfection of miR-622 mimics and then conducted DYRK2 3′UTR defect plasmid (pcDNA3.1-DYRK2-3′UTR-del). After 48 hours for transfection, Western blot and Transwell assay were executed again to detect the expression of DYRK2 and the ability of migration and invasion in CRC cells. Our results showed that the expression of DYRK2 in cotransfection of miR-622 mimics and the conducted DYRK2 3′UTR defect plasmid (pcDNA3.1-DYRK2-3′UTR-del) groups was higher than in miR-622 mimics group (DYRK2 mRNA was 0.116±0.011/mimics vs 0.732±0.081/cotransfection in SW1116 and 0.108±0.012/mimics vs 0.818±0.084/cotransfection in SW480, P<0.01) (Figure 4A–D). Correspondingly, the migration and invasion ability of CRC cells in the cotransfection group were also depressed (migration cells were 65.432±2.324/mimics vs 36.623±3.123/co-transfection in SW1116 and 72.593±3.241/mimics vs 38.645±2.747/cotransfection in SW480, P<0.01; invasion cells were 70.732±3.382/mimics vs 34.308±2.321/co-transfection in SW1116 and 78.386±3.917/mimics vs 31.326±2.834/cotransfection in SW480, P<0.01) (Figure 4E and F).
Figure 4.
DYRK2 suppressed the activity of miR-622 on migration and invasion of CRC cells. (A–D) Expression of DYRK2 was increased when cotransfection of miR-622 mimics and pcDNA3.1-DYRK2-3′UTR-del compared to single transfection of miR-622 mimics, determined by real time PCR and Western blot. (E, F) The stimulative effect of miR-622 on migration and invasion was reversed by up-regulation of DYRK2, detected by Transwell assay. All the data mentioned are shown as mean ± standard deviation. **P<0.01 vs miR-622 mimics + pcDNA3.1-DYRK2-3′UTR-del group.
Abbreviations: miR-622, microRNA-622; DYRK2, dual specificity tyrosine phosphorylation-regulated kinase 2; 3′UTR, 3′-untranslated region; CRC, colorectal cancer; PCR, polymerase chain reaction; del, deletion.
Discussion
DYRK2 is mainly localized in the cytoplasm and belongs to the second subclass of DYRK family. DYRK2 has a central kinase domain and an adjacent N-terminal DYRK homology (DH) box, it widely participates in various cell events including proliferation, apoptosis, differentiation and metastasis.25–28 It is reported that DYRK2 could phosphorylate snail – a zinc-finger transcription factor which acts in a key role in epithelial-mesenchymal transition – at Ser104 as a priming phosphorylation for GSK3, and inhibit snail-mediated tumor metastasis.29 Up to now, reports about DYRK2 in CRC are seldom seen. Yan et al reported the expression of DYRK2 declined in CRC tissue and concluded that DYRK2 could be a novel prognostic biomarker of CRC.16
In our study, compared to adjacent cancer tissues and normal human colon epithelial cell line NCM460, we found the same production trend of DYRK2 in CRC tissues and cell lines as Yan et al’s research.16 A further correlation analysis showed there is a close relation between DYRK2 and clinical stages of CRC. It is generally accepted that miRNAs participate in the progressions of various malignancies through regulation of multiple target genes. A series of our findings showed that miR-622 was over-expressed in CRC tissues and cell lines, and a remarkably negative relation between DYRK2 and miR-622 was found in our research. We conducted a functional experiment to clarify that miR-622 had intervened in migration and invasion of CRC cells, and the luciferase reporter assay verified that miR-622 could target the regulation of DYRK2 and inhibit its downstreams. The last antisense experiment confirmed miR-622 acted on aggressive phenotypes in CRC cells through DYRK2 and its downstream signal pathway. As a tumor-related miRNA, miR-622 was reported as an oncogene or tumor suppressor gene in different cancers. Our findings indicated miR-622 could be an oncogene in CRC. Decreasing the expression of miR-622 in CRC cells by targeting regulation of DYRK2 could suppress the migration and invasion.
Like in most other malignant tumors, the migration and invasion of CRC are very complicated biological processes involving multiple signal pathways and various mechanisms. miR-622 and its targeting of DYRK2 is only one element in a complex regulatory network. Our findings suggest that decreasing the production of miR-622 should be considered a promising strategy for targeted therapies in colon cancer.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (No 81502333), Technological innovation fund of Shenyang technology division (No F15-139-9-07), and The PhD Start-up Research foundation of Liaoning Province (No 201601225).
Footnotes
Disclosure
The authors declare no conflict of interest in this work.
References
- 1.Chen W, Zheng R, Zhang S, et al. Annual report on status of cancer in China, 2010. Chinese J Cancer Res. 2014;26(1):48–58. doi: 10.3978/j.issn.1000-9604.2014.01.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim DH. Colorectal cancer epidemic in Korea. Annals of Coloproctology. 2013;29(1):1–4. doi: 10.3393/ac.2013.29.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu Y, Liu Q, Chen G, et al. Outcome of rectal cancer surgery in obese and nonobese patients: a meta-analysis. World J Surg Oncol. 2016;14(1):23. doi: 10.1186/s12957-016-0775-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Signorini L, Delbue S, Ferrante P, Bregni M. Review on the immunotherapy strategies against metastatic colorectal carcinoma. Immunotherapy-UK. 2016;8(10):1245–1261. doi: 10.2217/imt-2016-0045. [DOI] [PubMed] [Google Scholar]
- 5.Zong Z, Zhou T, Rao L, et al. Musashi2 as a novel predictive biomarker for liver metastasis and poor prognosis in colorectal cancer. Cancer Med-US. 2016;5(4):623–630. doi: 10.1002/cam4.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fabbri M, Croce CM, Calin GA. MicroRNAs. Cancer J. 2008;14(1):1–6. doi: 10.1097/PPO.0b013e318164145e. [DOI] [PubMed] [Google Scholar]
- 7.Ghanbari R, Mosakhani N, Asadi J, et al. Downregulation of Plasma MiR-142-3p and MiR-26a-5p in Patients With Colorectal Carcinoma. Iranian J can Prevention. 2015;8(3):e2329. doi: 10.17795/ijcp2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo XB, Jing CQ, Li LP, et al. Down-regulation of miR-622 in gastric cancer promotes cellular invasion and tumor metastasis by targeting ING1 gene. World J Gastroentero. 2011;17(14):1895–1902. doi: 10.3748/wjg.v17.i14.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Liu Y, Liu W, Zhang W, Xu J. EZH2-mediated loss of miR-622 determines CXCR4 activation in hepatocellular carcinoma. Nat Commun. 2015;6:8494. doi: 10.1038/ncomms9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song WH, Feng XJ, Gong SJ, et al. microRNA-622 acts as a tumor suppressor in hepatocellular carcinoma. Cancer Bio & The. 2015;16(12):1754–1763. doi: 10.1080/15384047.2015.1095402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang R, Luo H, Wang S, et al. MiR-622 suppresses proliferation, invasion and migration by directly targeting activating transcription factor 2 in glioma cells. J Neuro-onco. 2015;121(1):63–72. doi: 10.1007/s11060-014-1607-y. [DOI] [PubMed] [Google Scholar]
- 12.Fang Y, Sun B, Li Z, Chen Z, Xiang J. MiR-622 inhibited colorectal cancer occurrence and metastasis by suppressing K-Ras. Mol Carcinogen. 2016;55(9):1369–1377. doi: 10.1002/mc.22380. [DOI] [PubMed] [Google Scholar]
- 13.Balaguer F, Moreira L, Lozano JJ, et al. Colorectal cancers with microsatellite instability display unique miRNA profiles. Clin Cancer Res. 2011;17(19):6239–6249. doi: 10.1158/1078-0432.CCR-11-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma W, Yu J, Qi X, et al. Radiation-induced microRNA-622 causes radioresistance in colorectal cancer cells by down-regulating Rb. Oncotarget. 2015;6(18):15984–15994. doi: 10.18632/oncotarget.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nihira NT, Yoshida K. Engagement of DYRK2 in proper control for cell division. Cell Cycle. 2015;14(6):802–807. doi: 10.1080/15384101.2015.1007751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan H, Hu K, Wu W, et al. Low Expression of DYRK2 (Dual Specificity Tyrosine Phosphorylation Regulated Kinase 2) Correlates with Poor Prognosis in Colorectal Cancer. PloS One. 2016;11(8):e0159954. doi: 10.1371/journal.pone.0159954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enomoto Y, Yamashita S, Yoshinaga Y, et al. Downregulation of DYRK2 can be a predictor of recurrence in early stage breast cancer. Tumour Biol. 2014;35(11):11021–11025. doi: 10.1007/s13277-014-2413-z. [DOI] [PubMed] [Google Scholar]
- 18.Teng KY, Qiu MZ, Li ZH, et al. DNA polymerase eta protein expression predicts treatment response and survival of metastatic gastric adenocarcinoma patients treated with oxaliplatin-based chemotherapy. J Transl Med. 2010;8:126. doi: 10.1186/1479-5876-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C, Ridzon DA, Broomer AJ, et al. Real time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng J, Wang K, Liu X, Chen S, Chen J. The quantification of tomato microRNAs response to viral infection by stem-loop real time RT-PCR. Gene. 2009;437(1–2):14–21. doi: 10.1016/j.gene.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 21.Pfaffl MW. A new mathematical model for relative quantification in real time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Jia Z, Zhang C, et al. miR-499 protects cardiomyocytes from H 2O 2-induced apoptosis via its effects on Pdcd4 and Pacs2. RNA Biol. 2014;11(4):339–350. doi: 10.4161/rna.28300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Zhao W, Fu Q. miR-335 suppresses migration and invasion by targeting ROCK1 in osteosarcoma cells. Mol Cel Bio. 2013;384(1–2):105–111. doi: 10.1007/s11010-013-1786-4. [DOI] [PubMed] [Google Scholar]
- 24.Xu D, Takeshita F, Hino Y, et al. miR-22 represses cancer progression by inducing cellular senescence. J Cell Biol. 2011;193(2):409–424. doi: 10.1083/jcb.201010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He L, Hannon GJ. MicroRNAs small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Wu Y, Miao X, et al. Silencing of DYRK2 increases cell proliferation but reverses CAM-DR in Non-Hodgkin’s Lymphoma. Int J Biol Macromol. 2015;81:809–817. doi: 10.1016/j.ijbiomac.2015.08.067. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida K. Role for DYRK family kinases on regulation of apoptosis. Biochem Pharmacol. 2008;76(11):1389–1394. doi: 10.1016/j.bcp.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 28.Enomoto Y, Yamashita S, Yoshinaga Y, et al. Downregulation of DYRK2 can be a predictor of recurrence in early stage breast cancer. Tumour Biol. 2014;35(11):11021–11025. doi: 10.1007/s13277-014-2413-z. [DOI] [PubMed] [Google Scholar]
- 29.Mimoto R, Taira N, Takahashi H, et al. DYRK2 controls the epithelial-mesenchymal transition in breast cancer by degrading Snail. Cancer Lett. 2013;339:214–225. doi: 10.1016/j.canlet.2013.06.005. [DOI] [PubMed] [Google Scholar]