The ciliary kinesin KIF17 is a cargo, but not a motor, of the IFT-B complex. The ciliary entry of KIF17 through the transition zone depends on its binding to IFT-B, as well as on its nuclear localization signal.
Abstract
Cilia function as cellular antennae to sense and transduce extracellular signals. A number of proteins are specifically localized in cilia. Anterograde and retrograde ciliary protein trafficking are mediated by the IFT-B and IFT-A complexes in concert with kinesin-2 and dynein-2 motors, respectively. However, the role of KIF17, a homodimeric kinesin-2 protein, in protein trafficking has not been fully understood in vertebrate cilia. In this study, we demonstrated, by using the visible immunoprecipitation assay, that KIF17 interacts with the IFT46–IFT56 dimer in the IFT-B complex through its C-terminal sequence located immediately upstream of the nuclear localization signal (NLS). We then showed that KIF17 reaches the ciliary tip independently of its motor domain and requires IFT-B binding for its entry into cilia rather than for its intraciliary trafficking. We further showed that KIF17 ciliary entry depends not only on its binding to IFT-B but also on its NLS, to which importin α proteins bind. Taking the results together, we conclude that in mammalian cells, KIF17 is dispensable for ciliogenesis and IFT-B trafficking but requires IFT-B, as well as its NLS, for its ciliary entry across the permeability barrier located at the ciliary base.
INTRODUCTION
Cilia are microtubule-based structures protruding from the cell surface, which perform a variety of biological functions, such as the perception and transduction of physiological and developmental signals, including the Hedgehog (Hh) signaling pathway. To perform these functions, cilia contain a large number of unique proteins, including a variety of receptors and ion channels. The roles of cilia as cellular sensory antennae have been highlighted by findings that defects in ciliary assembly and function result in phenotypically diverse disorders, generally called ciliopathies (Schwartz et al., 2011; Madhivanan and Aguilar, 2014). Although the ciliary membrane is continuous with the plasma membrane, protein and lipid compositions of cilia differ greatly from those of the cell body because the ciliary transition zone (TZ) serves as a diffusion/permeability barrier (Sung and Leroux, 2013; Wei et al., 2015). Proteins required for the assembly and functions of cilia are transported from the cell body. The transport process includes entry of the proteins into cilia through the TZ and trafficking of proteins along axoneme microtubules within cilia.
The ciliary TZ is structurally characterized by the presence of transition fibers and Y-linked structures, which have been proposed to constitute a barrier similar to the nuclear pore complex (NPC; Takao and Verhey, 2016). Therefore some ciliary proteins, such as KIF17 and RP2, contain a nuclear localization signal (NLS) and enter the cilium by a mechanism dependent on the Ran GTPase and importin β2 (also known as transportin 1 [TNPO1]; Dishinger et al., 2010; Hurd et al., 2011).
On the other hand, bidirectional trafficking of ciliary proteins along the axoneme is mediated by intraflagellar transport (IFT) particles, which contain two large multisubunit complexes, IFT-A and IFT-B, composed of six and 16 subunits, respectively (Taschner et al., 2012; Lechtreck, 2015; Katoh et al., 2016). Anterograde protein trafficking from the ciliary base to the tip is mediated by the IFT-B complex with the aid of kinesin-2 motor proteins, whereas retrograde protein trafficking is mediated by the IFT-A complex in concert with dynein-2. Deficiency in any of the IFT-B subunits often leads to extremely short or no cilia, indicating a crucial role of protein trafficking in ciliary assembly.
Two different types of kinesin-2 motors have been implicated in ciliary anterograde protein trafficking: heterotrimeric kinesin-II, composed of KIF3A, KIF3B, and KIFAP3 (KLP-20, KLP-11, and KAP-1 in Caenorhabditis elegans), and homodimeric KIF17 (OSM-3 in C. elegans; Scholey, 2013). In C. elegans, heterotrimeric kinesin-II and the OSM-3 homodimer participate in IFT within the axoneme core (proximal/middle) segment and the distal segment, respectively (Pan et al., 2006; Scholey, 2013; Prevo et al., 2015). In contrast, in vertebrates, the contribution of KIF17 to ciliary assembly is unclear, as ciliogenesis appeared to be normal in KIF17-deficient mice and zebrafish (Yin et al., 2011; Pooranachandran and Malicki, 2016), although its involvement in the trafficking of some ciliary proteins has been implicated (Jenkins et al., 2006; Leaf and von Zastrow, 2015).
KIF17 accumulates at the ciliary tip of mammalian cells (Dishinger et al., 2010; Jiang et al., 2015). Verhey and colleagues showed that KIF17 gains entrance to the ciliary compartment by a mechanism resembling nuclear import (Dishinger et al., 2010; Kee et al., 2012; Takao and Verhey, 2016). Namely, it is translocated into cilia through the TZ by a mechanism dependent on its NLS and on the Ran GTPase and TNPO1. On the other hand, how KIF17 is transported to the ciliary tip along the axoneme in mammalian cells was unknown because a construct of KIF17 that lacked its motor domain was able to reach the ciliary tip (Williams et al., 2014; Jiang et al., 2015). This suggests that KIF17 is a cargo of the IFT machinery rather than being a motor for the machinery, at least in mammalian cells. Previous studies suggested that the IFT-B complex interacts with KIF17/OSM-3 (Snow et al., 2004; Ou et al., 2005; Insinna et al., 2008; Howard et al., 2013), although it was not clear which IFT-B subunit(s) are directly involved in the interaction.
We recently developed a novel and versatile strategy for studying protein–protein interactions, the visible immunoprecipitation (VIP) assay (Katoh et al., 2015, 2016); the VIP assay can visually detect binary protein interactions by fluorescence microscopy without performing electrophoresis and immunoblotting and can determine interactions between more than two proteins at a time. By using this strategy, we determined not only the overall architectures of the exocyst and BBSome complexes (Katoh et al., 2015), both of which are composed of eight subunits, but also those of the IFT-A and IFT-B complexes (Katoh et al., 2016; Hirano et al., 2017). In the present study, we applied our flexible VIP assay system to detect the interaction of KIF17 with the IFT-B complex and found that KIF17 interacts with the IFT46–IFT56 dimer in the IFT-B complex. We also found that a sequence conserved between vertebrate KIF17 and C. elegans OSM-3 located immediately upstream of the NLS is responsible for the interaction of KIF17 with IFT46–IFT56. Furthermore, we showed that both the NLS and the IFT-B–interacting sequence of KIF17 are required for its ciliary entry.
We further established IFT56-knockout (KO) and KIF17-KO lines of human telomerase reverse transcriptase–immortalized retinal pigmented epithelial (hTERT-RPE1) cells using a modified clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system and showed that IFT56-KO and KIF17-KO cells are normal with respect to ciliogenesis. However, we found that KIF17 was unable to enter cilia in IFT56-KO cells, whereas IFT-B localization was not affected by the KIF17 deficiency. These results unequivocally demonstrate that KIF17 requires IFT-B binding, as well as its NLS, for entry into cilia.
RESULTS
KIF17 interacts with the IFT46–IFT56 dimer of the IFT-B complex
Although previous studies suggested that KIF17/OSM-3 interacts with the IFT-B complex (Snow et al., 2004; Ou et al., 2005; Insinna et al., 2008; Howard et al., 2013), it was uncertain which subunit(s) were directly involved in the interaction between IFT-B and KIF17. To address this issue, we applied the VIP assay, which we recently developed as a simple and flexible strategy for detecting protein–protein interactions (Katoh et al., 2015, 2016). Using this assay, we recently determined the architectures of three multisubunit complexes: the exocyst, BBSome, and IFT-B complex, composed of eight, eight, and 16 subunits, respectively. In particular, our study unequivocally showed that the IFT-B complex comprises two parts—a core subcomplex composed of 10 subunits (IFT22/25/27/46/52/56/70/74/81/88) and a peripheral subcomplex composed of six subunits (IFT20/38/54/57/80/172; Katoh et al., 2016); these subcomplexes are linked by composite interactions involving IFT38/52/57/88 (Boldt et al., 2016; Katoh et al., 2016; Taschner et al., 2016; Figure 1E).
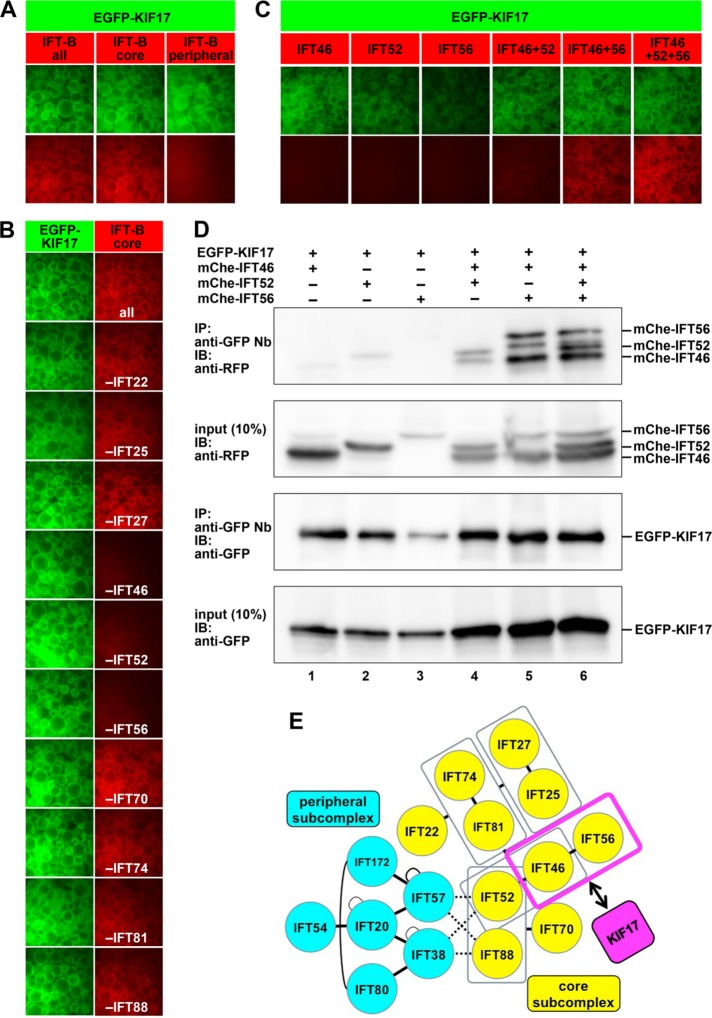
FIGURE 1:
Interaction of KIF17 with the IFT46–IFT56 dimer demonstrated by the VIP assay. (A) Interaction of KIF17 with the IFT-B core subcomplex. Lysates were prepared from HEK293T cells coexpressing EGFP-KIF17 and all IFT-B subunits, all core subunits, or all peripheral subunits fused to mChe/tRFP and processed for the VIP assay using GST-tagged anti-GFP Nb as described in Materials and Methods. (B) A subtractive VIP assay was performed to determine the IFT-B core subunits involved in its interaction with KIF17. Lysates prepared from HEK293T cells coexpressing EGFP-KIF17 and all but one (as indicated) of the core subunits fused to mChe/tRFP were processed for the VIP assay. (C, D) Confirmation of the involvement of the IFT46–IFT56 dimer in the KIF17–IFT-B interaction. Lysates prepared from HEK293T cells coexpressing EGFP-KIF17 and mChe-tagged IFT46 (lane 1), IFT52 (lane 2), IFT56 (lane 3), IFT46 + IFT52 (lane 4), IFT46 + IFT56 (lane 5), or IFT46 + IFT52 + IFT56 (lane 6) were processed for the VIP assay (C) or immunoblotting analysis (D) as described in Materials and Methods. (E) Schematic representation of the interaction of KIF17 with the IFT46–IFT56 dimer in the IFT-B complex.
We first examined whether KIF17 can interact with individual IFT-B subunits. When enhanced green fluorescent protein (EGFP)–KIF17 was coexpressed with any one of the 16 IFT-B subunits fused to mCherry (mChe) or TagRFP (tRFP) in HEK293T cells, and lysates prepared from the transfected cells were subjected to immunoprecipitation with glutathione S-transferase (GST)–tagged anti-GFP nanobody (Nb) prebound to glutathione-Sepharose beads. However, we were unable to detect the interaction of KIF17 with any of the IFT-B subunits (unpublished data). We then used one of the advantages of our flexible VIP assay; in our previous studies, we successfully found not only one-to-many but also many-to-many subunit interactions in the exocyst and IFT-B complexes (Katoh et al., 2015, 2016). When EGFP-KIF17 was coexpressed with all of the IFT-B subunits or all of the core or peripheral subunits as mChe/tRFP fusions, we were able to detect red fluorescence signals when all IFT-B subunits and all core subunits, but not all peripheral subunits, were coexpressed with EGFP-KIF17 (Figure 1A). Therefore data from the VIP assay indicate that the core subcomplex of IFT-B contains subunits responsible for its interaction with KIF17.
We then applied another advantage of our flexible assay to find the core subunit(s) of IFT-B responsible for its interaction with KIF17—namely, the subtractive VIP assay, in which omitting one or more of the core subunits is expected to abolish red fluorescence signals if those core subunit(s) are crucial for the interaction of KIF17 with the IFT-B complex. The red fluorescence signals were attenuated considerably by omitting IFT46 or IFT56 from the 10 core subunits and weakly by omitting IFT52 but not by omitting any of the other core subunits (Figure 1B). To examine whether IFT46, IFT52, and/or IFT56 are indeed involved in the interaction of the IFT-B complex with KIF17, we then performed VIP assays for one-to-many protein interactions. As shown in Figure 1C, red signals were detected robustly when mChe-IFT46 + IFT56 or mChe-IFT46 + IFT52 + IFT56 were coexpressed with EGFP-KIF17 and detected weakly when mChe-IFT46 + IFT52 were coexpressed. We confirmed the VIP data by subjecting the immunoprecipitates to conventional immunoblotting analysis (Figure 1D); bands for mChe-tagged proteins were detected when mChe-IFT46 + IFT56 (lane 5) or mChe-IFT46 + IFT52 + IFT56 (lane 6) were coexpressed with EGFP-KIF17, and weak bands were detected when mChe-IFT46 + IFT52 were coexpressed (lane 4). We therefore concluded that the IFT46–IFT56 dimer mainly participates in interaction of the IFT-B complex with KIF17, as schematically shown in Figure 1E, although IFT52 may play an auxiliary role in the interaction. The substantial attenuation of the red fluorescence signal observed when IFT52 was omitted in the subtractive VIP assay (Figure 1B) was likely due to the critical role of IFT52 in connecting IFT46–IFT56 to the other seven core subunits (Figure 1E).
KIF17 interacts with IFT46–IFT56 via a conserved sequence immediately upstream of its NLS
We then set out to determine the region of the KIF17 protein that is responsible for its interaction with the IFT46–IFT56 dimer. To this end, we constructed a series of C-terminal truncation mutants of KIF17 because its NLS is located in the C-terminal region (Figure 2A) and because KIF17 lacking its motor domain retains its ability to reach the ciliary tip (Williams et al., 2014; Jiang et al., 2015). As seen in the VIP data in Figure 2B, not only KIF17(1-1018) (second row), which contains the NLS (Figure 2A), but also KIF17(1-1014) (third row), which lacks the NLS, interacted with the IFT46–IFT56 dimer. In support of these results, substitution of all the basic residues in the NLS (1015KRKK1018) to Ala did not affect the interaction of KIF17 with IFT46–IFT56 (AAAA, fifth row). A further truncation up to residue 999, however, abolished the interaction of KIF17 with IFT46–IFT56 (fourth row).
FIGURE 2:
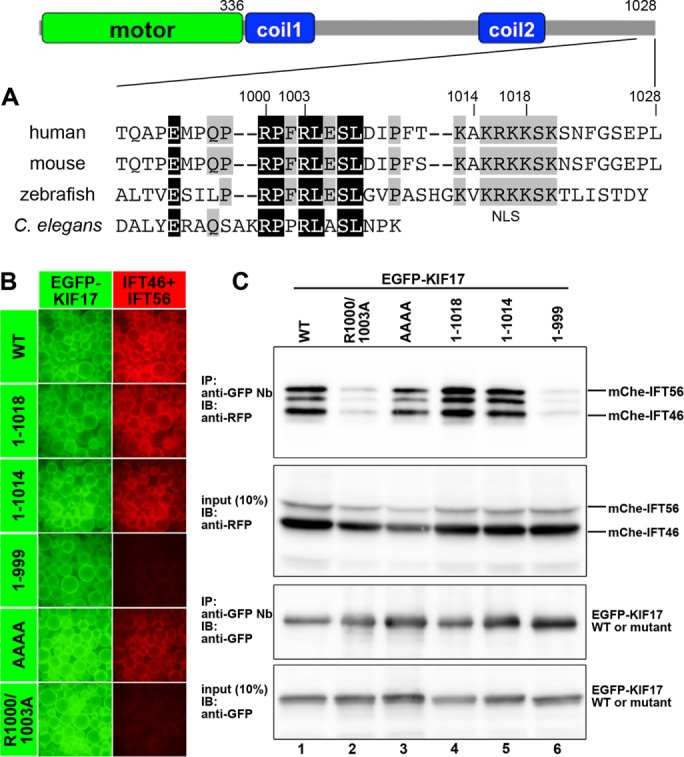
Interaction of KIF17 with the IFT46–IFT56 dimer through its conserved C-terminal sequence. (A) Schematic representation of the overall structure and domain organization of KIF17 and alignment of the C-terminal sequences of human, mouse, and zebrafish KIF17 and C. elegans OSM-3. Coil1 and Coil2 are coiled-coil regions. Residues conserved in all and three members are shown in black and gray boxes, respectively. (B, C) Involvement of the conserved C-terminal sequence of KIF17 in its interaction with the IFT46–IFT56 dimer. Lysates prepared from HEK293T cells coexpressing mChe-IFT46 and mChe-IFT56 and EGFP-KIF17(WT), EGFP-KIF17(1-1018), EGFP-KIF17(1-1014), EGFP-KIF17(1-999), EGFP-KIF17(AAAA), or EGFP-KIF17(R1000/1003A) were subjected to the VIP assay (B) or immunoblotting analysis (C).
By comparing the sequences of various vertebrate KIF17s, we noticed that the sequence (RPxRLxSL; residues 1000–1007 in human KIF17) immediately upstream of the NLS is highly conserved (Figure 2A). A similar sequence is found at the C-terminus of C. elegans OSM-3, although it lacks the NLS. We therefore substituted the conserved Arg residues at amino acids 1000 and 1003 to Ala (R1000/1003A) and found that KIF17(R1000/1003A) did not interact with the IFT46–IFT56 dimer in the VIP assay (Figure 2B, bottom row). We confirmed the VIP data by subjecting the immunoprecipitates to conventional immunoblotting analysis (Figure 2C); EGFP-KIF17(1-999) and EGFP-KIF17(R1000/1003A) had very low ability to coimmunoprecipitate mChe-IFT46 and mChe-IFT56 (lanes 6 and 2, respectively). Note that we consistently observed an extra band between those of mChe-IFT46 and mChe-IFT56 after coimmunoprecipitation of EGFP-KIF17 with mChe-IFT46 and mChe-IFT56 (Figures 1D and 2C, top), although we did not detect the extra band when we used a reciprocal combination of fluorescent fusion proteins (Figure 6D, lane 1). We do not know the origin of the extra band, but mChe-IFT56 may be liable to partial degradation when it forms a complex with other IFT proteins.
FIGURE 6:
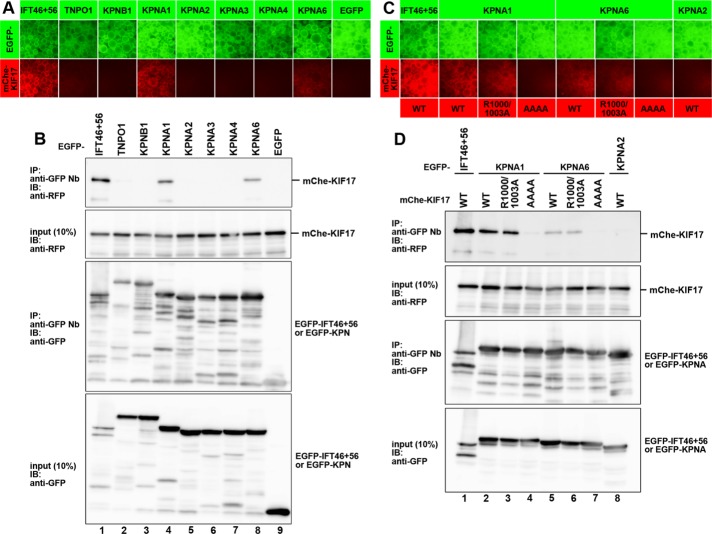
KIF17 NLS interacts with importin α proteins. (A, B) KIF17 interacts with KPNA1 and KPNA6. Lysates prepared from HEK293T cells coexpressing mChe-KIF17 and EGFP-IFT46 + GFP-IFT56, EGFP-TNPO1, EGFP-KPNB1, EGFP-KPNA1, EGFP-KPNA2, EGFP-KPNA3, EGFP-KPNA4, EGFP-KPNA6, or EGFP were processed for the VIP assay (A) or immunoblotting analysis (B). (C, D) Binding of KPNA1 and KPNA6 to the KIF17 NLS. Lysates prepared from HEK293T cells coexpressing mChe-KIF17(WT), mChe-KIF17(R1000/1003A), or mChe-KIF17(AAAA) and EGFP-IFT46 + EGFP-IFT56, EGFP-KPNA1, EGFP-KPNA6, or EGFP-KPNA2 as indicated were processed for the VIP assay (C) or immunoblotting analysis (D).
Both the NLS and IFT-B–binding sequence are required for the ciliary entry of KIF17
The long-standing question regarding vertebrate KIF17 is whether this motor protein functions as a motor or is a cargo of the IFT machinery. To obtain an answer to this question, we examined the localization of various KIF17 constructs. After transfection of an expression vector for the EGFP-KIF17 construct into hTERT-RPE1 cells, we cultured the cells under serum-starvation conditions for 24 h to induce ciliogenesis (see Materials and Methods) and processed them for immunostaining for acetylated α-tubulin (Ac-α-tubulin; a marker for the ciliary axoneme) and γ-tubulin (a marker for the basal body). As shown in Figure 3A and Supplemental Figure S1A, EGFP-KIF17(WT) accumulated at the ciliary tips. In addition, a weak EGFP signal was also observed in the nucleus. Essentially the same localization pattern was observed for a motor-less construct, EGFP-KIF17(337-1028) (Supplemental Figure S1C); this observation is consistent with previous studies (Williams et al., 2014; Jiang et al., 2015). In considerable contrast, a motor-domain construct, EGFP-KIF17(1-336), was not found at the ciliary tip or in the nucleus (Supplemental Figure S1B). Thus the C-terminal tail region determines localization of KIF17 at the ciliary tip.
FIGURE 3:
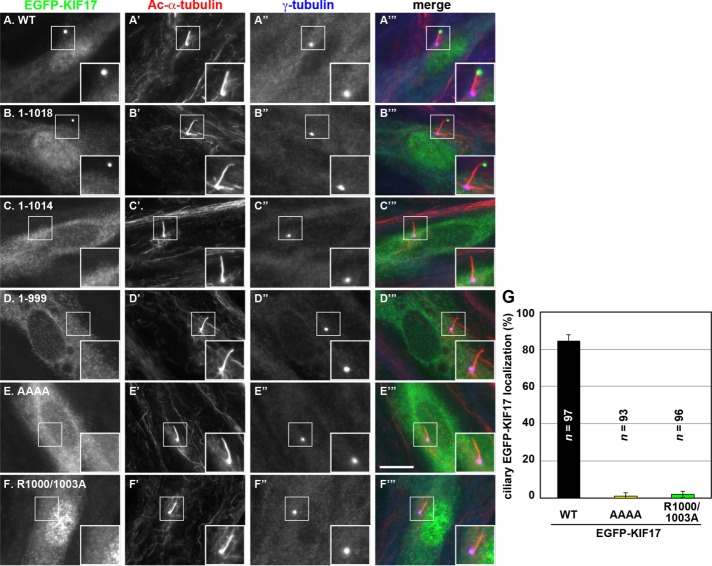
Ciliary entry of KIF17 is dependent on both its binding to IFT-B and its NLS. hTERT-RPE1 cells were transfected with an expression vector for (A) EGFP-KIF17(WT), (B) EGFP-KIF17(1-1018), (C) EGFP-KIF17(1-1014), (D) EGFP-KIF17(1-999), (E) EGFP-KIF17(AAAA), or (F) EGFP-KIF17(R1000/1003A). The transfected cells were cultured for 24 h under normal conditions and for a further 24 h under serum-starvation conditions, as described in Materials and Methods. The cells were immunostained for Ac-α-tubulin (A′–F′) and γ-tubulin (A′′–F′′). Merged images are shown in A′′′–F′′′. Insets, enlarged images of the boxed. Scale bar, 10 µm. (G) Cells positive for ciliary signals of EGFP-KIF17(WT), EGFP-KIF17(AAAA), or EGFP-KIF17(R1000/1003A) were counted; percentages of positive ciliated cells. Values are means ± SE (error bars) of three independent experiments. In each set of experiments, 30–34 ciliated cells were observed, and the total numbers of ciliated cells observed (n) are shown.
We then examined localization of KIF17 constructs used in the experiments shown in Figure 2. EGFP-KIF17(1-1018), which contains both the NLS and the IFT-B–binding sequence, localized, like EGFP-KIF17(WT), at the ciliary tip and in the nucleus (Figure 3B). In striking contrast, no signal was observed at the ciliary tip or in the nucleus for EGFP-KIF17(1-1014) (Figure 3C), indicating that the NLS is required for entry into both the cilium and nucleus. This is supported by the observation that EGFP-KIF17(AAAA), an NLS mutant, failed to enter the cilium and nucleus (Figure 3E; also see Figure 3G).
EGFP-KIF17(1-999), which lacks both the NLS and the IFT-B–binding sequence, also failed to enter the cilium and nucleus (Figure 3D). By contrast, EGFP-KIF17(R1000/1003A), an IFT-B-binding–defective mutant, retained the ability to enter the nucleus, although it could not enter the cilium (Figure 3F; also see Figure 3G). These observations together indicate that the NLS serves as a signal for entry into the cilium as well as the nucleus, consistent with previous reports (Dishinger et al., 2010; Jiang et al., 2015). On the other hand, IFT-B binding is required for KIF17 entry into the cilium. This is compatible with the notion that KIF17 is a cargo of the IFT-B complex. However, the observation that KIF17(R1000/1003A) cannot enter the cilium was somewhat unexpected because the IFT-B complex mediates anterograde ciliary protein trafficking, but there has been no report suggesting the involvement of IFT-B in the ciliary entry of proteins (see Discussion).
IFT56 is essential for the entry of KIF17 into cilia
To support the idea that the ciliary entry of KIF17 is mediated by the IFT-B complex, we then attempted to establish hTERT-RPE1 cell lines defective in IFT56. We expected that the lack of IFT56 would inhibit the binding of IFT-B to KIF17 but would not severely affect IFT-B assembly or ciliogenesis in human cells for the following reasons: 1) our model of the architecture of IFT-B (Katoh et al., 2016; Figure 1E) predicts that IFT56 has an auxiliary role in the assembly of the IFT-B complex; 2) Taschner et al. (2016) purified the recombinant IFT-B complex in the absence of IFT56; and 3) mutations of DYF13/TTC26/IFT56 in Chlamydomonas and mice were reported to marginally affect IFT-B assembly and ciliogenesis (Ishikawa et al., 2014; Swiderski et al., 2014).
Because hTERT-RPE1 cells retain the characteristics of normal primary cells and are believed to have low homologous recombination activity, the establishment of biallelic KO lines from hTERT-RPE1 cells was reported to be difficult, although not impossible. Miyamoto et al. (2015) established biallelic KIF2A-KO lines from hTERT-RPE1 cells via sequential gene targeting with two distinct targeting vectors containing neomycin-resistance and puromycin-resistance genes. To overcome this problem, we recently developed an efficient genome editing strategy applicable to hTERT-RPE1 cells by modification of the CRISPR/Cas9 system (Materials and Methods; Supplemental Figure S2A) and established IFT88-KO and IFT20-KO cell lines, which are both defective in ciliogenesis (Katoh et al., 2017).
Using two different target sequences (Supplemental Table S1), we established five independent IFT56-KO cell lines. Among them, we selected two cell lines (56-1-7 and 56-2-6) established using different target sequences for the following detailed analyses. PCR analysis of genomic DNAs demonstrated that clones 56-1-7 and 56-2-6 have monoallelic reverse and forward integration, respectively, of the knock-in donor vector in the IFT56 gene (Supplemental Figure S2B). Direct sequencing of the PCR products demonstrated that, in the other allele, clones 56-1-7 and 56-2-6 have a 1–base pair insertion (c.244_245 insC, p.Leu45Profs*15) and a 7–base pair deletion (c.139_145 delGCTGTAG, p.Val9Glufs*19), respectively (Supplemental Figure S2, D and E), probably resulting from error-prone nonhomologous end joining after CRISPR/Cas9-mediated DNA cleavage.
We then examined the phenotype of the IFT56-KO cell lines. After incubation of control hTERT-RPE1 cells and the two IFT56-KO cell lines under serum-starvation conditions to induce ciliogenesis, we triple immunostained the cells with antibodies against Arl13b, Ac-α-tubulin, and γ-tubulin (Figure 4, A–C). In control RPE1 cells, Arl13b and Ac-α-tubulin mark the ciliary membrane and axoneme, respectively, and γ-tubulin is localized to the basal body (Figure 4, A–A′′′). The staining pattern for these marker proteins in the IFT56-KO cell lines (Figure 4, B–B′′′ and C–C′′′) was similar to that in control cells, indicating that ciliogenesis or the integrity of cilia is not affected by the absence of IFT56. This was in striking contrast to the phenotypes of IFT88-KO and IFT20-KO cell lines, in which ciliogenesis is abolished (Katoh et al., 2017).
FIGURE 4:
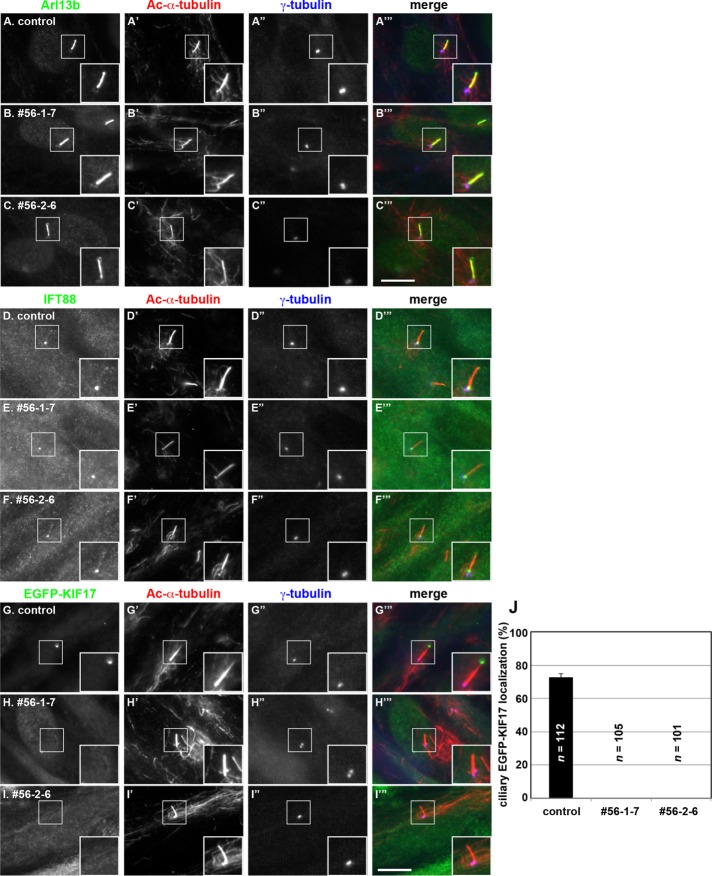
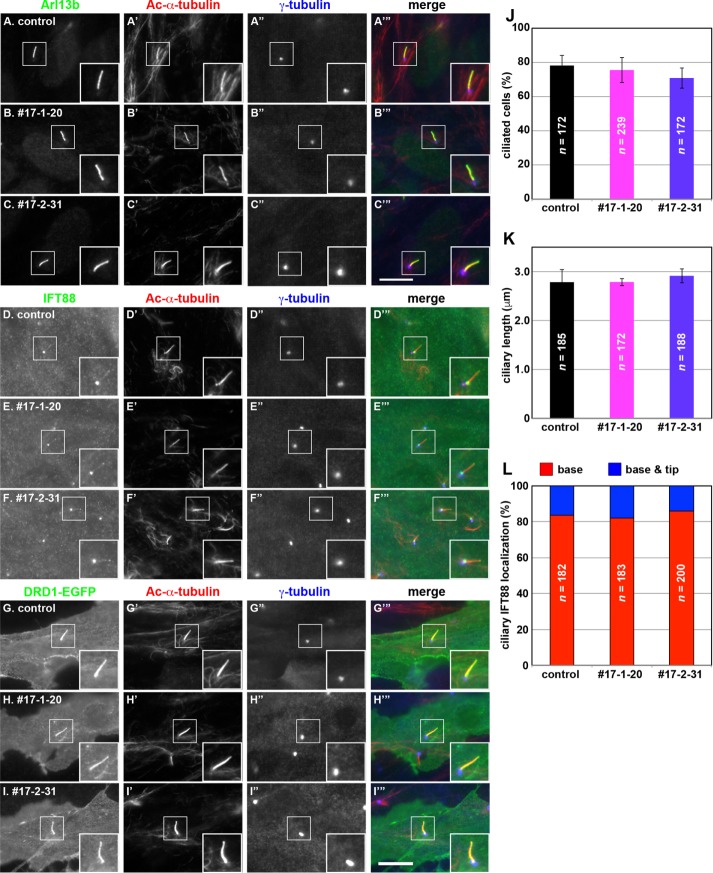
Localization of ciliary proteins in IFT56-KO cells. (A–F) Control RPE1 cells (A, D) and IFT56-KO cell lines 56-1-7 (B, E) and 56-2-6 (C–F) were cultured under serum starvation conditions for 24 h to induce ciliogenesis and triple immunostained for Arl13b (A–C) or IFT88 (D–F), Ac-α-tubulin (A′–F′), and γ-tubulin (A′′–F′′). (G–I) Control RPE1 cells (G) and the IFT56-KO cell lines 56-1-7 (H) and 56-2-6 (I), which stably express EGFP-KIF17 (G–I) and were established by infection of a lentiviral expression vector, were cultured under serum starvation conditions and immunostained for Ac-α-tubulin (G′–I′) and γ-tubulin (G′′–I′′). Merged images are shown in A′′′–I′′′. Insets, enlarged images of the boxed regions. Scale bar, 10 µm. (J) Control, 56-1-7, and 56-2-6 cells positive for ciliary EGFP-KIF17 signals were counted; percentages of positive ciliated cells. Values are means ± SE (error bars) of three independent experiments. In each set of experiments, 50–55 ciliated cells were observed, and the total numbers of ciliated cells observed (n) are shown . Note that no ciliary localization of EGFP-KIF17 was observed in the two IFT56-KO cell lines.
We then examined the localization of Smoothened (Smo), which is a G protein–coupled receptor (GPCR) involved in Hh signaling. Smo is known to enter cilia when the Hh signaling pathway is activated by Smoothened agonist (SAG), as observed in control RPE1 cells (Supplemental Figure S3, compare A and B). As in control cells, Smo was not found within cilia under basal conditions (Supplemental Figure S3, C and E), whereas it entered cilia when stimulated with SAG (Supplemental Figure S3, D and F) in the IFT56-KO cell lines 56-1-7 and 56-2-6. The phenotype of IFT56-KO cells is different from that of cells derived from IFT25-KO and IFT27-KO mice, in which a substantial fraction of Smo accumulates within cilia even under basal conditions, resulting in aberrant Hh signaling, although ciliogenesis appears to be unaffected (Keady et al., 2012; Eguether et al., 2014).
Staining for IFT88, an IFT-B core subunit, was observed mainly around the basal body and faintly along the cilium and at the tip in both control RPE1 cells and the IFT56-KO cell lines (Figure 4, D–F). Taken together with the fact that ciliogenesis or Smo trafficking is not affected in IFT56-KO cell lines, we conclude that IFT56 is not necessary for assembly or major functions of the IFT-B complex.
We then sought to compare KIF17 localization between control and IFT56-KO cells. When stably expressed in control RPE1 cells, EGFP-KIF17 was found at the ciliary tip (Figure 4G). In striking contrast to control cells, no ciliary signals of EGFP-KIF17 were observed in IFT56-KO cell lines (Figure 4, H and I; also see Figure 4J). These observations together unequivocally demonstrate that, for its entry into cilia, KIF17 requires binding to the IFT-B complex via the IFT46–IFT56 dimer.
KIF17 is dispensable for ciliogenesis and IFT-B trafficking
We then established KIF17-KO lines of hTERT-RPE1 cells to examine whether KIF17 is required for ciliogenesis and IFT-B trafficking. Using two different target sequences (one in exon 2 and the other in exon 3; Supplemental Table S1), we established two independent KIF17-KO cell lines, 17-1-20 and 17-2-31, both of which have monoallelic forward integration of the knock-in donor vector and a 1–base pair insertion in the other allele (c.763_764 insT, p.Gly152Trpfs*3, and c.640_641 insA, p.Arg111Glufs*22, respectively; Supplemental Figure S2, C, F, and G). Thus, for both alleles in both KO cell lines, the KIF17 protein is disrupted in the middle of the motor domain (Figure 2A). Although our attempts to demonstrate the absence of the KIF17 protein in the KIF17-KO cell lines using three different antibodies that were used in previous studies have been unsuccessful, disruption of the motor domain in both alleles of the two KO cell lines established using different target sequences makes it unlikely that the KIF17 protein with the functional motor domain is expressed in the KO cell lines.
When cilia were immunostained with antibodies against Arl13b and Ac-α-tubulin after induction of ciliogenesis (Figure 5, A–C), no significant difference was observed in the proportion of ciliated cells (Figure 5J) or ciliary length (Figure 5K) between control RPE1 cells and the two KIF17-KO cell lines. When immunostained for IFT88, the majority of both control RPE1 cells and the two KIF17-KO cell lines exhibited IFT88 staining at the ciliary base and minor populations at both the base and tip (Figure 5, D–F and L). Given previous reports showing that tubulins and outer dynein arm proteins are trafficked through binding to the IFT-B complex (Hou et al., 2007; Ahmed et al., 2008; Bhogaraju et al., 2013), normal ciliary assembly and normal IFT88 localization in KIF17-KO cells indicate that KIF17 is not essential for IFT-B function.
FIGURE 5:
Localization of ciliary proteins in KIF17-KO cells. (A–F) Control RPE1 cells (A, D) and KIF17-KO cell lines 17-1-20 (B, E) and 17-2-31 (C, F) were cultured under serum starvation conditions for 24 h to induce ciliogenesis and triple immunostained for Arl13b (A–C) or IFT88 (D–F), Ac-α-tubulin (A′–F′), and γ-tubulin (A′′–F′′). (G–I) Control RPE1 cells (G) and KIF17-KO cell lines 17-1-20 (H) and 17-2-31 (I), which stably express DRD1-EGFP (G–I) and were established by infection of a lentiviral expression vector, were cultured under serum starvation conditions and immunostained for Ac-α-tubulin (G′–I′) and γ-tubulin (G′′–I′′). Merged images are shown in A′′′–I′′′. Insets, enlarged images of the boxed regions. Scale bar, 10 µm. (J) Ciliated control and KIF17-KO cells were counted; percentages of ciliated cells. Values are means ± SE (error bars) of three independent experiments. In each experiment, 55–81 cells were observed, and the total numbers of cells observed (n) are shown. (K) Ciliary lengths (lengths of Ac-α-tubulin staining) of individual control cells and KIF17-KO cells were measured. Values are means ± SE (error bars) of three independent experiments. In each set of experiments, 54–69 ciliated cells were observed, and the total numbers of ciliated cells observed (n) are shown. (L), Localization of IFT88 in individual control and KIF17-KO cells was classified as “ciliary base,” “ciliary tip,” and “base and tip” and counted. The percentages of these populations are expressed as stacked bar graphs. In each set of experiments, 55–71 ciliated cells were observed, and the total numbers of ciliated cells observed (n) are shown. Note that cells with IFT88 signals at only the ciliary tip were not observed.
Staining for Smo was rarely observed within cilia under basal conditions in control RPE1 cells and the two KIF17-KO cell lines (–SAG), whereas the majority of control and KIF17-KO cells exhibited ciliary Smo staining after SAG treatment (+SAG; Supplemental Figure S4). Leaf and von Zastrow (2015) recently reported that ciliary localization of dopamine receptor D1 (DRD1) was greatly reduced by exogenous expression of a KIF17 mutant lacking motor activity, although ciliary localization of another GPCR, SSTR3, was not affected by expression of the KIF17 mutant. We therefore examined the localization of DRD1-EGFP stably expressed in control RPE1 cells and the two KIF17-KO cell lines. As shown in Figure 5G, DRD1-EGFP was uniformly distributed along cilia in control cells. The uniform ciliary distribution of DRD1-EGFP was not changed in the two KIF17-KO cell lines (Figure 5, H and I). Although we do not know the exact reason for the apparent difference between the result obtained using cells with exogenous expression of a motor-defective KIF17 mutant (Leaf and von Zastrow, 2015) and our result using KIF17-KO cell lines, expression of such a dominant-negative KIF17 mutant could indirectly affect the DRD1 trafficking.
In any event, our results indicate that, at least in mammalian cells, KIF17 is dispensable for ciliogenesis and IFT-B trafficking. This is compatible with previous studies showing that KIF17-deficent mice and zebrafish are normal with respect to ciliary assembly (Yin et al., 2011; Pooranachandran and Malicki, 2016).
Interaction of KIF17 NLS with importin α
A previous study indicated that KIF17 interacts with importin β2 (TNPO1; because the nomenclature of importins is confusing, varying among species (Miyamoto et al., 2016), we also include the official symbols of the corresponding importins), and mutation of its NLS (KRKK) to AAAA abolished the interaction (Dishinger et al., 2010). We therefore sought to confirm the potential interaction between KIF17 and TNPO1 by the VIP assay. However, we were unable to detect an interaction between EGFP-TNPO1 and mChe-KIF17 (Figure 6A, second column). Furthermore, we did not detect an interaction between EGFP-KPNB1 (importin β1) and mChe-KIF17 (third column). Because the sequence KRKK is typical of a classical monopartite-type NLS, which is recognized by importin α family proteins (Miyamoto et al., 2016), we then used the VIP assay to examine whether some importin α proteins can bind to KIF17. Data shown in Figure 6A suggest that KIF17 interacts with KPNA1/importin α1 (fourth column) and KPNA6/importin α6 (eighth column); KPNA1 and KPNA6 belong to the same importin α1 subfamily and show 82% amino acid identity in their primary structure (Miyamoto et al., 2016). The VIP data were confirmed by immunoblotting; as shown in Figure 6B, KIF17 interacted robustly with KPNA1 (lane 4) and weakly with KPNA6 (lane 8) but not with any other KPNA examined.
We then addressed whether KPNA1 and KPNA6 interact with the NLS (1015KRKK1018) of KIF17, using the VIP assay and conventional immunoblotting. As shown in Figure 6, C and D, the interaction of KIF17 with KPNA1 or KPNA6 was abolished by the AAAA mutation. In contrast, mutation of the IFT-B–binding sequence (R1000/1003A) did not affect the KIF17–KPNA interaction.
DISCUSSION
Before this study, it was been controversial whether KIF17 is a motor for or a cargo of the IFT-B complex in vertebrate cilia because KIF17-deficient mice and zebrafish are normal with respect to ciliary assembly (Yin et al., 2011; Pooranachandran and Malicki, 2016) and motor-less KIF17 can reach the ciliary tips (Williams et al., 2014; Jiang et al., 2015; also see Supplemental Figure S1). By taking advantage of the VIP assay, we first demonstrated the interaction of KIF17 with the IFT-B complex via the IFT46–IFT56 dimer and identified a C-terminal IFT-B–binding sequence that is conserved in vertebrate KIF17 and likely also in C. elegans OSM-3. Our subsequent analyses unequivocally showed that KIF17 entry into the cilium depends on its binding to IFT-B in addition to the NLS. Further experiments using IFT56-KO and KIF17-KO cells revealed that KIF17 cannot localize within cilia in IFT56-KO cells, whereas ciliary assembly and localization of the IFT-B complex and ciliary GPCRs examined were unaffected by the KIF17 deficiency. The most plausible explanation for these results is that KIF17 enters the cilium, most likely across the TZ, by binding to the IFT-B complex via the IFT46–IFT56 dimer and is then transported along the axoneme toward the ciliary tip by continuous binding to IFT-B. As proposed previously, we believe that heterotrimeric kinesin-II mainly functions as an anterograde motor for the IFT-B complex in vertebrate cells. This is indirectly supported by our recent finding that KIF3B, a subunit of heterotrimeric kinesin-II, interacts with subunits of IFT-B other than IFT46 and IFT56 (Funabashi et al., unpublished data).
How can we explain the apparently different roles of vertebrate KIF17 and C. elegans OSM-3? The axoneme of the sensory cilia of C. elegans has a bipartite structure with the proximal segment composed of nine microtubule doublets and the distal segment composed of nine singlets (Scholey, 2013; Mijalkovic et al., 2016); the proximal segment corresponds to the typical axoneme of vertebrate cilia, whereas the distal segment structure is not generally found in vertebrate cilia. Recent studies provided data demonstrating a model of the concerted actions of heterotrimeric kinesin-II and OSM-3 in C. elegans cilia (Prevo et al., 2015; Mijalkovic et al., 2016) that was proposed by Scholey (2013); heterotrimeric kinesin-II transports IFT particles along the proximal segment toward a “handover zone” where OSM-3 gradually replaces kinesin-II and drives the transport of IFT particles along the distal segment toward the ciliary tip (Prevo et al., 2015; Mijalkovic et al., 2016). Thus one can speculate that the transport powered by heterotrimeric kinesin-II is a common event in both vertebrate and C. elegans cilia, whereas OSM-3–mediated transport along the distal segment is specific for C. elegans cilia. If this is the case, OSM-3 needs to be trafficked toward the handover zone as a cargo of IFT particles along the proximal segment of C. elegans cilia. From another perspective, a future issue that should be addressed is the role of KIF17 at the ciliary tip of vertebrate cells.
How does KIF17 gain entry into cilia in an IFT-B–dependent manner? At the ciliary base, there is a permeability barrier with properties resembling those of the NPC (Takao and Verhey, 2016; Verhey and Yang, 2016); similar to the NPC, the permeability barrier of the TZ contains nucleoporin proteins (Kee et al., 2012). A study by Takao et al. (2014) showed that the cross-linking of a nucleoporin blocked the ciliary entry of various soluble proteins, including KIF17 and IFT88, whereas it did not affect the entry of membrane proteins. On the other hand, Wei et al. (2013) showed that FBF1, a protein located at the transition fiber and the distal appendage of the basal body, interacts directly with IFT54 and is required for transit of assembled IFT particles across the barrier. Furthermore, a recent superresolution microscopic study suggested the assembly of IFT particles around the TZ before entry into the cilium (Yang et al., 2015). Overall, it is likeliest that ciliary entry of KIF17 requires binding to assembled IFT particles as well as to an NPC-like machinery around the TZ.
Our study showed that the NLS of KIF17 is essential for its entry into both the nucleus and cilium, although OSM-3 lacks an NLS in the corresponding region. We showed binding of the KIF17 NLS to KPNA1 (importin α1) and KPNA6 (importin α6), although we were unable to confirm the previously proposed binding to TNPO1 (importin β2); a recent study suggested that binding of KIF17 to TNPO1 is enhanced by regulatory proteins, such as Rab23 (Lim and Tang, 2015), but our attempts to show the involvement of Rab23 in KIF17–TNPO1 binding have been unsuccessful. However, our results are compatible with the fact that the KIF17 NLS is typical of classical monopartite-type NLSs, which are recognized by importin α proteins (Miyamoto et al., 2015), but does not fit PY-NLSs, which are recognized by transportin/importin β2 proteins (Twyffels et al., 2014). Further studies are required toward understanding of the roles of specific importins in the ciliary entry of proteins.
MATERIALS AND METHODS
Plasmids
Construction of expression vectors for IFT-B proteins was described previously (Katoh et al., 2016). Human KIF17 cDNA (clone ID IRAK140P09) was obtained from the RIKEN BioResource Center through the National BioResource Project of the Ministry of Education, Culture, Sports, Science and Technology, Japan, and subcloned into the pCAG-EGFP-C or pCAG-mCherry-C vector. KIF17 mutants were constructed using the SLiCE cloning method (Motohashi, 2015). Mouse importin and transportin cDNAs were kindly provided by Yoshihiro Yoneda, Noriko Yasuhara, Masahiro Oka, and Tetsuji Moriyama (National Institutes of Biomedical Innovation, Health and Nutrition, Japan; Tsuji et al., 1997) and subcloned into pEGFP-C1 or pEGFP-C2.
Plasmids for production of the replication-defective, self-inactivating lentiviral vector pRRLsinPPT-EGFP and packaging plasmids (pRSV-REV, pMD2.g, and pMDLg/pRRE) were kindly provided by Peter McPherson (McGill University, Montreal, Canada; Thomas et al., 2009). Human KIF17 or DRD1 (obtained from Kazusa DNA Research Institute, Chiba, Japan; Product ID FXC00007) cDNA was subcloned into pRRLsinPPT-EGFP.
Antibodies and reagents
The following antibodies were obtained from the indicated vendors: monoclonal mouse anti–Ac-α-tubulin (1:500; 6-11B-1; Sigma-Aldrich); monoclonal rabbit anti–Ac-α-tubulin (1:300; D20G3; Cell Signaling Technology); monoclonal mouse anti–γ-tubulin (1:1000; GTU88; Sigma-Aldrich); polyclonal rabbit anti-Arl13b (1:1000; 17711-1-AP; ProteinTech); polyclonal rabbit anti-IFT88 (1:200; 13967-1-AP; ProteinTech); polyclonal rabbit anti-Smo (1:200; ab38686; Abcam); monoclonal mouse anti-GFP (1:1000; JL-8; BD Biosciences); polyclonal rabbit anti-RFP (1:1000; PM005; MBL Life Science); Alexa Fluor–conjugated secondary antibodies (Molecular Probes); and peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories). SAG and Polyethylenimine Max were purchased from Enzo Life Sciences and Polysciences, respectively. GST-tagged anti-GFP Nb prebound to glutathione–Sepharose 4B beads were prepared as described previously (Katoh et al., 2015).
Establishment of KO cell lines using the CRISPR/Cas9 system
The strategy for the KO of genes in hTERT-RPE1 cells (CRL-4000; ATCC) using the CRISPR/Cas9 system and homology-independent repair is described elsewhere in detail (Katoh et al., 2017). Single guide RNA (sgRNA) sequences targeting the human IFT56 or KIF17 gene (Supplemental Table S1) were designed using CRISPRscan (www.crisprscan.org; Moreno-Mateos et al., 2015). Double-stranded oligonucleotides for these sequences were separately inserted into the knock-in donor vector pDonor-tBFP-NLS-Neo (deposited in Addgene, ID 80766) and the all-in-one sgRNA expression vector pSpCas9(BB)-2A-Puro (Addgene plasmid 48139). hTERT-RPE1 cells cultured to ∼3.0 × 105 cells on a 12-well plate were transfected with 1 µg of the pSpCas9(BB)-2A-Puro and 0.25 µg of the donor vector using X-tremeGENE9 DNA Transfection Reagent (Roche Applied Science). After selection in medium containing G418 (600 µg/ml), the cells with nuclear tBFP fluorescence were isolated. To confirm KO of the IFT56 or KIF17 gene, genomic DNA was extracted from isolated cells and subjected to PCR using KOD FX Neo DNA polymerase (Toyobo). Three sets of primers (Supplemental Table S1) were used to distinguish the following three states of integration of the donor vector: forward integration, reverse integration, and no integration with a small insertion or deletion (Supplemental Figure S2A). Direct sequencing of the PCR products ensured KO of both alleles of the IFT56 or KIF17 gene, with integration of the donor vector and/or small deletion/insertion causing a frameshift.
Preparation of cells stably expressing EGFP-KIF17 or DRD1-EGFP
Lentiviral vectors were prepared as described previously (Takahashi et al., 2012). Briefly, pRRLsinPPT-EGFP-KIF17 or pRRLsinPPT-DRD1-EGFP was transfected into HEK293T cells using Polyethylenimine Max with the packaging plasmids (pRSV-REV, pMD2.g, and pMDL/pRRE). Culture medium was replaced 8 h after transfection. Culture medium containing lentiviral particles were collected at 24, 36, and 48 h after transfection, passed through a 0.45-µm filter (Sartorius), and centrifuged at 32,000 × g at 4°C for 4 h using an R15A rotor and Himac CR22G centrifuge (Hitachi Koki, Japan). Precipitated viral particles were resuspended in Opti-MEM (Invitrogen). Control RPE1 cells and IFT56-KO or KIF17-KO cells that express EGFP-KIF17 or DRD1-EGFP were prepared by addition of the lentiviral suspension to the culture medium, followed by a 24-h incubation. These cells were used for immunofluorescence analysis.
VIP assay
The VIP assay was performed as described previously (Katoh et al., 2015, 2016). Briefly, HEK293T cells grown to ∼1.6 × 106 cells on a six-well plate in DMEM with high glucose (Nacalai Tesque) supplemented with 5% fetal bovine serum (FBS) were transfected with expression vectors for EGFP and mChe/tRFP fusions (2 µg each) using Polyethylenimine Max (20 µg). After 24 h, the cells were lysed in 250 µl of lysis buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid–KOH, pH 7.4, 150 mM NaCl, 0.1% Triton X-100, and 10% glycerol) containing a protease inhibitor cocktail (EDTA-free; Nacalai Tesque). The lysates were mixed with 5 µl of GST–anti-GFP Nb beads at 4°C for 1 h. After washing three times with lysis buffer, the precipitated beads bearing fluorescent proteins were observed using an all-in-one-type fluorescence microscope (Biozero BZ-8000, Keyence) with a 20×/0.75 objective lens under constant conditions (sensitivity ISO 400, exposure 1/30 s for green; and sensitivity ISO 800, exposure 1/10 s for red) unless otherwise noted.
Immunofluorescence analysis
hTERT-RPE1 cells were cultured in DMEM/F-12 (Nacalai Tesque) supplemented with 10% FBS and 0.348% sodium bicarbonate. To induce ciliogenesis, cells were grown up to 100% confluence on coverslips and starved for 24 h in Opti-MEM containing 0.2% bovine serum albumin. Expression vectors were transfected into the cells using X-tremeGENE9 DNA Transfection Reagent (Roche Applied Science).
For immunofluorescence analysis, cells were fixed and permeabilized with 3% paraformaldehyde at 37°C for 5 min and subsequently in 100% methanol for 5 min at −20°C, and washed three times with phosphate-buffered saline. The fixed/permeabilized cells were blocked with 10% FBS and stained with antibodies diluted in 5% FBS. The stained cells were observed using an Axiovert 200 M microscope (Carl Zeiss).
Supplementary Material
Acknowledgments
We are grateful to Yoshihiro Yoneda, Noriko Yasuhara, Masahiro Oka, and Tetsuji Moriyama for providing importin cDNAs and Yoshihide Hayashizaki and Sumio Sugano for providing the KIF17 cDNA. We also thank Helena Akiko Popiel and Masahiro Oka for critical reading of the manuscript. This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas “Cilia and Centrosome” from the Ministry of Education, Culture, Sports, Science and Technology, Japan (Grants 25113514 and 15H01211 to K.N.); grants from the Japan Society for the Promotion of Science (Grants 22390013, 15H04370, and 15K14456 to K.N. and 25860044 and 15K07929 to Y.K.); and grants from the Uehara Memorial Foundation to K.N. and from the Takeda Science Foundation to Y.K.
Abbreviations used:
- GST
glutathione S-transferase
- Hh
Hedgehog
- IFT
intraflagellar transport
- KO
knockout
- Nb
nanobody
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- SAG
Smoothened agonist
- Smo
Smoothened
- TZ
transition zone
- VIP
visible immunoprecipitation.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-09-0648) on January 11, 2017.
REFERENCES
- Ahmed NT, Gao C, Lucker BF, Cole DG, Mitchell DR. ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J Cell Biol. 2008;183:313–322. doi: 10.1083/jcb.200802025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Cajánek L, Fort C, Blisnick T, Weber K, Taschner M, Mizuno N, Lamla S, Bastin P, Nigg EA, Lorentzen E. Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science. 2013;341:1009–1012. doi: 10.1126/science.1240985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldt K, van Reeuwijk J, Lu Q, Koutroumpas K, Nguyen TM, Texier Y, van Beersum SEC, Horn N, Willer JR, Mans D, et al. An organelle-specific protein landscape identifies novel diseases and molecular mechanisms. Nat Commun. 2016;7:11491. doi: 10.1038/ncomms11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dishinger JF, Kee HL, Jenkins PM, Fan S, Hurd TW, Hammond JW, Truong YN-T, Margolis B, Martens JR, Verhey KJ. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-β2 and RanGTP. Nat Cell Biol. 2010;12:703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguether T, San Agustin JT, Keady BT, Jonassen JA, Liang Y, Francis R, Tobita K, Johnson CA, Abdelhamed ZA, Lo CW, Pazour GJ. IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev Cell. 2014;21:279–290. doi: 10.1016/j.devcel.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Katoh Y, Nakayama K. Intraflagellar transport-A complex mediates ciliary entry as well as retrograde trafficking of ciliary G protein-coupled receptors. Mol Biol Cell. 2017;28:429–439. doi: 10.1091/mbc.E16-11-0813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Qin H, Follit JA, Pazour GJ, Rosenbaum JL, Witman GB. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J Cell Biol. 2007;176:653–665. doi: 10.1083/jcb.200608041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard PW, Jue SF, Maurer RA. Interaction of mouse TTC30/DYF-1 with multiple intraflagellar transport complex B proteins and KIF17. Exp Cell Res. 2013;319:2275–2281. doi: 10.1016/j.yexcr.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd TW, Fan S, Margolis BL. Localization of retinitis pigmentosa 2 to cilia is regulated by importin β2. J Cell Sci. 2011;124:718–726. doi: 10.1242/jcs.070839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insinna C, Pathak N, Perkins B, Drummond I, Besharse JC. The homodimeric kinesin, Kif17, is essential for vertebrate photoreceptor sensory outer segment development. Dev Biol. 2008;316:160–170. doi: 10.1016/j.ydbio.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ide T, Yagi T, Jiang X, Hirono M, Sasaki H, Yanagisawa H, Wemmer KA, Stainier DYR, Qin H, et al. TTC26/DYF13 is an intraflagellar transport protein required for transport of motility-related proteins into flagella. Elife. 2014;3:e01566. doi: 10.7554/eLife.01566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins PM, Hurd TW, Zhang L, McEwen DP, Brown RL, Margolis B, Verhey KJ, Martens JR. Ciliary targeting of olfactory CNG channels requires the CNGB1b subunit and the kinesin-2 motor protein, KIF17. Curr Biol. 2006;16:1211–1216. doi: 10.1016/j.cub.2006.04.034. [DOI] [PubMed] [Google Scholar]
- Jiang L, Tam BM, Ying G, Wu S, Hauswirth WW, Frederick JM, Moritz OL, Baehr W. Kinesin family 17 (osmotic avoidance abnormal-3) is dispensable for photoreceptor morphology and function. FASEB J. 2015;29:4866–4880. doi: 10.1096/fj.15-275677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Michisaka S, Nozaki S, Funabashi T, Hirano T, Takei R, Nakayama K. Practical method for targeted disruption of cilia-related genes by using CRISPR/Cas9-mediated homology-independent knock-in system. Mol Biol Cell. 2017;28 doi: 10.1091/mbc.E17-01-0051. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Nozaki S, Hartanto D, Miyano R, Nakayama K. Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. J Cell Sci. 2015;128:2351–2362. doi: 10.1242/jcs.168740. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Terada M, Nishijima Y, Takei R, Nozaki S, Hamada H, Nakayama K. Overall architecture of the intraflagellar transport (IFT)-B complex containing Cluap1/IFT38 as an essential component of the IFT-B peripheral subcomplex. J Biol Chem. 2016;291:10962–10975. doi: 10.1074/jbc.M116.713883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keady BT, Samtani R, Tobita K, Tsuchiya M, San Augstin JT, Follit JA, Jonassen JA, Subramanian R, Lo CW, Pazour GJ. IFT25 links the signal-dependent movement of hedgehog components to intraflagellar transport. Dev Cell. 2012;22:940–951. doi: 10.1016/j.devcel.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee HL, Dishinger JF, Blasius TL, Liu C-J, Margolis B, Verhey KJ. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol. 2012;14:431–437. doi: 10.1038/ncb2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaf A, von Zastrow M. Dopamine receptors reveal an essential role of IFT-B, KIF17, and Rab23 in delivering specific receptors to primary cilia. Elife. 2015;4:e06996. doi: 10.7554/eLife.06996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF. IFT-cargo interactions and protein transport in cilia. Trends Biochem Sci. 2015;40:765–778. doi: 10.1016/j.tibs.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YS, Tang BL. A role for Rab23 in the trafficking of Kif17 to the primary cilium. J Cell Sci. 2015;128:2996–3008. doi: 10.1242/jcs.163964. [DOI] [PubMed] [Google Scholar]
- Madhivanan K, Aguilar RC. Ciliopathies: the trafficking connection. Traffic. 2014;15:1031–1056. doi: 10.1111/tra.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijalkovic J, Prevo B, Peterman EJG. Why motor proteins team up: intraflagellar transport in C. elegans cilia. Worm. 2016;5:e1170275. doi: 10.1080/21624054.2016.1170275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Hosoba K, Ochiai H, Royba E, Izumi H, Sakuma T, Yamamoto T, Dynlacht BD, Matsuura S. The microtubule-depolymerizing activity of a mitotic kinesin protein KIF2A drives primary cilia disassembly coupled with cell proliferation. Cell Rep. 2015;10:664–673. doi: 10.1016/j.celrep.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Yamada K, Yoneda Y. Importin α: a key molecule in nuclear transport and non-transport functions. J Biochem. 2016;160:69–75. doi: 10.1093/jb/mvw036. [DOI] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Vejnar CE, Beaudoin J-D, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K. A simple and efficient seamless DNA cloning method using SLiCE from Escherichia coli laboratory strains and its application to SLiP site-directed mutagenesis. BMC Biotechnol. 2015;15:47. doi: 10.1186/s12896-015-0162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G, Blacque OE, Snow JJ, Leroux MR, Scholey JM. Functional coordination of intraflagellar transport motors. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- Pan X, Ou G, Civelekoglu-Scholey G, Blacque OE, Endres NF, Tao L, Mogilner A, Leroux MR, Vale RD, Scholey JM. Mechanism of transport of IFT particles in C. elegans cilia by the concerted action of kinesin-II and OSM-3 motors. J Cell Biol. 2006;174:1035–1045. doi: 10.1083/jcb.200606003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooranachandran N, Malicki JJ. Unexpected roles for ciliary kinesins and intraflagellar transport proteins. Genetics. 2016;203:771–785. doi: 10.1534/genetics.115.180943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevo B, Mangeol P, Oswald F, Scholey JM, Peterman EJ. Functional differentiation of cooperating kinesin-2 motors orchestrates cargo import and transport in C. elegans cilia. Nat Cell Biol. 2015;17:1536–1545. doi: 10.1038/ncb3263. [DOI] [PubMed] [Google Scholar]
- Scholey JM. Kinesin-2: a family of heterotrimeric and homodimeric motors with diverse intracellular transport functions. Annu Rev Cell Dev Biol. 2013;29:443–469. doi: 10.1146/annurev-cellbio-101512-122335. [DOI] [PubMed] [Google Scholar]
- Schwartz R, Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow JJ, Ou G, Gunnarson AL, Walker MR, Zhou HM, Brust-Mascher I, Scholey JM. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol. 2004;6:1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- Sung C-H, Leroux MR. The roles of evolutionary conserved functional modules in cilia-related trafficking. Nat Cell Biol. 2013;15:1387–1397. doi: 10.1038/ncb2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiderski RE, Nakano Y, Mullins RF, Seo S, Bánfi B. A mutation in the mouse Ttc26 gene leads to impaired hedgehog signaling. PLoS Genet. 2014;10:e1004689. doi: 10.1371/journal.pgen.1004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Kubo K, Waguri S, Yabashi A, Shin H-W, Katoh Y, Nakayama K. Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J Cell Sci. 2012;125:4049–4057. doi: 10.1242/jcs.102913. [DOI] [PubMed] [Google Scholar]
- Takao D, Dishinger JF, Kee HL, Pinskey JM, Allen BL, Verhey KJ. An assay for clogging the ciliary pore complex distinguishes mechanisms of cytosolic and membrane protein entry. Curr Biol. 2014;24:2288–2294. doi: 10.1016/j.cub.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao D, Verhey KJ. Gated entry into the ciliary compartment. Cell Mol Life Sci. 2016;73:119–127. doi: 10.1007/s00018-015-2058-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Bhogaraju S, Lorentzen E. Architecture and function of IFT complex proteins in ciliogenesis. Differentiation. 2012;83:S12–S22. [Google Scholar]
- Taschner M, Weber K, Mourão A, Vetter M, Awasthi M, Stiegler M, Bhogaraju S, Lorentzen E. Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin-binding IFT-B2 complex. EMBO J. 2016;35:773–790. doi: 10.15252/embj.201593164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S, Ritter B, Verbich D, Sanson C, Bourbonnière L, McKinney RA, McPherson PS. Intersectin regulates dendritic spine development and somatodendritic endocytosis but not synaptic vesicle recycling in hippocampal neurons. J Biol Chem. 2009;284:12410–12419. doi: 10.1074/jbc.M809746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji L, Takumi T, Imamoto N, Yoneda Y. Identification of novel homologues of mouse importin α, the α subunit of the nuclear pore-targeting complex, and their tissue-specific expression. FEBS Lett. 1997;416:30–34. doi: 10.1016/s0014-5793(97)01092-2. [DOI] [PubMed] [Google Scholar]
- Twyffels L, Gueydan C, Kruys V. Transportin-1 and Transportin-2: protein nuclear import and beyond. FEBS Lett. 2014;588:1857–1868. doi: 10.1016/j.febslet.2014.04.023. [DOI] [PubMed] [Google Scholar]
- Verhey KJ, Yang W. Permeability barriers for generating a unique ciliary protein and lipid composition. Curr Opin Cell Biol. 2016;41:109–116. doi: 10.1016/j.ceb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Ling K, Hu J. The essential roles of transition fibers in the context of cilia. Curr Opin Cell Biol. 2015;35:98–105. doi: 10.1016/j.ceb.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Xu Q, Zhang Y, Li Y, Zhang Q, Hu Z, Harris PC, Torres VE, Ling K, Hu J. Transition fibre protein FBF1 is required for the ciliary entry of assembled intraflagellar transport complexes. Nat Commun. 2013;4:2750. doi: 10.1038/ncomms3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CL, McIntyre JC, Norris SR, Jenkins PM, Zhang L, Pei Q, Verhey K, Martens JR. Direct evidence for BBSome-associated intraflagellar transport reveals distinct properties of native mammalian cilia. Nat Commun. 2014;5:5813. doi: 10.1038/ncomms6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TT, Su J, Wang W-J, Craige B, Witman GB, Tsou M-FB, Liao J-C. Superresolution pattern recognition reveals the architectural map of the ciliary transition zone. Sci Rep. 2015;5:14096. doi: 10.1038/srep14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Takei Y, Kido MA, Hirokawa N. Molecular motor KIF17 is fundamental for memory and learning via differential support of synaptic NR2A/2B levels. Neuron. 2011;70:310–325. doi: 10.1016/j.neuron.2011.02.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.