A major question in cytokinesis research is the precise architecture of actin and nonmuscle myosin II filaments in the animal cell contractile ring. Superresolution and transmission electron microscopy support an actomyosin organization consistent with the sliding-filament/purse-string model of cytokinesis.
Abstract
Despite recent advances in our understanding of the components and spatial regulation of the contractile ring (CR), the precise ultrastructure of actin and myosin II within the animal cell CR remains an unanswered question. We used superresolution light microscopy and platinum replica transmission electron microscopy (TEM) to determine the structural organization of actin and myosin II in isolated cortical cytoskeletons prepared from dividing sea urchin embryos. Three-dimensional structured illumination microscopy indicated that within the CR, actin and myosin II filaments were organized into tightly packed linear arrays oriented along the axis of constriction and restricted to a narrow zone within the furrow. In contrast, myosin II filaments in earlier stages of cytokinesis were organized into small, discrete, and regularly spaced clusters. TEM showed that actin within the CR formed a dense and anisotropic array of elongate, antiparallel filaments, whereas myosin II was organized into laterally associated, head-to-head filament chains highly reminiscent of mammalian cell stress fibers. Together these results not only support the canonical “purse-string” model for contractile ring constriction, but also suggest that the CR may be derived from foci of myosin II filaments in a manner similar to what has been demonstrated in fission yeast.
INTRODUCTION
Early thin-section transmission electron microscopy (TEM) studies by Schroeder (1970, 1972, 1973) and others (Perry et al., 1971; Sanger and Sanger, 1980; Maupin and Pollard, 1986; Mabuchi et al., 1988) helped establish that cytokinesis in animal cells was mediated by a circumferential contractile ring (CR) of actin and putative nonmuscle myosin II filaments. Many of these studies examining the morphology of the CR hypothesized that the organization of actin and myosin II was similar to that seen in muscle sarcomeres and that this arrangement facilitated a sliding filament–based purse-string contraction mechanism for ring constriction. However, clear evidence of the exact architecture of actin and myosin II in the CR was lacking in this earlier work, and more recent studies called into question the purse-string CR model and suggested that myosin II–based actin filament translocation may not be necessary (Ma et al., 2012) and that a circumferentially organized CR is not responsible for cytokinesis in certain animal cell types (Fishkind and Wang, 1993; Debiasio et al., 1996; Reichl et al., 2008). Although the ultrastructure of the constituent actin filaments in the CR of fission yeast has been demonstrated (Kamasaki et al., 2007), the precise arrangement of actin and myosin II filaments in the CR of animal cells has remained elusive and a crucial unanswered question (see reviews by Eggert et al., 2006; Pollard, 2010; Green et al., 2012). Recent work by Fenix et al. (2016) using superresolution light microscopy in live and fixed HeLa cells suggested that myosin II filament assemblages or stacks are present in the CR and apparently oriented appropriately for a purse-string contraction mechanism.
Echinoderm embryos have served as the basis for many foundational studies on cytokinesis, including some of the initial TEM-based characterizations of the CR (Schroeder, 1972) and on the role of the mitotic apparatus in cleavage plane specification (Rappaport, 1961), as well as in providing evidence that myosin II is essential for the contractile force for cytokinesis (Mabuchi and Okuno, 1977) and demonstrating the involvement of RhoA in the regulation of cytokinesis (Mabuchi et al., 1993; Bement et al., 2005). Contractile rings of first-division-cycle sea urchin embryos may be isolated by adhering dividing embryos to poly-l-lysine–coated coverslips and then applying a stream of buffer to isolate cortices. This method affords the unusual opportunity to examine the CR from the vantage point of the cytoplasmic face of the plasma membrane, and a number of investigators have used these isolated cleavage cortices to examine CR component structure and function (Yonemura and Kinoshita, 1986; Schroeder and Otto, 1988; Otto and Schroeder, 1990; Bonder et al., 1988; Mabuchi, 1994; Nishimura and Mabuchi, 2003; Uehara et al., 2008).
In the present study, we combined superresolution three-dimensional (3D) structured illumination light microscopy (SIM; Gustafsson et al., 2008; Kner et al., 2009) with critical-point dry and rotary shadow platinum replica TEM (Svitkina et al., 1995; Svitkina, 2007) to reveal the ultrastructural organization of CR actin and myosin II filaments in the contractile rings of first-division sea urchin embryos. Our SIM-based localization of myosin II filaments shows that their structural organization undergoes a transformation in concert with the maturation of the ring, transitioning from a wide zone of separate clusters to a narrower band of linear aligned filaments—a reorganization that appears to depend on the presence of actin filaments. Using platinum replica TEM, we show for the first time that the ultrastructural arrangement of CR myosin II filaments corresponds to bundles of longitudinally connected filaments oriented parallel to the cleavage plane. We also demonstrate that actin filaments in the CR are arrayed in a dense, aligned mat of elongate, unbranched filaments that comprise a precisely defined band oriented parallel to the plane of division. Taken together, these results extend our knowledge of CR actin and myosin II filament ultrastructure and support the canonical purse-spring contraction model for the mechanism of cytokinesis in large spherical and suspended cells such as echinoderm embryos.
RESULTS
The CRs in isolated cortices contain characteristic protein constituents
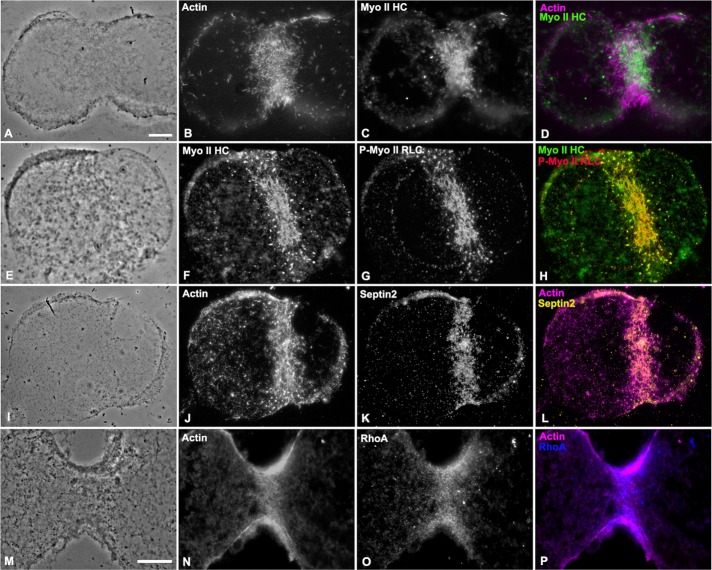
The CR contains a suite of characteristic protein constituents, including three essential filament systems consisting of the contractile machinery of actin and myosin II filaments, as well as septin filaments that serve as a membrane scaffold and CR constituent recruiting system (reviewed in Eggert et al., 2006; Green et al., 2012; Bridges and Gladfelter, 2015). Previous studies using cortices isolated from cleaving sea urchin embryos demonstrated the presence of actin and myosin II in these preparations (Schroeder and Otto, 1988; Uehara et al., 2008). We confirmed and extended these studies by using immunolabeling and wide-field fluorescence microscopy to show that isolated cleavage cortices contained actin, myosin II, and septin 2 filaments (Figure 1, A–L), along with the master regulator of CR assembly, RhoA (Figure 1, M–P). Cortices isolated from dividing blastomeres tended to take on a characteristic butterfly shape, particularly with later stages of cytokinesis, as has been previously reported (Yonemura and Kinoshita, 1986; Schroeder and Otto, 1988; Uehara et al., 2008). These preliminary characterization experiments showed that a number of the major expected components of the CR were preserved in cortical preparations and suggested that these cortices therefore represent a reasonable model of the in situ CR. However, as illustrated in Figure 1, conventional, diffraction-limited microscopy does not allow for a determination of the precise structural organization of CR components.
FIGURE 1:
The CRs in isolated cortices contain characteristic protein constituents. Wide-field imaging of immunofluorescently labeled cleavage cortices reveals that the CR regions contain the expected protein constituents, namely actin (B, J, N; magenta in D, L, P), myosin II heavy chain (Myo II HC in C, F; green in D, H), Ser-19–phosphorylated myosin II regulatory light chain (P-Myo II RLC in G; red in H), septin 2 (K; yellow in L), and RhoA (O; blue in P). Phase contrast images (A, E, I, M) show the indented/butterfly-like appearance of cortices isolated during cytokinesis. Bars, 10 µm (A, M); magnification equivalent in A–L.
Characterization of two myosin II antibodies that allow for visualization of filament orientation
Owing to the resolution limitations associated with conventional microscopy, we used 3D SIM imaging in combination with two antibody-based fluorescent probes for myosin II filaments to investigate the architecture of myosin II filaments during cortical CR assembly. Previous work demonstrated that SIM can resolve the motor domain/head and tail/rod regions of myosin II bipolar filaments fluorescently labeled with appropriate probes (Beach et al., 2014; Burnette et al., 2014; Fenix et al., 2016). The myosin II–specific antibodies we used consisted of a commercial monoclonal anti–phospho Ser-19 myosin II regulatory light chain (RLC) antibody that we used as a tag for the myosin II head/motor domain. Past studies using immuno–electron microscopy with phospho-myosin II RLC antibodies (Shutova et al., 2014), as well as fluorescence-based SIM localization with a myosin II RLC antibody (Fenix et al., 2016), demonstrated that these types of antibodies label the head domains of myosin II bipolar filaments. The other anti–myosin II antibody we used was a rabbit polyclonal antibody raised against the purified heavy chain of sea urchin egg myosin II (Henson et al., 1999). We characterized the labeling patterns of these two myosin II antibodies by immunolabeling the relatively simple array of myosin II filaments that exists in the cytoskeleton of sea urchin coelomocytes (Supplemental Figure S1). As we previously demonstrated (Henson et al., 1999), myosin II filaments in platinum replica TEM of coelomocytes treated with gelsolin to remove actin filaments were restricted to a sparse distribution near the nucleus (Supplemental Figure S1, A and B). These myosin II filaments displayed a characteristic spectrum of filament organizations, including end-to-end chains, laterally associated stacks, and/or networks, with the occasional isolated individual bipolar filament (Supplemental Figure S1, A and B). The 3D SIM imaging of coelomocytes labeled with the two myosin II antibodies demonstrated that in both multifilament assemblages as well as in individual filaments, the phospho–myosin II RLC antibody (red in Supplemental Figure S1, C–F) labeled the head domain ends, whereas the myosin II heavy chain antibody (green in Supplemental Figure S1, C–F) tended to label the entire filament and therefore allowed for the discrimination of the self-associated tails/rods region. This staining pattern of red head and green tail—with overlapping regions appearing yellow—is apparent in myosin II filaments organized in longitudinal chains (Supplemental Figure S1, D and F), in networks (Supplemental Figure S1D), and as individual filaments (Supplemental Figure S1F, arrow). Direct comparison of TEM and SIM images of individual myosin II filaments (Supplemental Figure S1, G–J) strongly suggests that the antibody localization patterns are a direct reflection of myosin II filament organization, and we provide a diagram of our interpretation of the distribution of the antibody probes in Supplemental Figure S1K.
The 3D SIM imaging reveals that myosin II filament CR organization transitions from clusters, to patches, to a linearly aligned band during the cytokinesis process
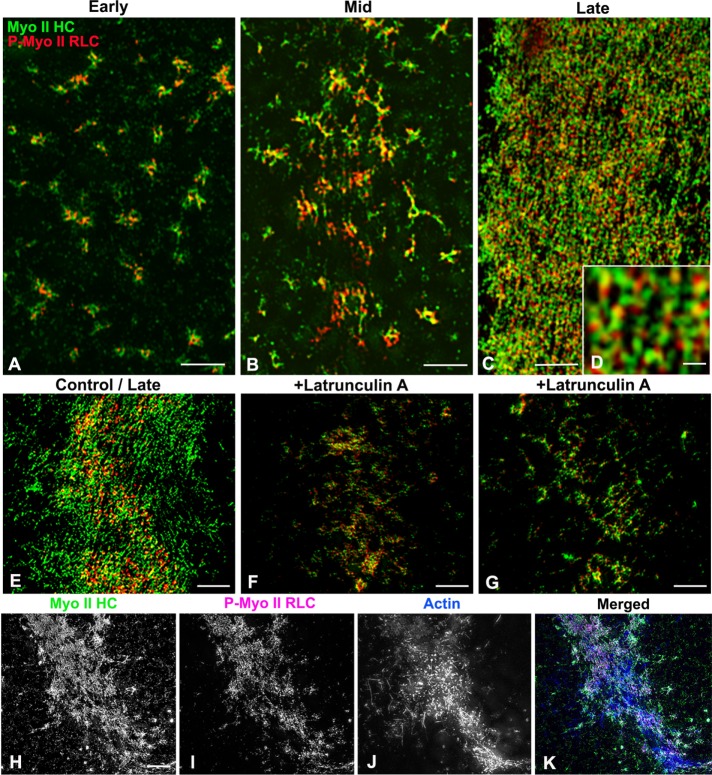
A series of cortices was isolated over the time course of cytokinesis from synchronously dividing embryos that allowed us to examine the development of myosin II CR organization over time (Figures 2 and 3). The 3D SIM imaging of isolated cleavage cortices stained with the two myosin II antibodies revealed a transformation of myosin II filament organization coincident with the development of the CR. Early cleavage cortices generally lacked the butterfly shape/indented morphology of later stages, and myosin II filaments were arranged in a broad stripe of clusters (Figures 2, A–C, and 3A). Nearest-neighbor index (NNI) analysis (Figure 2J; Clarke and Evans, 1954) indicated that the spacing of the cluster centers demonstrated a significant deviation from randomness in the direction of uniform spacing (p < 0.01). Within these clusters, a common arrangement of myosin II filaments was difficult to determine because circular, network, stellate, and linear chain patterns of filament assemblages were all present, although ring-like structures did tend to predominate (Figures 2, A–C, and 3A). In samples further along in cytokinesis, cleavage constrictions in the cortices were more noticeable, with myosin II clusters replaced by patches of more extensive myosin II staining that appeared as a network or a more linear arrangement (Figures 2, D–F, and 3B). Linear extensions of myosin II filaments from clusters were apparent at this mid stage (Figure 3B). In cortical CRs from late-stage cleavage embryos, the myosin II filaments formed an extensive and well-delineated band consisting of aligned linear arrays of myosin II filaments oriented parallel to the long axis of the CR (Figures 2, G–I, and 3C). Analysis of the width change between the distribution of the clusters in the early cortices and the linear band pattern in later cortices suggested that the CR zone became significantly narrower over time (Figure 2K). Within the linear pattern of staining in the CR of late cortices, the green and red dots associated with tails and heads of myosin II filaments, respectively, appeared to often alternate (Figures 2I, inset, and 3, C and D), suggestive of laterally associated chains of head-to-head arranged myosin II filaments, although the precise orientation of these filaments was difficult to determine, given the superposition of staining that occurs over the ∼300-nm axial resolution of the 3D SIM focal plane.
FIGURE 2:
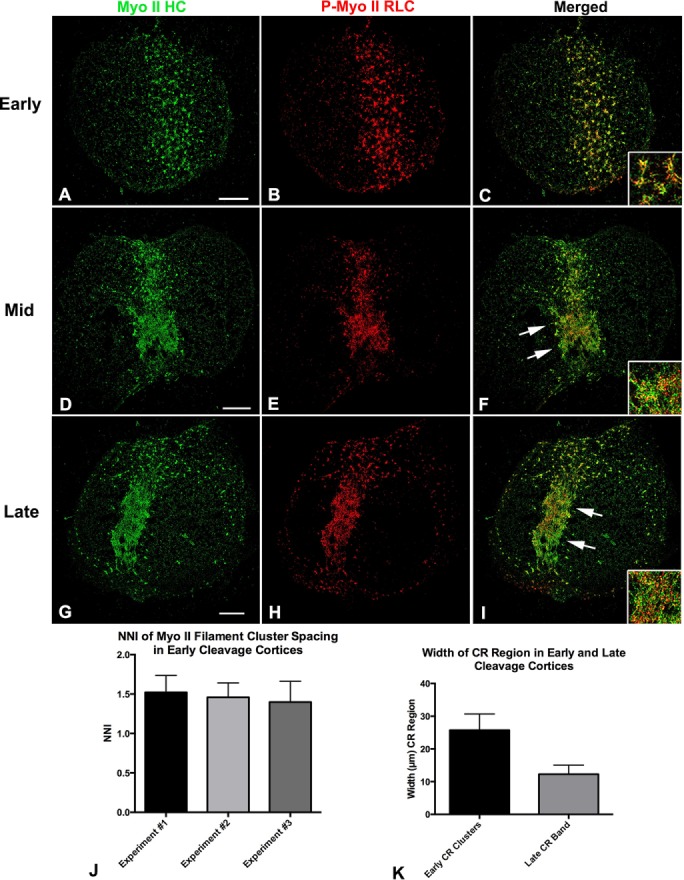
SIM imaging indicates that CR region myosin II filaments undergo a transition from clusters to linear arrays during cytokinesis. The 3D SIM through-focus projections (120 nm/slice over ∼600 nm of Z) of early-stage cortical CR regions stained for myosin II HC (A; green in C) and P-Myo II RLC (B; red in C) reveal the presence of a broad band of clusters of myosin II filaments. Higher magnification (inset in C) reveals that clusters often appear as rings with linear extensions. NNI analysis of cluster spacing in early cortices from three experiments indicates that they are distributed in a uniform/regular manner (J). Mid-stage cleavage cortices (D–F) show the presence of myosin II clusters in larger patches associated with faint linear elements (arrows). At higher magnification (inset in F). the myosin II filaments appeared to be arranged into networks. CRs in late-cleavage-stage cortices (G–I) are characterized by striking linear arrays (arrows) of aligned myosin II filaments (inset in I) in a narrower band. Measurement of the width of the overall CR region myosin II staining over time in three separate experiments (K) indicates a progressive narrowing coincident with progression from early to late-cleavage-stage cortices. Bar, 10 µm.
FIGURE 3:
SIM imaging of myosin II filament orientation in the CR emphasizes a change from clusters to linear arrays and indicates an association with actin filaments. The 3D SIM through-focus projections (120 nm/slice over ∼600 nm of Z) of early-stage (A), mid-stage (B), and late-stage (C) cleavage cortices stained with MyoII-HC (green) and P-Myo II RLC (red) antibodies. Early cortices (A) contain ring-shaped clusters of myosin II filaments, whereas in mid-stage cortices (B), the clusters appear to have linear extensions and have interconnected into networks. In late-stage cortices, the CR region contains an apparent linear arrangement of myosin II filaments that at high magnification (D) resolves into an alternating pattern of green and red staining, suggesting that myosin II filaments are in longitudinal chains of head-to-head–oriented filaments. Treatment with the actin-disrupting drug latrunculin A allows myosin II to assemble in the CR region (F, G), but the myosin arrangement does not reach the linearized pattern seen in the cortices isolated from equivalent-time-point untreated embryos (E). Triple labeling of a cortical CR with fluorescent phalloidin (J, blue in K), Myo II HC (H, green in K), and P-Myo II RLC (I, magenta in K) reveals that actin filaments concentrate in the CR in concert with myosin II filaments. Bars, 2 μm (A–C), 200 nm (D), 5 μm (E–K).
With regard to the relationship of myosin II and actin filaments in the CR, we also performed 3D SIM imaging on cleavage cortices triple labeled for myosin II heavy chain, the phospho-myosin II RLC, and actin filaments and noted that all three labels concentrated in the CR region (Figures 3, H–K), although actin filament imaging in the CR is complicated by the presence of actin filaments in the core bundles of surface microvilli. Furthermore, we used treatment with the actin filament–disrupting agent latrunculin A before cytokinesis to test the effect of the lack of actin filaments on myosin II organization. Unlike the linearized array of myosin II apparent in untreated control cortices (Figure 3E), latrunculin A–treated cortices isolated at equivalent time points displayed a band of myosin II clusters and patches (Figure 3, F and G). This suggests that myosin II filaments are recruited and can partially organize in the CR region in an actin filament–independent manner, but that actin filaments may be required for the linearized organization seen in late-stage cortices.
Platinum replica TEM of cortical CRs reveals an ordered, circumferentially arranged filament array
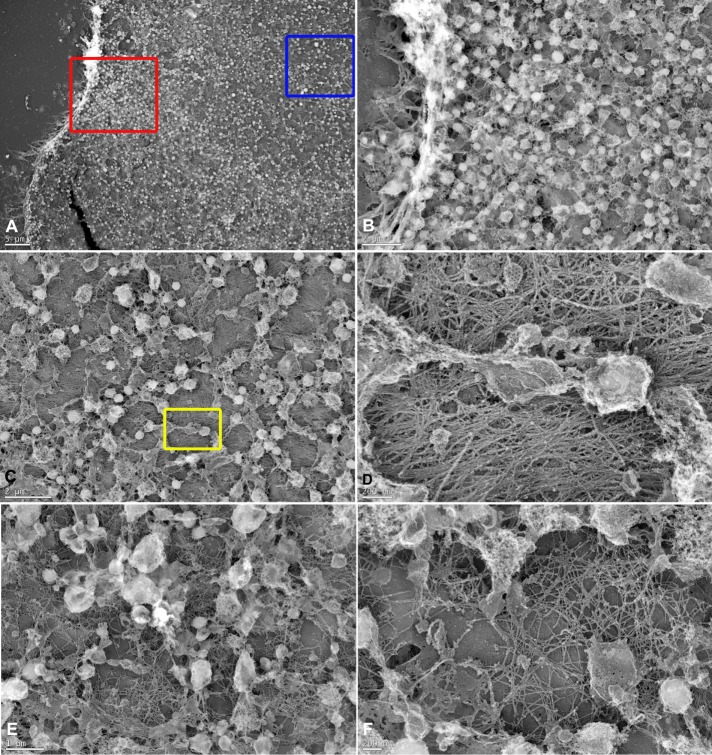
The unusual accessibility of the CR in isolated sea urchin embryo cortices allowed us to perform platinum replica TEM on these preparations to obtain further insight into the arrangement of actin and myosin II filaments. We initially examined cortices that had been isolated and then fixed in the absence of detergent extraction (Figure 4). In these cortices, numerous membranous organelles (acidic vesicles/pigment granules, segments of the endoplasmic reticulum [ER], and clathrin-coated endocytic vesicles) remained associated with and tended to obscure the cytoplasmic face of the plasma membrane (Figure 4, A–F). However, high-magnification images in the region of a presumptive CR showed the presence of a dense mat of roughly aligned parallel filaments, many of which had the ultrastructural appearance of actin filaments (Figure 4D). We also noted that the submembranous filaments present in areas outside of the CR region tended to be isotropic in orientation (Figure 4, E and F).
FIGURE 4:
TEM of non–detergent-extracted cortices reveals submembranous actin-like filaments and membranous organelles in the CR region. (A) Low-magnification view of an indented cortex with microvilli on the edge and the surface covered with membranous vesicles and ER remnants. The red box appears at higher magnification in B, and the blue box appears at higher magnification in C. (B) Microvilli and numerous vesicles are evident in this image with some limited views of the cytoplasmic face of the plasma membrane. (C) Examination of an area with fewer vesicles at higher magnification reveals an underlying mat of actin-like filaments with an overall parallel alignment that we interpret as the presumptive CR. The yellow box appears at higher magnification in D. (D) High-magnification view of the submembranous actin-like filaments in putative contractile ring, showing that the majority are aligned into uniform bundles, although there are filaments running in other directions. Note the vesicles, remnants of the ER (center), and a clathrin-coated vesicle (top, center). (E, F) In regions not associated with the CR, the cortex still contains many submembranous actin-like filaments (E, low magnification; F, high magnification), although they assume a more isotropic arrangement. Scale bars as indicated.
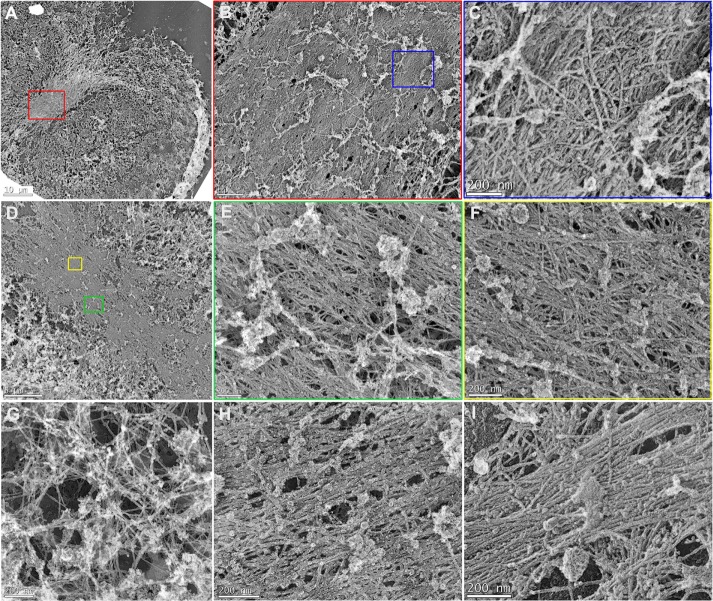
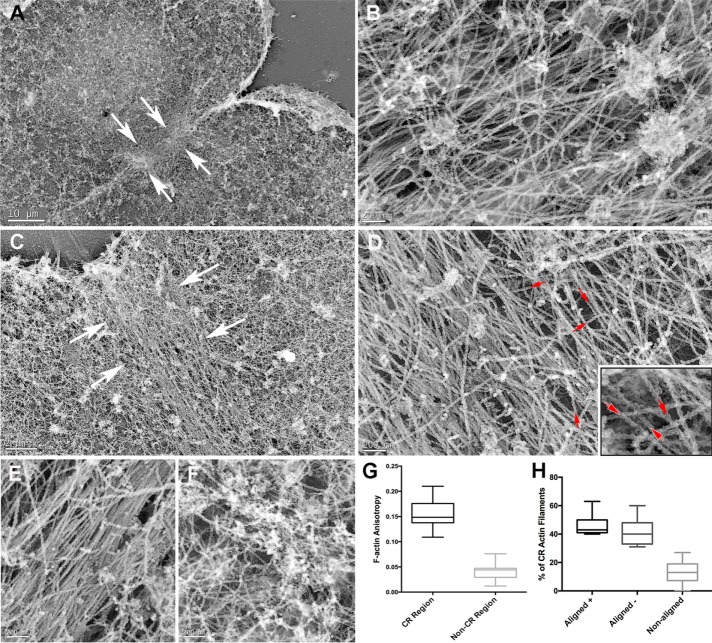
Combining fixation with detergent extraction allowed for a clearer view of actin-like filaments in the CR of isolated cortices (Figure 5). The filament arrays were quite pronounced in later–cleavage stage cortices (Figure 5A), with well-defined margins and a width range of 10–15 µm, and resembled the 3D SIM–based images of the CR in late-stage cortices (Figures 2 and 3). The dense concentration of filaments within these CRs appeared unbranched, elongate and aligned in parallel arrays (Figure 5, C, E, F, and H). However, the orientation of filaments was not completely uniform, and intercalated within the packed assemblage of actin-like filaments were other filamentous and globular structures that did not appear actin like (Figure 5, C, E, F, and H), as well as numerous clathrin-coated structures (Figure 5, C and F, and Supplemental Figure S2, D and E). The overall orientation of all the filaments tended to be parallel with the cleavage plane, suggesting a circumferential alignment of filaments within the CR, as previously suggested by earlier-thin section TEM studies of sea urchin embryos (Schroeder, 1972; Usui and Yoneda, 1982). Comparison of CR filaments in sea urchin embryo cortices (Figure 5H) with filaments in the stress fibers of LLC-PK1 mammalian cultured cells (Figure 5I) emphasizes the similarity of the appearance of the filaments in these two contractile structures from significantly different cell types.
FIGURE 5:
TEM of detergent-extracted cortices shows that the CR consists of a closely packed array of actin-like filaments. A dense mat of filaments is apparent in the CR region of buttery shaped (A) and detergent-extracted cleavage cortices seen at low (A, D), medium (B, from red box in A; E, from green box in D), and high (C, from blue box in B; F, from yellow box in D) magnification. The CR region consists of tightly packed, elongate, largely unbranched, and aligned actin-like filaments, with other filament types and globular structures (H) intercalated into the structure. Note that the majority of CR filaments appear aligned parallel to the axis of constriction of the cleavage furrow and that clathrin-coated vesicles and membranous remnants are also present. In areas of cortices not associated with the CR, isotropic arrangements of actin-like filaments predominate (G). Comparison of the actin-like filament assemblage in sea urchin CR (H) with a mammalian LLC-PK1 cell stress fiber (I) emphasizes the similarity between these two structures. Scale bars as indicated.
TEM ultrastructure of CR actin filaments revealed by myosin S1 decoration
To definitively identify actin filaments within the CRs, we treated isolated, detergent-extracted cortices with myosin S1 before fixation and platinum replica processing. In these cortices (Figure 6), CR filaments displayed the characteristic “twisted-rope” structure of myosin S1–decorated actin filaments (Svitkina et al., 1995). These densely packed CR actin filaments concentrated in a well-defined band within the furrow (Figure 6, A and C) and appeared elongate, unbranched, and aligned parallel to the cleavage plane (Figure 6, B and D). Myosin S1 decoration allowed us to quantify the actin filament anisotropy in regions within the CR (Figure 6E) and outside of it where filament orientations appeared random (Figure 6F). This analysis showed that the extent of F-actin anisotropy was statistically significantly greater (p < 0.001) within the CR than in other regions of the cortex (Figure 6G). Analysis of the polarity of S1-decorated actin filaments within the CR revealed an apparent mixed polarity (red arrows in Figure 6D, inset). Quantitative analysis of filament polarity on ∼300 S1-decorated actin filaments in the CR region of 10 different cortices from two separate experiments (Figure 6H) showed roughly similar percentages of plus and minus end–oriented filaments that were aligned with the CR axis (44 vs. 40%) with fewer off-axis filaments (16%).
FIGURE 6:
TEM of myosin S1–labeled cortices allows for the definitive identification of CR actin filaments. Low- (A, C), and high- (B, D, inset in D) magnification images of the CR regions (demarcated by white arrows in A, C) of two indented cleavage cortices shows the characteristic twisted-rope appearance of myosin S1–decorated actin filaments. Within the CR regions, these actin filaments appear elongate, unbranched, and densely packed, with the majority aligned parallel to the cleavage plane. The actin filament orientation appears mixed, as demonstrated by the red arrows in D and the inset in D. (G) Quantitative analysis of F-actin anisotropy in five different cortices measured within (E) and outside (F) of the CR region. Note that F-actin anisotropy is statistically significantly higher (p < 0.0001) within the CR region. (H) The roughly similar percentages of plus and minus end–oriented filaments that were aligned with the CR axis, with a smaller number of off-axis filaments. Scale bars as indicated.
Dividing sea urchin embryos contain microvilli (Schroeder, 1978) with core bundles of elongate actin filaments (Burgess and Schroeder, 1977). One potential complication of performing platinum replica TEM on detergent-extracted cleavage cortices was trying to distinguish between CR actin filaments and actin filaments in the core bundles of microvilli trapped between the embryo and the coverslip. However, fortuitous tears in the CR that occurred during the cortex isolation allowed for imaging beneath the mat of CR filaments and revealed the underlying presence of microvillar core actin bundles (Supplemental Figure S2, A–C). In addition, the microvillar core actin bundles have a distinct tightly packed structure due to cross-linking via the bundling protein fascin (Otto et al., 1980) and therefore do not closely resemble the more loosely packed CR actin filaments. However, at this point, we cannot rule out the possibility that MV core actin rootlets may integrate into the CR actin filament assemblage.
TEM ultrastructure of CR myosin II filaments revealed by gelsolin treatment
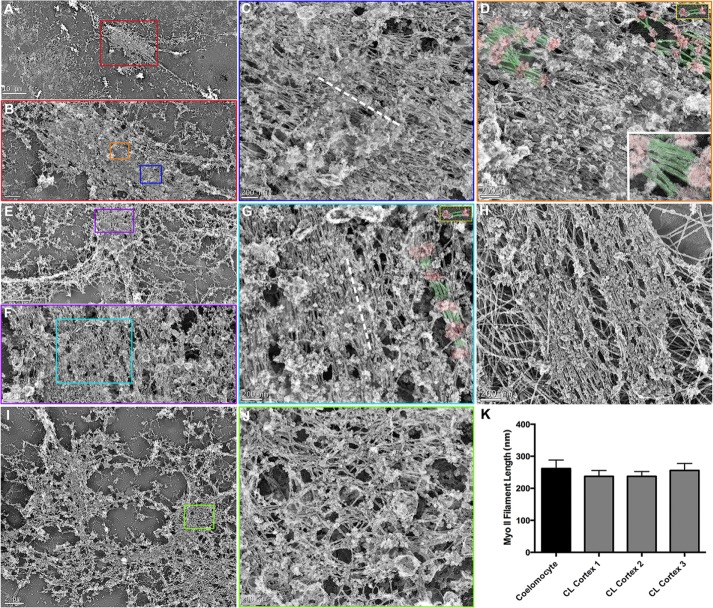
As we showed earlier with platinum replica TEM of coelomocyte cytoskeletons (Supplemental Figure S2), myosin II filaments can be clearly identified after the removal of actin filaments by treatment with the actin-severing protein gelsolin (Svitkina et al., 1989; Henson et al., 1999). Of importance, Otto and Schroeder (1988) showed and we confirmed that gelsolin treatment does not noticeably alter the light microscopic organization of myosin II filaments seen in the CRs of immunolabeled cortices. In addition, Verkhovsky et al. (1995) established through correlative light and electron microscopy that myosin II organization imaged in live cells is identical to the TEM images of the same cells after gelsolin treatment. We used gelsolin treatment on isolated and detergent-extracted cortices and discovered CR-like structures that consisted of dense assemblages of aligned myosin II bipolar filaments (Figure 7). The identity of these myosin II filaments was based on their well-established ultrastructural signature in platinum replica TEM (Svitkina et al., 1989, 1997; Henson et al., 1999, Hoelzle and Svitkina, 2012; Shutova et al., 2012, 2014), with an overall dumbbell appearance with a smooth middle zone of rods/tails separating groups of globular heads at either end (Supplemental Figure S1, H and I, and Figure 7, D, inset in D, and G) and a length range of 200–300 nm (Figure 7K). Comparison of myosin II filament lengths between coelomocytes and three cortices from two separate experiments (n = 20 filaments in each condition) indicates a similar length distribution (Figure 7K). The myosin II filaments tended to align in parallel with the long axis of the CR/cleavage plane (dashed white lines in Figure 7, C and G) and in regions where the filaments were sparse enough that the structural organization appeared to be a head-to-head longitudinal arrangement (Figures 7, D and G), although this was not always clearly defined. We saw longitudinal chains of myosin II filaments in coelomocytes (Supplemental Figure S1; Henson et al., 1999), and others reported myosin II filaments in this type of arrangement in different cell types (Svitkina et al., 1989, 1995, 1997; Shutova et al., 2012). In addition, the correlation between the SIM images of CR myosin II filament linear arrays (Figures 2, G–I, and 3C) and the TEM images (Figure 7) reinforces the conclusion that these are myosin II bipolar filaments. Of interest, the structural pattern of myosin II filaments seen in the TEM of cortical CRs (Figure 7G) was difficult to distinguish from the arrangement present in stress fibers of gelsolin-treated LLC-PK1 cells (Figure 7H) and other mammalian cell types (Verkhovsky et al., 1987; Svitkina et al., 1989; Shutova et al., 2012). Given the contractile nature of the actomyosin assemblage in stress fibers (Katoh et al., 2001), it is not surprising that the filament organization in these structures has some resemblance to the CR.
FIGURE 7:
TEM of detergent-extracted, gelsolin-treated cortices reveals tightly packed bundles of myosin II filaments within the CR. Linear arrays of myosin II filaments are evident in detergent-extracted and gelsolin-treated cleavage cortices imaged at low (A, E), medium (B, from red box in A; F, from purple box in E), and high (C, from blue box in B; D, from orange box in B; G, from cyan box in F) magnification. The myosin II filaments are recognized by their characteristic appearance of globular heads and smooth rod/tail regions, their 200–300 nm length (K), and their alignment into head-to-head–associated longitudinal chains. In D and G, a subset of myosin II filament heads have been colored red and tails/rods green, and the gold-bordered insets at the top right show similarly colored isolated myosin II filaments from coelomocytes for comparison. The bottom right inset in D is a high-magnification view of an array of myosin II filaments from a similarly colored cortical CR. The myosin II filaments within the CR region tend to align parallel to the long axis of the CR/cleavage plane (axis indicated by dashed white line in C and G). Myosin II filament organization in the CRs is very similar in appearance to the array of myosin II filaments seen in TEM of gelsolin-treated stress fibers from cultured mammalian LLC-PK1 cells (H; the large filaments in the background are intermediate filaments). In some cortices, myosin II filaments are arrayed in a more isotropic network organization (I, low magnification; J, high magnification from green box in I), which are reminiscent of the patches of myosin II filament networks seen in SIM images of mid-stage cortices (Figures 2 and 3). Scale bars as indicated.
SIM imaging revealed the presence of distinct myosin II clusters or foci in early furrows (Figures 2 and 3). Distinguishing between early and later CR structures in platinum replica TEM was more difficult, given that the recognition of a cortex from a dividing embryo in the TEM was based primarily on its butterfly shape that is typically only apparent later in cytokinesis. However, we were able to observe early-stage, gelsolin-treated CRs using TEM where patches of myosin II filaments (Figure 7, I and J) were observed similar to the patches seen in mid-stage cortices imaged by 3D SIM (Figures 2, D–F, and 3B). It is important to note that both the SIM imaging and the platinum replica TEM indicated that the myosin II filaments in these patches were not aligned into the parallel linear arrays seen in CRs of late-stage cortices (Figures 2I, 3C, and 7, A–H) but instead tended to form isotropic networks (Figure 7, I and J).
DISCUSSION
The combined efforts of many investigators have led to a near-complete parts list for the CR and a consensus on how the cleavage plane is specified (reviewed in Eggert et al., 2006; Green et al., 2012). However, achieving a mechanistic understanding of how the ring assembles and constricts in animal cells requires that the precise arrangement of contractile proteins be determined. A number of studies using thin-section TEM demonstrated the presence of aligned arrays of actin-like filaments—in some cases, conclusively identified using myosin S1 decoration—in the CR of mammalian cells (Schroeder, 1970, 1973; Sanger and Sanger, 1980; Maupin and Pollard, 1986) and sea urchin embryos (Schroeder, 1972; Usui and Yoneda, 1982; Yonemura et al., 1991). A subset of these works has documented that the CR actin filament ultrastructure is associated with arrays of other filaments tentatively identified as nonmuscle myosin II (Schroeder, 1973; Sanger and Sanger, 1980; Maupin and Pollard, 1986). The majority of these studies concluded that actin and myosin II filaments were structured in a manner consistent with a sliding-filament/purse-string mechanism for cytokinesis. However, despite the high quality of the thin-section TEM in these studies, this previous work was unable to clearly demonstrate the structural organization of actin and myosin II filaments within the CR.
In the present study, we used isolated cortices from synchronously dividing sea urchin embryos combined with superresolution 3D SIM light microscopy and platinum replica TEM to extend our knowledge of the architecture of actin and myosin II filaments in the CR. With regard to CR actin filaments, our TEM images demonstrate that actin filaments are structured into a well-defined, dense mat of tightly packed, largely anisotropic, unbranched, and elongate filaments (Figures 4– 6). The long axis of these actin filaments aligned parallel to the axis of cleavage (Figures 5 and 6), and myosin S1 decoration indicated the expected presence of antiparallel actin filaments (Figure 6D) as previously reported in the CR (Schroeder, 1973; Sanger and Sanger, 1980). It was interesting to note the significant similarity between the platinum replica TEM images of actin filaments within the sea urchin CR and those in stress fibers of cultured mammalian cells (Figure 5, H and I), two structures that depend on the formin-based nucleation of elongated actin filaments for their formation (reviewed in Pollard, 2010; Tojkander et al., 2012).
In terms of myosin II filament organization, our 3D SIM images of sea urchin embryo cortex CRs suggest a progression of myosin II filament arrangement from regularly spaced clusters in early stages, to patches of networks in mid stages, to tight bands of linearly aligned end-to-end chains in late stages (Figures 2 and 3). This overall progression from a broad to a narrow equatorial distribution has been identified as a general feature of CR myosin II organization in a wide range of cell types (Werner et al., 2007; Lewellyn et al., 2011; Szafer-Glusman et al., 2011; reviewed in Green et al., 2012), including sea urchin embryos (Mabuchi, 1994; Foe and von Dassow, 2008), with the implication being that it is driven by the interaction of myosin II filaments with other constituents of the CR, such as actin filaments. Our demonstration that treatment with latrunculin A does not interfere with myosin II filament recruitment to the CR region but does inhibit the transition to a linearized organization corroborates and extends previous work in sea urchin embryos (Foe and von Dassow, 2008).
With regard to the clusters present in the CR regions of early-cleavage-stage sea urchin cortices, previous studies using confocal imaging of whole embryos (Foe and von Dassow, 2008) as well as confocal (Maupin et al., 1994; Werner et al., 2007; Wollrab et al., 2016) and total internal reflection fluorescence (Zhou and Wang, 2008) microscopy in other animal cell types have reported the presence of punctate myosin II staining in the CR region. In fission yeast, nodes of the myosin II protein Myo2 contribute to the assembly of the CR (Laplante et al., 2015; reviewed in Pollard and Wu, 2010; Lee et al., 2012), and the intriguing possibility exists that the clusters we see in sea urchin cortex CR regions and the puncta present in other animal cells correspond to similar structures. It remains to be demonstrated in the sea urchin CR whether the myosin II clusters are organized similarly to what was recently reported for fission yeast nodes (Laplante et al., 2016) and whether they coalesce into the linear CR structure over time as is the case in fission yeast. However, the intermediate levels of organization observed in mid-stage sea urchin CR regions (Figure 3B) suggest that this may be occurring.
In the CRs of late-cleavage-stage cortices, our SIM and TEM images suggest that myosin II filaments are arranged in bundles of laterally associated head-to-head chains with their long axis running parallel to the cleavage plane (Figures 2, 3, and 7). The structuring of myosin II filaments into head-to-head longitudinal chains that can be laterally aggregated into stacks was previously demonstrated by TEM imaging in other cell types (Svitkina et al., 1989, 1995; Shutova et al., 2012), as well as in vitro (Billington et al., 2013). The arrangement of CR myosin II bipolar filaments that we report is in general agreement with the recent SIM-based studies of Beach et al. (2014) and Fenix et al. (2016), which both showed arrays of aligned myosin II filaments in the CR of cultured mammalian cells. In Fenix et al. (2016), the myosin II filaments are grouped into stacks, whereas in our SIM and TEM imaging, it appeared that the myosin II arrangement was not as well ordered with regard to the registration of the head and rod/tail domains of the bipolar filament assemblages. We hypothesize that the differences between these CR myosin II organizations are due to the differences inherent in cytokinesis in a large, spherical, and suspended embryo versus a small, substrate-attached tissue culture cell. As was the case with actin filaments, we noted a remarkable resemblance between the myosin II filament organization in the sea urchin CR and that present in the mammalian cell stress fibers that we imaged by TEM (Figures 7, G and H), as well as the previously published TEM images of stress fibers (Verkhovsky et al., 1987; Svitkina et al., 1989, 1995). This suggests a level of conservation of actin and myosin II architecture in contractile structures present in widely disparate cell types and supports the early thin-section TEM work by Sanger and Sanger (1980) and more recent SIM-based studies by Burnette and coworkers (Burnette et al., 2014; Fenix et al., 2016) that emphasize the similar structural organization of the CR and stress fibers.
The improved view of CR actin and myosin II filament structure that we provide in the present study for sea urchin embryos is consistent with the canonical sliding-filament/purse-string mechanism for cytokinesis hypothesized by a large number of earlier studies. In this model, assemblages of actin and myosin II filaments aligned with their long axes parallel to one another and to the plane of the cleavage furrow form sarcomere-like units that allow for the sliding of actin filaments via the motor activity of the myosin II proteins. We acknowledge that imaging of CR organization in fixed samples does not absolutely prove the mechanism of cytokinesis and are developing imaging methodologies to test this model in live embryos. We also recognize that the existence of the CR organization that we describe in a large, spherical, and suspended embryo does not exclude the possibility that other cell types may engage in alternative cytokinesis mechanisms (reviewed in Uyeda and Nagasaki, 2004; Wang, 2005). This may be particularly true for smaller cells that perform cytokinesis when attached to a substrate, in which CRs containing circumferentially oriented actomyosin arrays may not be present (Fishkind and Wang, 1993; Debiasio et al., 1996; Reichl et al., 2008), although the recent work by Fenix et al. (2016) on substrate-attached HeLa cells does suggest an arrangement of myosin II filaments consistent with purse-string contraction. We are confident that our results provide clear structural evidence for a sliding filament–based mechanism of cell division in sea urchin embryos and that this finding may be applicable to cytokinesis in a wide variety of other cell types.
MATERIALS AND METHODS
Animals, antibodies, proteins, and reagents
Strongylocentrotus droebachiensis sea urchins were obtained from the Marine Biological Laboratory (MBL) Marine Resources Center (Woods Hole, MA), Lytechinus pictus sea urchins were purchased from Marinus Biological Supply Company (Long Beach, CA), and Strongylocentrotus purpuratus sea urchins were collected from the waters surrounding Port Townsend, WA, and maintained at the Friday Harbor Laboratories (Friday Harbor, WA). All animals were kept in either running seawater or closed artificial sea water systems at 10–15°C. Primary antibodies used included a rabbit polyclonal antibody raised against sea urchin egg myosin II heavy chain precipitated from egg extracts and electrophoretically purified (Henson et al., 1999), a mouse monoclonal antibody (mAb) against the Ser-19 phosphorylated form of myosin II RLC from Cell Signaling Technology (Danvers, MA), a rabbit polyclonal antibody against a C-terminal actin peptide from Sigma-Aldrich (St. Louis, MO), a rabbit mAb against a peptide from human septin 2 from Abcam (Cambridge, MA), and a mouse mAb against RhoA from Upstate Biotechnology (Lake Placid, NY). Appropriate secondary antibodies conjugated to Alexa Fluor 488 and 568, as well as Alexa Fluor 488– and 633–phalloidin, were obtained from Molecular Probes (Eugene, OR). Purified recombinant human plasma gelsolin and rabbit muscle myosin II subfragment 1 (S1) were purchased from Cytoskeleton (Denver, CO). TEM supplies and reagents were obtained from Electron Microscopy Sciences (Hatfield, PA), and the majority of other reagents were purchased from Sigma-Aldrich.
Gamete and coelomocyte collection, cleavage cortex isolation, and cell culture
Sea urchin gametes were collected via intracoelomic injection with 0.5 M KCl, with sperm collected dry and eggs spawned in either natural sea water or MBL artificial sea water (ASW; 423 mM NaCl, 9 mM KCl, 9.27 mM CaCl2, 22.94 mM MgCl2, 25.5 mM MgSO4, 2.14 mM NaHCO3, pH 8.0) and subsequently dejellied by multiple washing with ASW. Eggs were fertilized by addition of dilute sperm, and the fertilization envelopes were removed using either 1 M glycerol or 1 M urea (both at pH 8.0) and then washed into and reared in MBL calcium-free seawater (MBL ASW minus CaCl2 and plus 1 mM ethylene glycol tetraacetic acid [EGTA]). For experiments involving actin filament disruption, embryos were treated with calcium-free seawater containing 1 µM latrunculin A 30 min before cell division. Sea urchin coelomocytes were isolated and maintained as described in Henson et al. (1992) with the coelomocyte culture medium (CCM) consisting of 0.5 M NaCl, 5 mM MgCl2, 1 mM EGTA, and 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.2. Cleavage cortices were generated using the methods of Henson and Begg (1988) and Schroeder and Otto (1988). In brief, embryos at the appropriate stage of cell division were allowed to quickly settle onto poly-l-lysine (2 mg/ml)–coated coverslips and then exposed to fluid shear force from a pipette containing an isotonic cortex isolation buffer (CIB) consisting of 0.8 M mannitol, 5 mM MgCl2, 10 mM EGTA, and 10 mM HEPES, pH 7.4. Isolated cortices were rinsed twice in CIB before being further processed for light microscopic fluorescence localization or TEM platinum replica generation. LLC-PK1 cells were obtained from the American Type Culture Collection (Manassas, VA) and cultured according to the methods of Beach et al. (2014).
Fluorescence light microscopy
For fixation, isolated cortices were treated for 5 min in 2–3% formaldehyde in CIB; coelomocytes were fixed following the methods of Henson et al. (1999). For immunolocalization of RhoA, cortices were fixed using 10% trichloroacetic acid after Yonemura et al. (2004). For blocking buffer, phosphate-buffered saline plus 2% goat serum and 1% bovine serum albumin was used, followed by incubation in appropriate primary and secondary antibodies. Cells were mounted in nonhardening Vectashield antifade mounting medium (Vector Laboratories, Burlingame, CA), and conventional epifluorescence microscopic screening of samples was performed on a Nikon (Tokyo, Japan) 80i microscope using a 60×/1.4 numerical aperture (NA) Plan Apochromatic phase contrast objective lens with digital images captured using a Photometrics CoolSnap Cf cooled charge-coupled device (CCD) camera (Roper Scientific, Tucson, AZ). For superresolution 3D SIM (Kner et al., 2009), we used a DeltaVision OMX 3D-SIM Imaging System (GE Healthcare Bio-Sciences, Pittsburgh, PA) with an Olympus (Tokyo, Japan) 60×/1.42 NA objective lens. Captured images were reconstructed using SoftWoRx software and additional linear adjustments and manipulations made using ImageJ and Photoshop (Adobe, San Jose, CA).
Platinum replica TEM
The generation of critical point-dried and rotary-shadowed platinum replicas of coelomocyte and LLC-PK1 cell cytoskeletons and isolated cleavage cortices followed methods described in Henson et al. (1999) and Svitkina (2007). Coelomocytes were prefixed in 0.001% glutaraldehyde in CCM and then extracted for 2 min in 0.5% Triton X-100 in a buffer containing 75 mM KCl, 2 mM MgCl2, 320 mM sucrose, 20 mM EGTA, and 20 mM 1,4-piperazinediethanesulfonic acid (PIPES)–KOH, pH 7.0. Cortices were extracted in CIB plus 1% Triton X-100. LLC-PK1 cells were extracted for 2 min in 0.5% Triton X-100 in PEM buffer (100 mM PIPES-KOH, pH 6.9, 1 mM MgCl2, and 1 mM EGTA) containing 4% polyethylene glycol (molecular weight, 35,000). For gelsolin treatment of cells or cortices to remove actin filaments, samples were incubated for 15 min in 0.4 µg/ml gelsolin in either CIB or PEM minus EGTA and plus 1 mM CaCl2. For myosin S1 decoration of actin filaments, isolated cortices were treated for 15 min with 0.2 mg/ml myosin S1 in CIB. Gelsolin and myosin S1 treatment conditions were based on Hoelzle and Svitkina (2012). After the foregoing treatments, coelomocytes, isolated cortices, and LLC-PK11 cells were fixed in 2.5% glutaraldehyde in 0.1 mM sodium cacodylate, pH 7.3, for >30 min.
After fixation, all samples were treated with aqueous 0.1% tannic acid, followed by aqueous 0.2% uranyl acetate. Then the samples were dehydrated in a graded ethanol series, critical point dried, and rotary shadowed with platinum and carbon. The platinum replicas were separated from glass coverslips using hydrofluoric acid, mounted on Formvar-coated grids, and observed on a JEM 1011 TEM (JEOL, Peabody, MA) operated at 100 kV. Digital images were captured with an ORIUS 832.10W CCD camera (Gatan, Warrendale, PA) and are presented in inverted contrast.
Quantification of micrographs
For calculation of the NNI of the immunofluorescently labeled myosin II clusters in early CR regions, conventional and SIM images of three cortices each from three separate experiments were first thresholded to accentuate the brightest centers of the clusters. Then distances between nearest neighbors were measured using ImageJ, and the NNI was calculated and its statistical significance determined using the method of Clarke and Evans (1954). For CR region widths, conventional and SIM images from 10 early- and late-stage cortices from three separate experiments were measured using ImageJ. For the determination of filament anisotropy in TEM images of myosin S1–decorated actin filaments, we used FibrilTool, an ImageJ plug-in that quantifies fibrillar structures (Boudanoud et al., 2014). Five total cortices from two separate experiments were measured by performing an anisotropy measurement both within and outside of the CR region. Statistical significance was then based on a t test. For an analysis of actin filament bidirectionality in the CR, we determined the orientation of ∼300 S1 decorated actin filaments in TEM images taken from 10 different cortices from two separate experiments. For each image, we calculated the percentage of filaments that were aligned with the contractile ring and oriented in either a plus or a minus direction relative to one end of the CR assemblage. We also calculated the percentage of filaments for which the orientation was discernible but not aligned with the long axis of the CR. We used ImageJ to measure the lengths of myosin II filaments in TEM images of gelsolin-treated coelomocytes and cortices. Twenty myosin II filament lengths were determined in images of two coelomocytes and three separate cortices from two experiments.
Supplementary Material
Acknowledgments
We thank Tanya Svitkina and Jonathan Chia (University of Pennsylvania, Philadelphia, PA) for expert assistance with the generation of platinum replicas and TEM imaging; Billie Swalla (University of Washington, Seattle, WA) for access to instrumentation and reagents at Friday Harbor Laboratories; Rodney Jones, Jesse Bissell, Courtney Gamache, Zoe Irons, and Erik Williams (Dickinson College, Carlisle, PA) for help with experimentation and image analysis; and the late Ray Rappaport, memory of whom continues to inspire our work on cytokinesis. This research was supported by National Science Foundation STEP Grant 0856704 to Dickinson College and collaborative research grants to J.H.H. (MCB-1412688) and C.B.S. (MCB-1412734).
Abbreviations:
- CR
contractile ring
- SIM
structured illumination microscopy
- TEM
transmission electron microscopy
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-06-0466) on January 5, 2017.
REFERENCES
- Beach JR, Shao L, Remmert K, Li D, Betzig E, Hammer JA. Nonmuscle myosin II isoforms coassemble in living cells. Curr Biol. 2014;24:1160–1166. doi: 10.1016/j.cub.2014.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement WM, Benink HA, von Dassow G. A microtubule-dependent zone of active RhoA during cleavage plane specification. J Cell Biol. 2005;170:91–101. doi: 10.1083/jcb.200501131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billington N, Wang A, Mao J, Adelstein RS, Sellers JR. Characterization of three full-length human nonmuscle myosin II paralogs. J Biol Chem. 2013;288:33398–33410. doi: 10.1074/jbc.M113.499848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonder EM, Fishkind DJ, Henson JH, Cotran NM, Begg DA. Actin in cytokinesis: formation of the contractile apparatus. Zool Sci. 1988;5:699–711. [Google Scholar]
- Boudanoud A, Burian A, Borowska-Wykre˛t D, Uyttewaal M, Wrzalik R, Kwiatkowska D, Hamant O. FibrilTool, an ImageJ plug-in to quantify fibrillar structures in raw microscopy images. Nat Protoc. 2014;9:457–463. doi: 10.1038/nprot.2014.024. [DOI] [PubMed] [Google Scholar]
- Bridges AA, Gladfelter AS. Septin form and function at the cell cortex. J Biol Chem. 2015;290:17173–17180. doi: 10.1074/jbc.R114.634444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess DR, Schroeder TE. Polarized bundles of actin filaments within microvilli of fertilized sea urchin eggs. J Cell Biol. 1977;74:1032–1037. doi: 10.1083/jcb.74.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette DT, Shao L, Ott C, Pasapera AM, Fischer RS, Baird MA, Der Loughian C, Delanoe-Ayari H, Paszek MJ, Davidson MW, Lippincott-Schwartz J. A contractile and counterbalancing adhesion system controls the 3D shape of crawling cells. J Cell Biol. 2014;205:83–96. doi: 10.1083/jcb.201311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke PJ, Evans FC. Distance to nearest neighbor as a measure of spatial relationships in populations. Ecology. 1954;35:445–453. [Google Scholar]
- Debiasio RL, Larroca GM, Post PL, Taylor DL. Myosin II transport, organization, and phosphorylation: evidence for cortical flow/solation-contraction coupling during cytokinesis and cell locomotion. Mol Biol Cell. 1996;7:1259–1282. doi: 10.1091/mbc.7.8.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert US, Mitchison TJ, Field CM. Animal cytokinesis: from parts list to mechanisms. Annu Rev Biochem. 2006;75:543–566. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- Fenix A, Taneja N, Buttler CA, Lewis J, Van Engelenburg SB, Ohi R, Burnette DT. Expansion and concatenation of nonmuscle myosin IIA filaments drive cellular contractile system formation during interphase and mitosis. Mol Biol Cell. 2016;27:1465–1478. doi: 10.1091/mbc.E15-10-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishkind DJ, Wang Y-L. Orientation and three-dimensional organization of actin filaments in dividing cultured cells. J Cell Biol. 1993;123:837–848. doi: 10.1083/jcb.123.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE, von Dassow G. Stable and dynamic microtubules coordinately shape the myosin activation zone during cytokinetic furrow formation. J Cell Biol. 2008;183:457–470. doi: 10.1083/jcb.200807128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Annu Rev Cell Dev Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- Gustafsson MG, Shao L, Carlton PM, Wang CJ, Golubovskaya IN, Cande WZ, Agard DA, Sedat JW. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94:4957–497. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JH, Begg DA. Filamentous actin organization in the unfertilized sea urchin egg cortex. Dev Biol. 1988;127:338–348. doi: 10.1016/0012-1606(88)90320-x. [DOI] [PubMed] [Google Scholar]
- Henson JH, Nesbitt D, Wright BD, Scholey JS. Immuolocalization of kinesin in sea urchin coelomocytes: associations of kinesin with intracellular organelles. J Cell Sci. 1992;103:309–320. doi: 10.1242/jcs.103.2.309. [DOI] [PubMed] [Google Scholar]
- Henson JH, Svitkina TM, Burns AR, Hughes HE, MacPartland KJ, Nazarian R, Borisy GG. Two components of actin-based retrograde flow in sea urchin coelomocytes. Mol Biol Cell. 1999;10:4075–4090. doi: 10.1091/mbc.10.12.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzle MK, Svitkina T. The cytoskeletal mechanisms of cell–cell junction formation in endothelial cells. Mol Biol Cell. 2012;23:310–323. doi: 10.1091/mbc.E11-08-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamasaki T, Osumi M, Mabuchi I. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J Cell Biol. 2007;178:765–771. doi: 10.1083/jcb.200612018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Yumiko K, Amano M, Onishi H, Kaibuchi K, Fujiwara K. Rho-kinase-mediated contraction of isolated stress fibers. J Cell Biol. 2001;153:569–583. doi: 10.1083/jcb.153.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kner P, Chhun BB, Griffis ER, Winoto L, Gustafsson MG. Super-resolution video microscopy of live cells by structured illumination. Nat Methods. 2009;6:339–342. doi: 10.1038/nmeth.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante C, Berro J, Karatekin E, Hernadez-leyva A, Lee R, Pollard TD. Three myosins contribute uniquely to the assembly and constriction of the fission yeast cytokinetic contractile ring. Curr Biol. 2015;25:1–11. doi: 10.1016/j.cub.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante C, Huang F, Tebbs IR, Bewersdorf J, Pollard TD. Molecular organization of cytokinesis nodes and contractile rings by super-resolution fluorescence microscopy of live fission yeast. Proc Natl Acad Sci USA. 2016;113:E5876–E5885. doi: 10.1073/pnas.1608252113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I-J, Coffman VC, Wu J-Q. Contractile-ring assembly in fission yeast cytokinesis: recent advances and new perspectives. Cytoskeleton. 2012;69:751–763. doi: 10.1002/cm.21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewellyn L, Carvalho A, Desai A, Maddox AS, Oegema K. The chromosomal passenger complex and centralspindlin independently contribute to contractile ring assembly. J Cell Biol. 2011;193:155–169. doi: 10.1083/jcb.201008138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Kovács M, Conti MA, Wang A, Zhang Y, Sellers JR, Adelstein RS. Nonmuscle myosin II exerts tension but does not translocate actin in vertebrate cytokinesis. Proc Natl Acad Sci USA. 2012;109:4509–4514. doi: 10.1073/pnas.1116268109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi I. Cleavage furrow: timing of emergence of contractile ring actin filaments and establishment of the contractile ring by filament bundling in sea urchin eggs. J Cell Sci. 1994;107:1853–1862. doi: 10.1242/jcs.107.7.1853. [DOI] [PubMed] [Google Scholar]
- Mabuchi I, Hamaguchi Y, Fujimoto H, Morii N, Mishima M, Narumiya S. A rho-like protein is involved in the organisation of the contractile ring in dividing sand dollar eggs. Zygote. 1993;1:325331. doi: 10.1017/s0967199400001659. [DOI] [PubMed] [Google Scholar]
- Mabuchi I, Okuno M. The effect of myosin antibody on the division of starfish blastomeres. J Cell Biol. 1977;74:251–263. doi: 10.1083/jcb.74.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi I, Tsukita S, Tsukita S, Sawai T. Cleavage furrow isolated from newt eggs: contraction, organization of the actin filaments, and protein constituents of the furrow. Proc Natl Acad Sci USA. 1988;85:5966–5970. doi: 10.1073/pnas.85.16.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin P, Phillips CL, Adelstein RS, Pollard TD. Differential localization of myosin-II isozymes in human cultured cells and blood cells. J Cell Sci. 1994;107:3077–3090. doi: 10.1242/jcs.107.11.3077. [DOI] [PubMed] [Google Scholar]
- Maupin P, Pollard TD. Arrangement of actin filaments and myosin-like filaments in the contractile ring and actin-like filaments in the mitotic spindle of dividing HeLa cells. J Ultrastruct Mol Struct Res. 1986;94:92–103. doi: 10.1016/0889-1605(86)90055-8. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Mabuchi I. An IQGAP-like protein is involved in actin assembly together with Cdc42 in the sea urchin egg. Cell Motil Cytoskeleton. 2003;56:207–218. doi: 10.1002/cm.10146. [DOI] [PubMed] [Google Scholar]
- Otto JJ, Kane RE, Bryan J. Redistribution of actin and fascin in sea urchin eggs after fertilization. Cell Motil. 1980;1:31–40. doi: 10.1002/cm.970010104. [DOI] [PubMed] [Google Scholar]
- Otto JJ, Schroeder TE. Association of actin and myosin in the contractile ring. Ann NY Acad Sci. 1990;582:179–184. doi: 10.1111/j.1749-6632.1990.tb21678.x. [DOI] [PubMed] [Google Scholar]
- Perry MM, John HA, Thomas NST. Actin-like filaments in the cleavage furrow of newt egg. Exp Cell Res. 1971;65:249–253. doi: 10.1016/s0014-4827(71)80075-7. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol. 2010;22:50–56. doi: 10.1016/j.ceb.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Wu J-Q. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport R. Experiments concerning the cleavage stimulus in sand dollar eggs. J Exp Zool. 1961;148:81–89. doi: 10.1002/jez.1401480107. [DOI] [PubMed] [Google Scholar]
- Reichl EM, Ren Y, Morphew MK, Delannoy M, Effler JC, Girard KD, Divi S, Iglesias PA, Kuo SC, Robinson DN. Interactions between myosin and actin crosslinkers control cytokinesis contractility dynamics and mechanics. Curr Biol. 2008;18:471–480. doi: 10.1016/j.cub.2008.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger JM, Sanger JW. Banding and polarity of actin filaments in interphase and cleaving cells. J Cell Biol. 1980;86:568–575. doi: 10.1083/jcb.86.2.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TE. The contractile ring. I. Fine structure of dividing mammalian (HeLa) cells and the effects of cytochalasin B. Z Zellforsch Mikrosk Anat. 1970;109:431–449. [PubMed] [Google Scholar]
- Schroeder TE. The contractile ring. II. Determining its brief existence, volumetric changes, and vital role in cleaving Arbacia eggs. J Cell Biol. 1972;53:419–434. doi: 10.1083/jcb.53.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TE. Actin in dividing cells: contractile ring filaments bind heavy meromyosin. Proc Natl Acad Sci USA. 1973;70:1688–1692. doi: 10.1073/pnas.70.6.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TE. Microvilli on sea urchin eggs: a second burst of elongation. Dev Biol. 1978;64:342–346. doi: 10.1016/0012-1606(78)90085-4. [DOI] [PubMed] [Google Scholar]
- Schroeder TE, Otto JJ. Immunofluorescent analysis of actin and myosin in isolated contractile rings of sea urchin eggs. Zool Sci. 1988;5:713–725. [Google Scholar]
- Shutova M, Yang C, Vasiliev JM, Svitkina T. Functions of nonmuscle myosin II assembly of the cellular contractile system. PLoS One. 2012;7:e40814. doi: 10.1371/journal.pone.0040814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutova MS, Spessott WA, Giraudo CG, Svitkina T. Endogenous species of mammalian nonmuscle myosin IIA and IIB include activated monomers and heteropolymers. Curr Biol. 2014;24:1958–1968. doi: 10.1016/j.cub.2014.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM. Electron microscopic analysis of the leading edge of cells. Methods Cell Biol. 2007;79:295–319. doi: 10.1016/S0091-679X(06)79012-4. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Surguchova IG, Verkhovsky AB, Gelfand VI, Moeremans M, De Mey J. Direct visualization of bipolar myosin filaments in stress fibers of cultured fibroblasts. Cell Motil Cytoskeleton. 1989;12:150–156. doi: 10.1002/cm.970120304. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Verkhovsky AB, Borisy GG. Improved procedures for electron microscopic visualization of the cytoskeleton of cultured cells. J Struct Biol. 1995;115:290–303. doi: 10.1006/jsbi.1995.1054. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin-myosin system in fish epidermal keratocytes: mechanism of cell body translocation. J Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafer-Glusman E, Fuller MT, Giansanti MG. Role of Survivin in cytokinesis revealed by a separation-of-function allele. Mol Biol Cell. 2011;22:3779–3790. doi: 10.1091/mbc.E11-06-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojkander S, Gateva G, Lappalainen P. Actin stress fibers—assembly, dynamics and biological roles. J Cell Sci. 2012;125:1855–1864. doi: 10.1242/jcs.098087. [DOI] [PubMed] [Google Scholar]
- Uehara R, Hosoya H, Mabuchi I. In vivo phosphorylation of regulatory light chain of myosin II in sea urchin eggs and its role in controlling myosin localization and function during cytokinesis. Cell Motil Cytoskeleton. 2008;65:100–115. doi: 10.1002/cm.20246. [DOI] [PubMed] [Google Scholar]
- Usui N, Yoneda M. Ultrastructural basis of the tension increase in sea urchin eggs prior to cytokinesis. Dev Growth Diff. 1982;24:453–465. doi: 10.1111/j.1440-169X.1982.00453.x. [DOI] [PubMed] [Google Scholar]
- Uyeda TQP, Nagasaki A. Variations on a theme: the many modes of cytokinesis. Curr Opin Cell Biol. 2004;16:55–60. doi: 10.1016/j.ceb.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Verkhovsky AB, Surguchova IG, Svitkina TM, Tint IS, Gelfand VI. Organization of stress fibers in cultured fibroblasts after extraction of actin with bovine brain gelsolin-like protein. Exp Cell Res. 1987;173:244–255. doi: 10.1016/0014-4827(87)90349-1. [DOI] [PubMed] [Google Scholar]
- Verkhovsky AB, Svitkina TM, Borisy GG. Myosin II filament assemblages in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J Cell Biol. 1995;131:989–1002. doi: 10.1083/jcb.131.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-L. The mechanism of cortical ingression during early cytokinesis: thinking beyond the contractile ring hypothesis. Trends Cell Biol. 2005;15:581–588. doi: 10.1016/j.tcb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Werner M, Munro E, Glotzer M. Astral signals spatially bias cortical myosin recruitment to break symmetry and promote cytokinesis. Curr Biol. 2007;17:1286–1297. doi: 10.1016/j.cub.2007.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollrab V, Thiagarajan R, Wald A, Kruse K, Riveline D. Still and rotating myosin clusters determine cytokinetic ring constriction. Nat Commun. 2016;7:11860. doi: 10.1038/ncomms11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S, Hirao-Minakuchi K, Nishimura Y. Rho localization in cells and tissues. Exp Cell Res. 2004;295:300–314. doi: 10.1016/j.yexcr.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Yonemura S, Kinoshita S. Actin filament organization in the sand dollar egg cortex. Dev Biol. 1986;115:171–183. [Google Scholar]
- Yonemura S, Mabuchi I, Tsukita S. Mass isolation of cleavage furrows from dividing sea urchin eggs. J Cell Sci. 1991;100:73–84. [Google Scholar]
- Zhou M, Wang Y-L. Distinct pathways for the early recruitment of myosin II and actin to the cytokinetic furrow. Mol Biol Cell. 2008;19:318–326. doi: 10.1091/mbc.E07-08-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.