Ede1 is one of the earliest proteins recruited to sites of clathrin-mediated endocytosis. Truncation mutant analysis of Ede1 identified the minimal domain required for efficient localization to endocytic sites. Expression of the Eps15, the human homologue of Ede1, in yeast showed that it can be recruited with the same spatiotemporal dynamics as Ede1.
Abstract
During clathrin-mediated endocytosis (CME), endocytic-site maturation can be divided into two stages corresponding to the arrival of the early and late proteins at the plasma membrane. The early proteins are required to capture cargo and position the late machinery, which includes proteins involved in actin assembly and membrane scission. However, the mechanism by which early-arriving proteins select and stabilize endocytic sites is not known. Ede1, one of the earliest proteins recruited to endocytic sites, facilitates site initiation and stabilization. Deletion of EDE1 results in fewer CME initiations and defects in the timing of vesicle maturation. Here we made truncation mutants of Ede1 to better understand how different domains contribute to its recruitment to CME sites, site selection, and site maturation. We found that the minimal domains required for efficient Ede1 localization at CME sites are the third EH domain, the proline-rich region, and the coiled-coil region. We also found that many strains expressing ede1 truncations could support a normal rate of site initiation but still had defects in site-maturation timing, indicating separation of Ede1 functions. When expressed in yeast, human Eps15 localized to the plasma membrane, where it recruited late-phase CME proteins and supported productive endocytosis, identifying it as an Ede1 functional homologue.
INTRODUCTION
While some 60 proteins involved in clathrin-mediate endocytosis (CME) have been identified in the budding yeast Saccharomyces cerevisiae, how CME sites are initiated and stabilized is poorly understood (Godlee and Kaksonen, 2013; Lu et al., 2016). Among the earliest-arriving proteins that define the early phase of endocytic site maturation are Ede1, Syp1, Hrr25, clathrin, AP-2 complex, Yap1801/2, and Pal1. The lifetimes of these proteins at CME sites varies from tens of seconds to a few minutes (Carroll et al., 2009, 2011; Newpher et al., 2005; Stimpson et al., 2009; Peng et al., 2015). Early-phase proteins play an important role in cargo capture and efficient site initiation, although they are dispensable for vesicle formation and internalization, as depletion of these proteins does not prevent recruitment and internalization of coat proteins and actin (Brach et al., 2014). However, understanding endocytic-site initiation and stabilization will provide insight into a possible role for cargo capture in regulation of the endocytic pathway and whether and how other events, such as cell cycle progression, affect endocytosis.
Ede1 is among the earliest proteins to arrive at a nascent CME site, and it facilitates site initiation and maturation. EDE1 deletion impairs endocytosis by reducing the frequency of site initiations (Kaksonen et al., 2005) and decreasing the lifetimes of other endocytic proteins (Stimpson et al., 2009; Carroll et al., 2011). In ede1Δ cells, the rate of CME was reduced by ∼35% relative to wild-type cells (Gagny et al., 2000). When Ede1 is artificially tethered to cortical eisosomes, it recruits downstream coat and actin-associated proteins (Brach et al., 2014). Although it is clear that Ede1 plays a central role in site initiation and CME function, the mechanisms underlying Ede1 functions are poorly understood. Given the important and varied roles that Ede1 has in organizing endocytic sites, we systematically dissected the roles of each of its domains in site selection and maturation.
Ede1 is a large, multidomain protein believed to function as a clathrin adapter and scaffolding protein (Maldonado-Báez et al., 2008) and is required for efficient recruitment of other early-phase proteins to CME sites (Carroll et al., 2011). In humans, the endocytic protein epidermal growth factor receptor pathway substrate clone 15 (Eps15) has similar domain architecture to Ede1 (Confalonieri and Di Fiore, 2002; Reider et al., 2009). Similar to Ede1 in yeast, Eps15 is known to arrive at clathrin-coated pits in the early stages of site initiation along with AP-2 and FCHo1/2 in mammalian cells (Taylor et al., 2011). We sought to test whether Eps15 functions like Ede1 during endocytic-site initiation by expressing human Eps15 in yeast.
RESULTS AND DISCUSSION
The Ede1 coiled-coil domain is required but not sufficient for punctate, plasma membrane localization
Ede1 architecture is defined by several domains and motifs, including three N-terminal Eps15-homology (EH) domains that bind the NPF tripeptide motif (Confalonieri and Di Fiore, 2002; Miliaras and Wendland, 2004). These domains are followed by proline-rich (PP) and central coiled-coil (CC) domains (Reider et al., 2009). The Ede1 C-terminus is defined by a ubiquitin-associated (UBA) domain. The region between the coiled coils and the UBA domain is highly phosphorylated by the casein kinase Hrr25 (Peng et al., 2015).
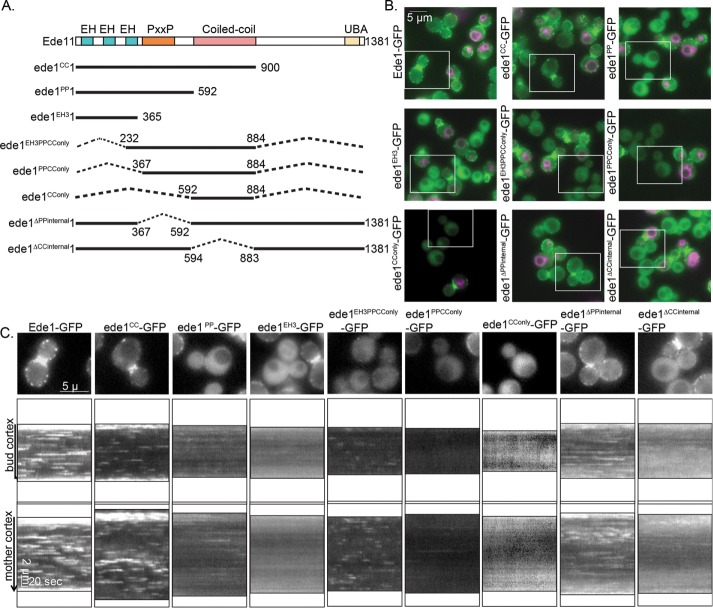
To better understand the functions of Ede1’s domains, we set out to determine which Ede1 domains contribute to endocytic-site localization by generating truncation mutants and integrating them into the endogenous locus fused to a C-terminal green fluorescent protein (GFP) tag (Figure 1A). We compared the expression and localization of these constructs by imaging the truncation strains in the same field as strains expressing full-length Ede1-GFP, which were distinguished from the truncation strains by the use of the lipophilic dye FM4-64 (Figure 1B). Ede1-GFP kymographs show many punctate patches that are clearly distinguishable from background and appear and disappear with variable lifetimes, as seen in kymographs drawn around the cell cortex (Figure 1, B and C). We found that every construct missing the coiled-coil domain (ede1PP-, ede1EH3-, and ede1∆CCinternal-GFP) resulted in kymographs that showed fewer, dimmer patches, which is consistent with previous studies (Boeke et al., 2014). In addition, we found that deletion of the UBA domain and a highly phosphorylated C-terminal region between amino acids 900 and 1341 (Peng et al., 2015) or deletion of the PP region did not have any effect on Ede1 localization or lifetime (Figure 1, B and C, and Supplemental Figure S1B, ede1CC-GFP, ede1ΔPPinternal-GFP). On the basis of our analysis of this panel of truncation mutants, we conclude that the coiled-coil domain is required for efficient Ede1 localization to the plasma membrane (PM). However, we found that expression of the coiled-coil domain alone was not sufficient for PM localization (Figure 1, B and C, ede1CConly-GFP). Although much of our data are consistent with previous studies, this result is contrary to that of Boeke et al. (2014). A possible explanation for this incongruence is that protein expression levels of ede1CConly-GFP are significantly lower than for Ede1-GFP (Supplemental Figure S1A), which may contribute to its lack of PM localization.
FIGURE 1:
Localization of Ede1-GFP truncation mutants. (A) Schematic diagram of full-length Ede1 and the truncation constructs that were integrated at the EDE1 locus. A C-terminal GFP tag, present on all constructs, is not shown. (B) Representative images from the first frame of movies of cells expressing the indicated constructs imaged in the same field as Ede1-GFP reference cells labeled with FM4-64 (magenta). Movies of the GFP channel were taken at 2 s/frame for 4 min, immediately followed by capture of a still image of FM4-64. Scale bar, 5 μm. (C) Magnification of the cell outlined in a white box from B followed by circular kymographs of the mother and bud cortex. Scale bars, 2 μm and 20 s.
We found a truncation containing the CC domain, the PP-rich region, and the third EH domain (Figure 1, B and C, ede1EH3PPCConly-GFP) was recruited to distinct PM patches despite also having low expression relative to Ede1-GFP (Supplemental Figure S1A). This construct also had much shorter lifetimes than Ede1-GFP (14.8 ± 13.0 vs. 51.5 ± 37.4 s, SD; Figure 2A). Recruitment of ede1EH3PPCConly-GFP to PM punctae is particularly striking because it lacks many of Ede1’s protein interaction domains and many of its phosphorylation sites.
FIGURE 2:
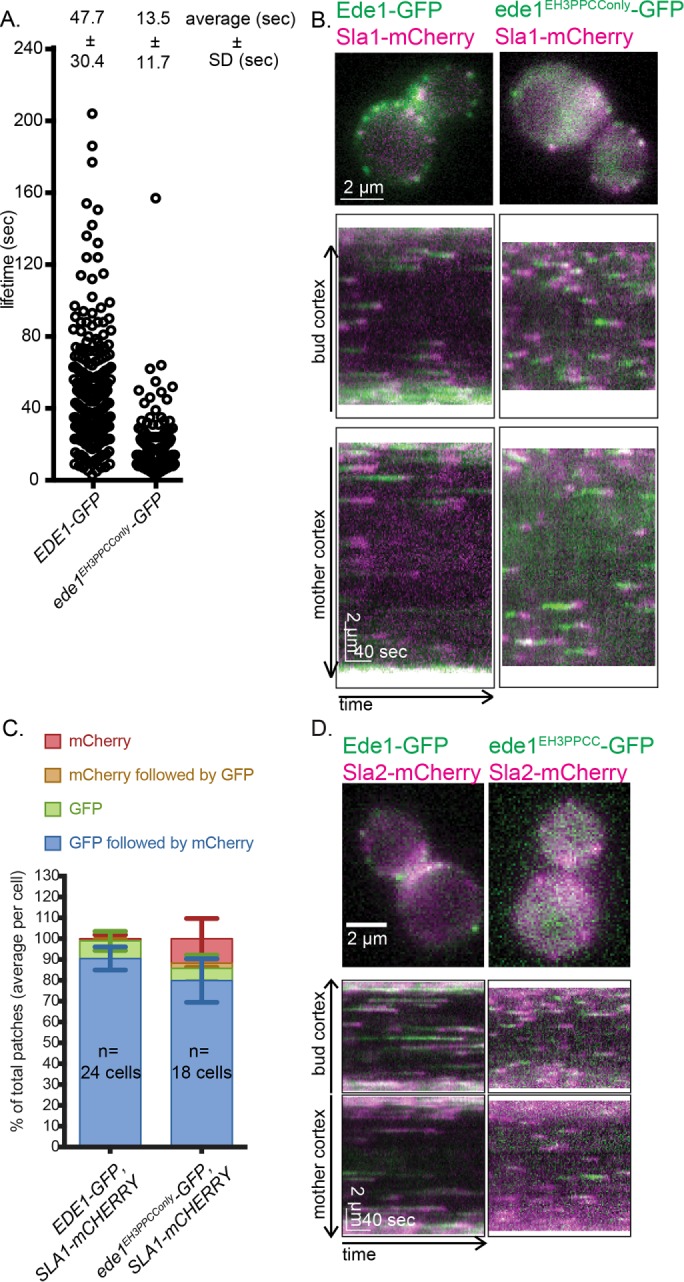
A construct containing the coiled-coil domain, third EH domain, and PP region is sufficient to recruit coat proteins. (A) Lifetimes of individual Ede1 patches from cells of the indicated genotype calculated from three independent experiments (EDE1-GFP, 328 patches; ede1EH3PPCConly-GFP, 480 patches). Mean ± SD. The p values were calculated using the Student’s t test. (B) Ede1-GFP variants were imaged with Sla1-mCherry for 4 min at 2 s/frame using simultaneous dual-color imaging. The first frame of a movie of a representative cell, along with circular kymographs of the mother and bud cortex. Scale bars, 2 μm and 40 s. (C) The patch traces identified in kymographs from three independent experiments were segregated into one of four categories depending on the relative arrival of GFP and mCherry: GFP or mCherry alone, GFP followed by mCherry, or mCherry followed by GFP. Mean percentage and SD of each category per cell. (D) Ede1-GFP variants were imaged with Sla2-mCherry for 4 min at 2 s/frame using simultaneous dual-color imaging. The first frame of a movie of a representative cell is shown along with circular kymographs of the mother and bud cortex. Scale bars, 2 μm and 40 s.
Because ede1EH3PPCConly-GFP is present on the PM, we next sought to determine whether ede1EH3PPCConly-GFP patches could recruit downstream endocytic machinery by expressing ede1EH3PPCConly-GFP with Sla1 tagged at the C-terminus with mCherry (Sla1-mCherry). Sla1 is a coat protein that arrives after Ede1 but before actin polymerization and internalization (Weinberg and Drubin, 2011; Goode et al., 2015), making it an ideal reporter for successful recruitment and function of downstream endocytic components. Using time-lapse imaging over 4 min, we found that in control cells (Ede1-GFP, Sla1-mCherry), on average, Sla1-mCherry was recruited to GFP patches in 90.5 ± 5.6% SD of kymograph traces analyzed. In the ede1EH3PPCConly-GFP strain, Sla1-mCherry was also recruited to GFP patches the majority of the time (Figure 2, B and C, 79.9 ± 10.5% SD). We also tested other CME coat proteins and found that the later-arriving coat protein Sla2-mCherry was also recruited to ede1EH3PPCConly-GFP patches (Figure 2D).
The ede1EH3PPCC and ede1ΔCCinternal strains have normal numbers of CME sites but altered maturation timing
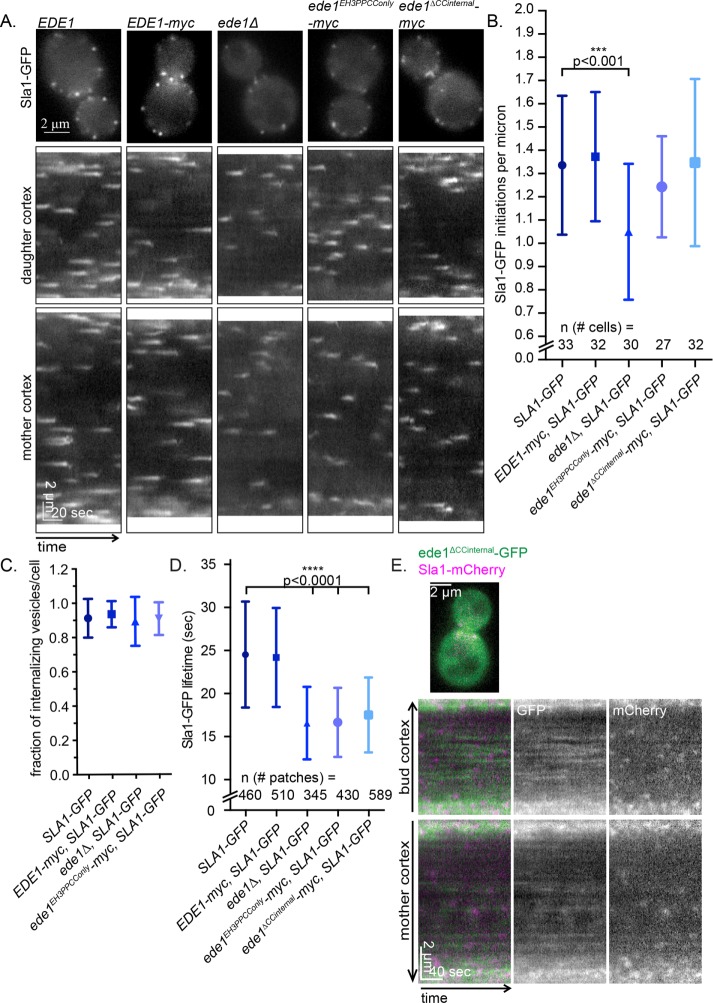
As a readout of endocytic function in the context of the localization phenotypes for different Ede1 truncations, we examined the number of CME sites formed and the timing of coat maturation. We C-terminally tagged the truncation constructs with a 13xMYC tag and used Sla1-GFP as a reporter for endocytosis because its localization and recruitment timing are highly sensitive to small perturbations in functions of other endocytic proteins (Kaksonen et al., 2003).
Consistent with results from previous studies, we observed a decreased number of endocytic sites in ede1Δ cells when imaging Sla1-GFP over 90 s (Figure 3B, SLA1-GFP vs. ede1Δ SLA1-GFP, 1.3 ± 0.3 vs. 1.0 ± 0.3 patches patches/μm, SD). We also observed a decrease in Sla1-GFP lifetime in ede1Δ cells (Figure 3D, SLA1-GFP vs. ede1Δ SLA1-GFP, 24.5 ± 6.2 vs. 15.9 ± 4.1 s.). We analyzed Sla1-GFP lifetimes separately for patches in the mother and the daughter cells of large-budded cells of control strains and did not find significant differences (Supplemental Figure S1C). Therefore the data that are shown are aggregates for lifetimes from the mother and daughter cells. Surprisingly, we found that ede1EH3PPCConly-myc expression facilitated the normal number of initiation events (Figure 3B, 1.2 ± 0.2 patches/μm). In addition, although we did not see a defect in the percentage internalizing Sla1-GFP patches (Figure 3C), we did find that Sla1-GFP patch lifetimes in ede1EH3PPCConly-myc cells were much shorter (Figure 3D, SLA1-GFP vs. ede1EH3PPCC-myc SLA1-GFP, 24.5 ± 6.2 vs. 16.6 ± 4.0 s, SD), indicating that this mutant does not fully execute all of Ede1’s regulatory functions. A possible explanation is that one of the removed domains recruits cargo or another endocytic coat protein, which in turn contributes to the characteristically longer and more variable lifetimes of endocytic events. Deletion of EDE1 is known to disrupt the localization of other early proteins, such as clathrin, AP-2, Syp1, and Pal1 (Carroll et al., 2011). In addition, certain protein–protein interactions have been mapped to regions that are deleted in the ede1EH3PPCC strain. For example, both Syp1 and Hrr25 specifically interact with the Ede1 C-terminal portion missing from ede1EH3PPCC (Reider et al., 2009; Peng et al., 2015). Ede1 and its EH domains in particular interact with both epsins and AP180 adapter proteins (Ent1/2, Yap1801/2; Maldonado-Báez et al., 2008; Dores et al., 2010) and play a role in coat maturation (Suzuki et al., 2012). Of interest, because the lifetimes of ede1EH3PPCC-GFP were significantly shorter and less variable than those of full-length Ede1-GFP, we hypothesize that such an interaction might participate in a proposed endocytic checkpoint that has been bypassed (Carroll et al., 2011; Layton et al. 2011).
FIGURE 3:
ede1EH3PPCConly and ede1ΔCCinternal facilitate endocytic initiation but not normal maturation timing. (A) The 90-s movies of Sla1-GFP in cells expressing 13xmyc-tagged Ede1 truncations were taken at 1 s/frame. First frame from a representative cell. Scale bar, 5 μm. This is followed by circular kymographs of the bud and mother cortex of the cells shown in A; scale bars, 2 μm and 20 s. (B) Quantification of kymograph analysis from three independent experiments of the number of patches that start during a 90-s movie in cells of the indicated genotype. The p values were calculated using Student’s t test. (C) Quantification of kymograph analysis from three independent experiments of the proportion of Sla1-GFP patches that internalize per cell (25 cells/strain). (D) Quantification of the lifetime of Sla1-GFP from three independent experiments in cells of the indicated genotype. (E) ede1ΔCCinternal-GFP and Sla1-mCherry were imaged for 4 min at 2 s/frame with ∼300-ms delay between channels. The first frame of a movie of a representative cell, in addition to a circular kymograph of the bud and mother cortex. Scale bars, 2 μm and 40 s.
We further focused our studies on ede1ΔCCinternal because the coiled-coil domain is necessary but not sufficient for efficient PM localization, and thus whereas the truncation retains many of its key domains, much of ede1ΔCCinternal-GFP is cytosolic. We tested whether this mutant can provide any endocytic function.
Surprisingly, we observed that, even though ede1ΔCCinternal is poorly recruited to the PM (Figure 1, B and C), it can also facilitate the normal number of initiation events (Figure 3B; SLA1-GFP vs. ede1ΔCCinternal SLA1-GFP, 1.3 ± 0.3 vs. 1.4 ± 0.4 patches patches/μm, SD). It is possible that patch initiation is rescued even though there is very little recruitment of ede1ΔCCinternal if later-arriving coat proteins are recruited to these patches by the partially localized truncation mutants. To test this possibility, we imaged ede1ΔCCinternal-GFP with Sla1-mCherry and found that ede1ΔCCinternal-GFP in faint patches on the PM. Sla1-mCherry was recruited to those patches in 83.5 ± 10.2% SD of kymographs analyzed (Figure 3E). However, Sla1-GFP lifetimes were also abnormal in strains that expressed ede1ΔCCinternal-myc (Figure 3D, SLA1-GFP vs. ede1ΔCCinternal-myc SLA1-GFP, 24.5 ± 6.2 vs. 17.5 ± 4.4 s, SD), indicating that proper maturation timing is more sensitive to perturbations in Ede1 than initiation efficiency.
It is striking that two very different Ede1 truncation mutants (ede1ΔCCinternal and ede1EH3PPCConly), each expressing complementary portions of the protein (N- and C- terminal ends vs. the middle), can each provide endocytic function in site initiation. It is very intriguing that ede1ΔCCinternal can support endocytic initiation while it seems much of the protein is cytosolic. This indicates that there may be some cytosolic interactions that are important for site initiation and/or a low threshold for the amount of Ede1 present to recruit downstream proteins at the cortex. It is also very surprising that ede1EH3PPCConly is able to recruit downstream proteins, even though it has a much shorter plasma membrane lifetime and is missing many protein–protein interaction domains. Together, these observations suggest that there is robustness and possibly redundancy in the endocytic-site initiation mechanism, potentially involving interactions both in the cytosol and at the cortex.
The Ede1 coiled-coil domain functions to coalesce Ede1 molecules
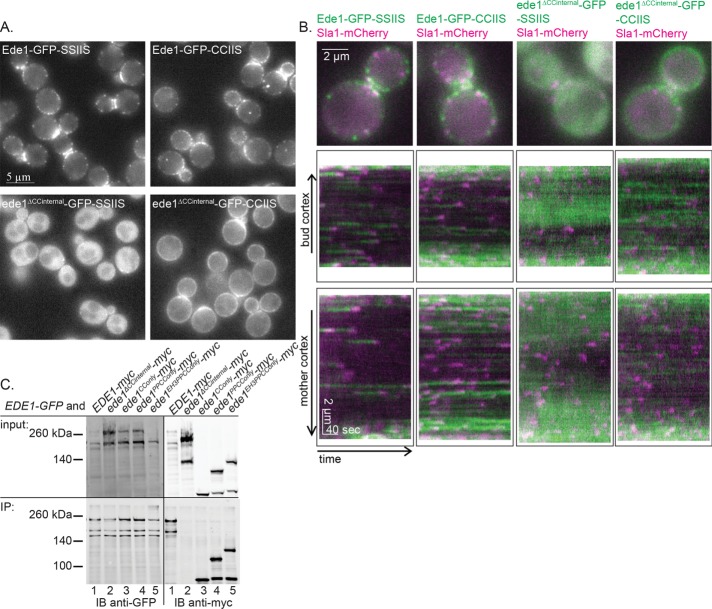
Because loss of the Ede1 coiled-coil domain results in significantly reduced localization of the truncation mutants to the PM, we tested whether artificially tethering ede1ΔCCinternal-GFP to the PM can restore normal Sla1 lifetimes. We recruited this mutant to the PM by appending the Ras2 CCaaX box (CCIIS), which is prenylated and palmitoylated and trafficked primarily to the PM, to the C-terminus (Chen and Thorner, 2007). As a control, we also generated a strain in which the prenyl and palmitoyl acceptor cysteines were mutated to serines (SSIIS). Compared to the SSIIS controls, more Ede1-GFP-CCIIS and ede1ΔCCinternal-GFP-CCIIS constructs were associated with the PM (Figure 4A). Of interest, whereas Ede1-GFP-CCIIS formed distinct punctae, ede1ΔCCinternal-GFP-CCIIS was recruited more uniformly around the cortex. Using kymograph analysis, we observed slightly faint punctae of ede1ΔCCinternal-GFP-CCIIS in addition to the halo of GFP around the PM and found that Sla1-mCherry was preferentially recruited to these punctae in ede1ΔCCinternal-GFP-CCIIS cells and not to the halo (Figure 4B). On the basis of these observations, we hypothesized that oligomerization by the coiled-coil domains is required for efficiently coalescing Ede1 into punctae at the PM.
FIGURE 4:
Coiled coils contribute to endocytic-site localization by aggregating Ede1 molecules. (A) Representative images of Ede1-GFP or ede1ΔCCinternal-GFP tagged with a prenylation motif (CCIIS) or a control sequence (SSIIS). Scale bar, 5 μm. (B) Constructs from A were expressed with Sla1-mCherry and imaged for 4 min at 2 s/frame with ∼300-ms delay between channels. The first frame of a movie of a representative cell, followed by a circular kymograph of the bud and mother cortex. Scale bars, 2 μm and 40 s. (C) Whole-cell lysates of diploid cells with the indicated genotype were subjected to immunoadsorption using mouse anti-GFP antibody, followed by SDS–PAGE and Western blotting, using rabbit anti-GFP antibody to detect Ede1-GFP and mouse anti-myc antibody to detect the Ede1-13xmyc-tagged truncations.
To test for oligomerization mediated by the coiled-coil domains, we performed an immunoadsorption experiment in whole-cell lysates from diploid cells expressing Ede1-GFP and an Ede1-myc truncation construct. After immunoadsorbing Ede1-GFP, we probed immunoblots to detect the presence of the different Ede1 truncation constructs. We found that in every case in which the truncation mutant contains the coiled-coils, including the coiled-coils alone (ede1CConly-myc), we observed an interaction with Ede1-GFP (Figure 4C, lanes 1 and 3–5). However, the ede1ΔCCinternal-myc truncation could not be coisolated with the full-length Ede1-GFP bait (Figure 4C, lane 2). Therefore it is likely that ede1ΔCCinternal-GFP-CCIIS localizes on the PM in a diffuse manner rather than in distinct punctae because the protein is unable to oligomerize. Although these observations may be the result of indirect binding between the Ede1 coiled coils and a bridging molecule, our interpretation is consistent with previous reports that the coiled coils are necessary for multimerization (Boeke et al., 2014). This result adds to previous evidence suggesting that Ede1 oligomerization is a key step in CME site initiation. The mechanism by which this oligomerization step, whether in the cytosol or at the plasma membrane, organizes downstream adapters will be an important question to answer in understanding how initiation of CME sites occurs.
Human Eps15 can provide Ede1 functions in yeast
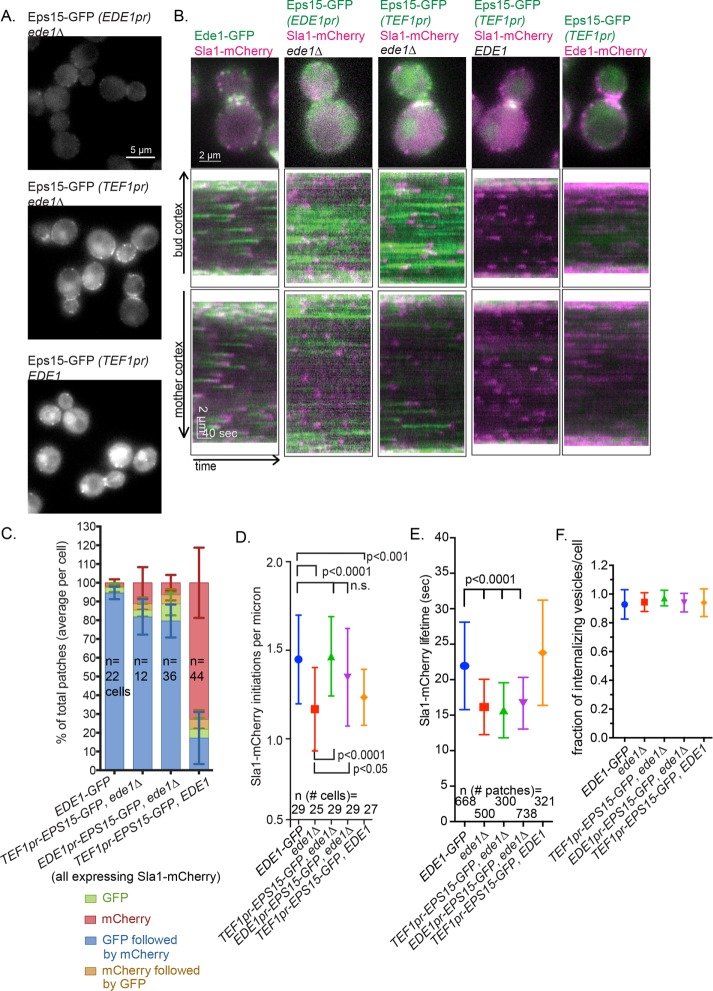
The ability to translate principles learned from studies in yeast to mammalian cells requires faithful identification of functional homologues. The early arrival of Ede1 and Eps15 in yeast and mammals, respectively, hinted that they might share functional homology (Reider et al., 2009; Stimpson et al., 2009; Taylor et al., 2011). Human Eps15 is an important adapter protein in clathrin-mediated endocytosis. Like Ede1, it arrives at the endocytic site early in the CME pathway. Because Eps15 has a domain structure very similar to Ede1, containing central coiled-coils for dimerization, three EH domains, and ubiquitin-interacting motifs (Cupers et al., 1997), we tested whether full-length Eps15 can function in the place of Ede1 in yeast. We first inserted the human EPS15 cDNA, with a C-terminal GFP tag, into the EDE1 locus, using the EDE1 promoter to drive expression. Western blot analysis showed that Eps15-GFP expression is ∼20% that of Ede1-GFP (Supplemental Figure S2). However, even though it was expressed at a much lower level than Ede1, Eps15-GFP localized to endocytic sites at the PM (Figure 5, A and B). In addition, Eps15-GFP appeared at the PM with similar timing to Ede1, arriving before Sla1. On average, an Eps15-GFP patch was joined by Sla1-mCherry in 78.5 ± 8.3% SD of kymograph traces analyzed compared with 94.5 ± 3.3% SD for Ede1-GFP (Figure 5C).
FIGURE 5:
Eps15 can function as an endocytic-site initiator in the absence of Ede1. (A) Representative still images of cells expressing the Eps15-GFP using the indicated promoter. (B) GFP constructs from A were expressed with Sla1-mCherry and imaged for 4 min at 2 s/frame with ∼300-ms delay between channels. The first frame of a movie of a representative cell, followed by a circular kymograph of the bud and mother cortex. Scale bars, 2 μm and 40 s. (C) The patch traces identified in kymographs from B in two independent experiments were segregated into one of four categories depending on the relative arrival of GFP and mCherry: GFP or mCherry alone, GFP followed by mCherry, or mCherry followed by GFP. The mean percentage and SD of each category per cell is indicated. (D) Quantification of kymograph analysis from two independent experiments of the number of patches that start during a 90-s movie in cells of the indicated genotype. The p values were calculated using Student’s t test. (E) Quantification of the Sla1-mCherry lifetime from two independent experiments in cells of the indicated genotype. (F) Quantification of kymograph analysis of the proportion of Sla1-mCherry patches that internalize from two independent experiments per cell (25 cells/strain).
Because Eps15 was expressed at a low level from the EDE1 promoter, we tested whether increasing expression to normal Ede1 levels would increase the frequency of Sla1 recruitment. We generated a construct expressing the EPS15 cDNA, tagged with GFP, under the translation elongation factor EF1-α (TEF1) promoter, which expresses genes at a constitutively high level (Mumberg et al., 1995). This construct was inserted at the URA3 locus, which allowed Eps15 expression in the presence or absence of Ede1, so we could test for competition in recruitment to endocytic sites. Use of the TEF1 promoter increased Eps15 expression approximately twofold (Supplemental Figure S2 and Figure 5A).
Ede1 presence prevented the majority of Eps15 from being recruited to the PM (Figure 5, A and B). When Ede1-mCherry and Eps15-GFP were imaged in the same cell, only Ede1-mCherry was detectable in the majority of cortical patches, indicating that these two proteins likely compete for binding sites in the endocytic machinery (Figure 5B). Furthermore, in this context, Eps15-GFP preceded Sla1-mCherry in very few patches (Figure 5C, 17.2 ± 13.9% SD, “mCherry only”). When EDE1 was deleted and Eps15-GFP was expressed from the TEF1 promoter, Eps15-GFP formed more distinct patches than when Eps15-GFP was expressed from the EDE1 promoter (Figure 5, A and B). However, the increased expression of Eps15 under the TEF1 promoter did not increase the frequency of patches in which Eps15-GFP precedes Sla1-mCherry (Figure 5C, EDE1pr-EPS15-GFP ede1Δ vs. TEF1pr-EPS15-GFP ede1Δ, 79.6 ± 8.8% vs. 81.8 ± 9.5% SD). Accordingly, the number of initiation events counted by imaging Sla1-mCherry in the ede1Δ cells was restored to normal levels when Eps15-GFP was expressed from either the EDE1 promoter or the TEF1 promoter (Figure 5D, EDE1 vs. EDE1pr-EPS15-GFP ede1Δ or TEF1pr-EPS15-GFP ede1Δ, 1.5 ± 0.3 vs. 1.4 ± 0.3 or 1.5 ± 0.2 patches/μm, SD). Of interest, when both Eps15-GFP and Ede1 are present, Sla1 initiations decrease, suggesting that they interfere with each other’s function in the cytosol, possibly via competition with downstream endocytic machinery (Figure 5D, EDE1-GFP vs. TEF1pr-EPS15-GFP, 1.5 ± 0.3 vs. 1.2 ± 0.2 patches/μm, SD). In addition, whereas Eps15 expression under either the EDE1 or the TEF1 promoter, in the absence of Ede1, can partially rescue site-initiation frequency, neither rescues decreased Sla1-mCherry lifetime at these sites (Figure 5E, EDE1 vs. TEF1pr-EPS15-GFP ede1Δ or EDE1pr-EPS15-GFP ede1Δ, 22.0 ± 6.2 vs. 15.7 ± 3.9 or 16.7 ± 3.6 s, SD). The decreased lifetime of Sla1-mCherry in these strains was not a result of a failure to internalize: we found no difference in the percentage of Sla1-mCherry sites that internalize from the cortex (Figure 5F).
These results underscore the separation of Ede1’s function in site initiation and site maturation. Determining whether Ede1 and Eps15 interact with similar homologous protein partners will be important to gaining deeper insights into how each protein functions in initiation and maturation. Similar to Ede1, we found that deletion of Eps15’s coiled coils in yeast greatly reduces its ability to be recruited to the plasma membrane regardless of the presence of Ede1, further emphasizing the functional similarities of the two proteins and their domain functions (Supplemental Figure S3, A and B). Expression of Eps15 in yeast, together with the demonstration that it is a functional Ede1 homologue, has important implications for our understanding of both proteins and how they function in CME in their respective organisms. Further studies on both proteins may shed light on the similarities and differences in the regulation of initiation events. The expression of a human endocytic protein in yeast may also provide an avenue for more complete dissection of its function.
MATERIALS AND METHODS
Yeast strains and plasmids
Cells were maintained on rich medium (yeast extract/peptone/dextrose) at 25 or 30°C. Table 1 lists the strains used in this work.
Table 1:
Strains used in this work.
| Strain | Genotype |
| DDY1102 | MATa/MATα, ade2-1/ADE2, his3∆200/ his3∆200, leu2-3,112/ leu2-3,112, ura3-52/ ura3-52, lys2-801/LYS2 |
| DDY904 | MATα, ade2-1, his3∆200, leu2-3,112, ura3-52, lys2-801 |
| DDY5380 | MATa, ede1PP-GFP(∆593-1381)::HIS3, his3∆200, leu2-3,112, ura3-52 |
| DDY5381 | MATα, EDE1-GFP::HIS, his3∆200, leu2-3,112, SLA1-mCherry::HIS, ura 3-52 |
| DDY5382 | MATa/MATα, ede1∆::HIS3/EDE1, his3∆200/ his3∆200, leu2-3/leu2-3, lys2-801/LYS, ura3-52/ura3-52 |
| DDY5383 | MATα, ede1∆::ede1CConly(∆6-586, ∆883-1376)-GFP::HIS3::LEU2, his3∆200, leu2-3,112, lys2-801, ura3-52 |
| DDY5384 | MATα, ede1∆::ede1PPCConly(∆6-366, ∆883-1376)-GFP::HIS3::LEU2, his3∆200, leu2-3,112, ura3-52 |
| DDY5385 | MATα, ede1∆::ede1∆ CCinternal(∆593-882)-GFP::HIS3::LEU2, his3∆200, leu2-3,112, LYS, ura3-52 |
| DDY5386 | MATα, ede1∆::EDE1-GFP::HIS3::LEU2, his3∆200, leu2-3,112, LYS2, ura3-52 |
| DDY5387 | MATa, ede1∆::ede1∆ CCinternal(∆593-882)-GFP::HIS3::LEU2, his3∆200, leu2-3,112, SLA1-mCherry::HIS3, ura3-52 |
| DDY5388 | MATa, ede1∆::ede1∆ PPinternal(∆366-592)-GFP::HIS3::LEU2, his3∆200, leu2-3,112, lys2-801, ura3-52 |
| DDY5389 | MATα, ede1∆::ede1EH3PPCConly(∆6-231, ∆883-1376)-GFP::HIS3::LEU2, his3∆200, leu2-3,112, LYS2, ura3-52 |
| DDY5390 | MATa, ede1∆::ede1EH3PPCConly(∆6-231, ∆883-1376)::LEU2, EDE1-GFP::HIS3, his3∆200, leu2-3,112, LYS, SLA1-mCherry::HIS3, ura3-52 |
| DDY5391 | MATa, ede1∆::ede1EH3PPCConly(∆6-231, D883-1376)-13xmyc::HIS3::LEU2, his3∆200, leu2-3,112, SLA1-GFP::HIS3, ura3-52 |
| DDY5392 | MATa, ede1∆::ede1∆ CCinternal(∆593-882)-13xmyc::HIS3::LEU2, his3∆200, leu2-3,112, SLA1-GFP::HIS3, ura3-52 |
| DDY5393 | MATα, ede1∆::EDE1::LEU, EDE1-myc::HIS3, his3∆200, leu2-3,112, SLA1-GFP::HIS3, ura3-52 |
| DDY3866 | MATa, EDE1-GFP::HIS3, his3∆200, leu2-3,112, ura3-52 |
| DDY2734 | MATa, his3Δ200, leu2-3,112, lys2-801, SLA1-GFP:HIS3, ura3-52 |
| DDY3798 | MATa, ede1∆::cgLEU2, his3∆200, leu2-3,112, SLA1-GFP::KanMX6, ura3-52 |
| DDY5394 | MATa, EDE1-GFP::HIS3::LEU2, his3∆200, leu2-3,112, LYS, SLA2-mCherry::KAN, ura3-52 |
| DDY5395 | MATa, ede1∆::ede1EH3PPCConly(∆6-231, ∆883-1376)-GFP::HIS3::LEU2, his3∆200, leu2-3,112, LYS, SLA2-mCherry::KAN, ura3-52 |
| DDY5396 | MATa, ede1∆::EDE1-GFP-CCIIS::LEU2, his3∆200, leu2-3, 112, LYS, ura3-52 |
| DDY5397 | MATa, ede1∆::EDE1-GFP-SSIIS::LEU2, his3∆200, leu2-3, 112, LYS, ura3-52 |
| DDY5398 | MATa, ede1∆::ede1∆CCinternal(∆562-883)-GFP-CCIIS::LEU2, his3∆200, leu2-3, 112, LYS, ura3-52 |
| DDY5399 | MATa, ede1∆::ede1∆CCinternal(∆562-883)-GFP-SSIIS::LEU2, his3∆200, leu2-3, 112, LYS, ura3-52 |
| DDY5400 | MATα, ede1∆::EDE1-GFP-SSIIS::LEU2, his3∆200, leu2-3, 112, LYS, SLA1-mCherry::HIS, ura3-52 |
| DDY5401 | MATα, ede1∆::EDE1-GFP-CCIIS::LEU2, his3∆200, leu2-3, 112, LYS, SLA1-mCherry::HIS, ura3-52 |
| DDY5402 | MATa, ede1∆::ede1∆CCinternal(∆562-883)-GFP-SSIIS::LEU2, his3∆200, leu2-3, 112, LYS, SLA1-mCherry::HIS, ura3-52 |
| DDY5403 | MATa, ede1∆::ede1∆CCinternal(∆562-883)-GFP-CCIIS::LEU2, his3∆200, leu2-3, 112, LYS, SLA1-mCherry::HIS, ura3-52 |
| DDY5404 | MATa, ede1∆::LEU2, his3∆200, leu2-3,112, LYS2, SLA1-mCherry::HIS3, ura3-52 |
| DDY5405 | MATa, ede1∆::Eps15-GFP::LEU2, his3∆200, leu2-3,112, LYS2, ura3-52 |
| DDY5406 | MATa, ede1∆::Eps15-GFP::LEU2, his3∆200, leu2-3,112, LYS2, SLA1-mCherry::HIS3, ura3-5 |
| DDY5407 | MATa, his3∆200, leu2-3,112, LYS2, ura3-52::TEF1p-Eps15-GFP::URA3 |
| DDY5408 | MATa, ede1∆::HIS3, his3∆200, leu2-3,112, LYS2, ura3-52::TEF1p-Eps15-GFP::URA3 |
| DDY5409 | MATa, ede1∆::HIS, his3∆200, leu2-3,112, LYS2, SLA1-mCherry::HIS3, ura3-52::TEF1p-Eps15-GFP::URA3 |
| DDY5410 | MATa, his3∆200, leu2-3,112, LYS2, SLA1-mCherry::HIS3, ura3-52::TEF1p-Eps15-GFP::URA3 |
| DDY5573 | MATa, EDE1-mCherry::KANMX, his3∆200, leu2-3,112, LYS2, ura3-52::TEF1p-Eps15-GFP::URA3 |
| DDY5574 | MATa, his3∆200, leu2-3,112, LYS2, ura3-52::TEF1p-eps15ΔCCinternal(321-520)-GFP::URA3 |
| DDY5575 | MATa, ede1∆::HIS3, his3∆200, leu2-3,112, LYS2, ura3-52:: TEF1p-eps15ΔCCinternal(321-520)-GFP::URA3 |
EDE1 truncations were constructed using either a PCR-based C-terminal truncation method described in Longtine et al. (1998) or pBluescript SK II containing the EDE1 open reading frame (ORF) with −385 base pairs upstream and +397 base pairs downstream from the genomic sequence (generously provided by Jiro Toshima, Tokyo University of Science; Suzuki et al., 2012). The downstream sequence is interrupted at +138 base pairs by the LEU2 ORF, which was used as a selectable marker, with 646 and 477 base pairs of upstream and downstream sequences, respectively, from the genomic sequence. Each truncation was then tagged with either 13xmyc or GFP using the C-terminal tagging method described in Longtine et al. (1998).
EDE1 prenylation constructs were generated from the foregoing pBluescript SKII EDE1 plasmid by replacing the stop codon with GGSGGS+GFP (S65T)+QSGDQISEPGTLDASAPGGNTSEASKSGSGG+CCIIS or GGSGGS+GFP (S65T) +QSGDQISEPGTLDASAPGGNTSEASKSGSGG+SSIIS. The DNA sequences used for these insertions were obtained from pRC285 and pRC286 (Chen et al., 2010), which were generously provided by Jeremy Thorner (University of California, Berkeley).
EPS15 cDNA was cloned from plasmid Eps15-pmCherryN1, a gift from Christien Merrifield (CNRS; Addgene plasmid 27696; Taylor et al., 2011) into the pBluescript SKII EDE1 plasmid described for EDE1pr-EPS15-GFP. For TEF1pr-EPS15-GFP, the same EPS15 cDNA was cloned into pRS306 with 408 base pairs of the upstream region of the TEF1 sequence. This was then integrated into the URA3 locus.
Live-cell imaging
Cells were grown to log phase at 25°C in imaging medium (synthetic medium lacking tryptophan) and immobilized on concanavalin A–coated coverslips. All images except those in Figure 2 were obtained using a Nikon Eclipse Ti microscope equipped with a Plan Apo VC 100×/numerical aperture (NA) 1.4 objective and a Neo scientific complementary metal-oxide semiconductor camera (Andor Technology). Images in Figure 2 were obtained using an Olympus IX81 microscope equipped with either a 100×/NA 1.4 objective (2B) or a 60×/NA 1.45 objective (2D) and an Orca-ER camera (Hamamatsu). Simultaneous two-color imaging for Figure 2 was performed on the Olympus IX81 using a 488-nm argon-ion laser (Melles Griot) and a mercury lamp filtered through a 575/20-nm filter. Exposure time and frequency of acquisition for time-lapse series are indicated in figure legends.
Images were processed using ImageJ (National Institutes of Health, Bethesda, MD) as previously described to threshold background noise (Kaksonen et al., 2003). Movies that had noticeable drift in the xy-axis were registered and corrected using StackReg (Thévenaz et al., 1998) Circular kymographs were generated using the segmented line tool to draw a circle around the cortex of the mother and daughter of a budding cell. Number of initiations was calculated from these circular kymographs by counting the number of patches that started past the first frame (those that also ended past the last frame were included). Lifetimes were measured by generating multiple kymographs around the cell (separated by 2°, for 180 kymographs total) that are perpendicular to the cortex and then measuring the pixel distance from the start to the finish of a patch. Persistent Ede1-GFP patches were not included in lifetime calculations for Figure 2 and Supplemental Figure S1. Sla1-GFP or Sla1-mCherry patches were considered to have internalized when the fluorescence centroid moved more >3 pixels (194 nm) away from the cortex. For each cell, 10 patches that were not at the bud neck were chosen for analysis.
GFP and mCherry correlation analysis was performed using the same type of kymograph analysis and manually notating each distinguishable patch as mCherry alone, GFP alone, mCherry followed by GFP, or GFP followed by mCherry. Each class was then calculated as a percentage of the total number of kymographs analyzed per cell (mother and daughter).
Immunoblotting
Yeast total cell extracts were prepared from log-phase cells using trichloroacetic acid (TCA) as previously described (Foiani et al., 1994). Briefly, cells were harvested from subsaturated cultures that were allowed to double at least twice in rich medium at either 25 or 30°C with shaking. These pellets were washed and resuspended in 20% TCA, and cells were disrupted by bead beating for at least 10 min at 4°C. The total extract was then spun at low speed at room temperature. The resulting pellets were resuspended and boiled in 2× Laemmli buffer and 1 M Tris to neutralize. These total cell extracts were subjected to SDS–PAGE and immunoblot analysis using mouse anti-GFP (Roche), rabbit anti-GFP (Torrey Pines), rabbit anti-Ede1 (generously provided by Linda Hicke, University of Texas at Austin), or mouse anti-Pgk1 (Invitrogen) antibodies.
Blots were exposed using an Odyssey imaging system (LI-COR). Bands were quantified using LI-COR imaging software.
Immunoadsorption
Approximately 70 OD of yeast cells was collected and kept frozen at −80°C overnight. Pellets were then resuspended in cold lysis buffer (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES]–KOH, pH 7.6, 150 mM KCl, 1 mM ethylene glycol tetraacetic acid [EGTA], 0.1% NP-40, and 1× Protease Inhibitor Cocktail [Roche]) and lysed using a mini–bead beater (Biospec) by beating at top speed for 30 s for two intervals at 4°C. Lysates were collected and clarified by spinning at 13,000 rpm in a bench-top microcentrifuge for 10 min at 4°C. Protein G–Sepharose beads (GE) were then added to the supernatant and incubated for 1 h at 4°C to preclear the clarified lysate. Beads were then removed, and GFP-tagged proteins were immunoadsorbed by adding 4.8 μg of mouse anti-GFP antibody (Roche) to the precleared, clarified lysate and incubating at 4°C for 2 h. Protein G–Sepharose beads were then added and incubated for another 1 h at 4°C. Beads were then collected and washed in 10× bead volume with wash buffer (50 mM HEPES–KOH, pH 7.6, 150 mM KCl, 1 mM EGTA, and 10% glycerol) four times. Proteins were then eluted from the beads by boiling in 2× Laemmli sample buffer and then subjected to SDS–PAGE and immunoblot analysis using rabbit anti-GFP antibody (Torrey Pines) and purified monoclonal anti–myc 9-E10 antibody.
Supplementary Material
Acknowledgments
We thank Jiro Toshima for sharing the full-length EDE1-GFP::LEU2 plasmid, Linda Hicke for sharing the anti-Ede1 antibody, and Jeremy Thorner for sharing prenylation motif plasmids. We are grateful to Georjana Barnes and all of the members of the Drubin/Barnes laboratory for helpful discussions, especially Emily Stoops for comments on the manuscript. This work was supported by National Institutes of Health Grant R35GM118149 to D.G.D.
Abbreviations used:
- CC
coiled-coil
- CME
clathrin-mediated endocytosis
- EH
Eps15 homology
- GFP
green fluorescent protein
- PM
plasma membrane
- PP
proline rich
- UBA
ubiquitin associated
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-06-0391) on January 5, 2017.
REFERENCES
- Boeke D, Trautmann S, Meurer M, Wachsmuth M, Godlee C, Knop M, Kaksonen M. Quantification of cytosolic interactions identifies Ede1 oligomers as key organizers of endocytosis. Mol Syst Biol. 2014;10:756. doi: 10.15252/msb.20145422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brach T, Godlee C, Moeller-Hansen I, Boeke D, Kaksonen M. The initiation of clathrin-mediated endocytosis is mechanistically highly flexible. Curr Biol. 2014;24:548–554. doi: 10.1016/j.cub.2014.01.048. [DOI] [PubMed] [Google Scholar]
- Carroll SY, Stimpson HEM, Weinberg J, Toret CP, Sun Y, Drubin DG. Analysis of yeast endocytic site formation and maturation through a regulatory transition point. Mol Biol Cell. 2011;23:657–668. doi: 10.1091/mbc.E11-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SY, Stirling PC, Stimpson HEM, Giesselmann E, Schmitt MJ, Drubin DG. A yeast killer toxin screen provides insights into a/b toxin entry, trafficking, and killing mechanisms. Dev Cell. 2009;17:552–560. doi: 10.1016/j.devcel.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RE, Patterson JC, Goupil LS, Thorner J. Dynamic localization of Fus3 mitogen-activated protein kinase is necessary to evoke appropriate responses and avoid cytotoxic effects. Mol Cell Biol. 2010;30:4293–4307. doi: 10.1128/MCB.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RE, Thorner J. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1773:1311–1340. doi: 10.1016/j.bbamcr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Confalonieri S, Di Fiore PP. The Eps15 homology (EH) domain. FEBS Lett. 2002;513:24–29. doi: 10.1016/s0014-5793(01)03241-0. [DOI] [PubMed] [Google Scholar]
- Cupers P, ter Haar E, Boll W, Kirchhausen T. Parallel dimers and anti-parallel tetramers formed by epidermal growth factor receptor pathway substrate clone 15. J Biol Chem. 1997;272:33430–33434. doi: 10.1074/jbc.272.52.33430. [DOI] [PubMed] [Google Scholar]
- Dores MR, Schnell JD, Maldonado-Baez L, Wendland B, Hicke L. The function of yeast epsin and Ede1 ubiquitin-binding domains during receptor internalization. Traffic. 2010;11:151–160. doi: 10.1111/j.1600-0854.2009.01003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M, Marini F, Gamba D, Lucchini G, Plevani P. The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol Cell Biol. 1994;14:923–933. doi: 10.1128/mcb.14.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagny B, Wiederkehr A, Dumoulin P, Winsor B, Riezman H, Haguenauer-Tsapis R. A novel EH domain protein of Saccharomyces cerevisiae, Ede1p, involved in endocytosis. J Cell Sci. 2000;113:3309–3319. doi: 10.1242/jcs.113.18.3309. [DOI] [PubMed] [Google Scholar]
- Godlee C, Kaksonen M. Review series: from uncertain beginnings: initiation mechanisms of clathrin-mediated endocytosis. J Cell Biol. 2013;203:717–725. doi: 10.1083/jcb.201307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL, Eskin JA, Wendland B. Actin and endocytosis in budding yeast. Genetics. 2015;199:315–358. doi: 10.1534/genetics.112.145540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Layton AT, Savage NS, Howell AS, Carroll SY, Drubin DG, Lew DJ. Modeling vesicle traffic reveals unexpected consequences for Cdc42p-mediated polarity establishment. Curr Biol. 2011;21:184–194. doi: 10.1016/j.cub.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lu R, Drubin DG, Sun Y. Clathrin-mediated endocytosis in budding yeast at a glance. J Cell Sci. 2016;129:1531–1536. doi: 10.1242/jcs.182303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Báez L, Dores MR, Perkins EM, Drivas TG, Hicke L, Wendland B. Interaction between Epsin/Yap180 adaptors and the scaffolds Ede1/Pan1 is required for endocytosis. Mol Biol Cell. 2008;19:2936–2948. doi: 10.1091/mbc.E07-10-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miliaras NB, Wendland B. EH proteins: multivalent regulators of endocytosis (and other pathways) Cell Biochem Biophys. 2004;41:295–318. doi: 10.1385/CBB:41:2:295. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Newpher TM, Smith RP, Lemmon V, Lemmon SK. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev Cell. 2005;9:87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Peng Y, Grassart A, Lu R, Wong CCL, Yates J, Barnes G, Drubin DG. Casein kinase 1 promotes initiation of clathrin-mediated endocytosis. Dev Cell. 2015;32:231–240. doi: 10.1016/j.devcel.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reider A, Barker SL, Mishra SK, Im YJ, Maldonado-Báez L, Hurley JH, Traub LM, Wendland B. Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J. 2009;28:3103–3116. doi: 10.1038/emboj.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson HEM, Toret CP, Cheng AT, Pauly BS, Drubin DG. Early-arriving Syp1p and Ede1p function in endocytic site placement and formation in budding yeast. Mol Biol Cell. 2009;20:4640–4651. doi: 10.1091/mbc.E09-05-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Toshima JY, Toshima J. Regulation of clathrin coat assembly by Eps15 homology domain–mediated interactions during endocytosis. Mol Biol Cell. 2012;23:687–700. doi: 10.1091/mbc.E11-04-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Processing. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- Toshima JY, Toshima J, Kaksonen M, Martin AC, King DS, Drubin DG. Spatial dynamics of receptor-mediated endocytic trafficking in budding yeast revealed by using fluorescent α-factor derivatives. Proc Natl Acad Sci USA. 2006;103:5793–5798. doi: 10.1073/pnas.0601042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg J, Drubin DG. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 2011;22:1–13. doi: 10.1016/j.tcb.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B, McCaffery JM, Xiao Q, Emr SD. A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J Cell Biol. 1996;135:1485–1500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.