Expression of the Drosophila slp1 gene depends on nonadditive interactions between two cis-regulatory enhancers. These enhancers are repressed by preventing either Pol II recruitment or release of promoter-proximal paused Pol II in a manner that is both enhancer and transcription factor specific and can account for their nonadditive interaction.
Abstract
The initial metameric expression of the Drosophila sloppy paired 1 (slp1) gene is controlled by two distinct cis-regulatory DNA elements that interact in a nonadditive manner to integrate inputs from transcription factors encoded by the pair-rule segmentation genes. We performed chromatin immunoprecipitation on reporter genes containing these elements in different embryonic genotypes to investigate the mechanism of their regulation. The distal early stripe element (DESE) mediates both activation and repression by Runt. We find that the differential response of DESE to Runt is due to an inhibitory effect of Fushi tarazu (Ftz) on P-TEFb recruitment and the regulation of RNA polymerase II (Pol II) pausing. The proximal early stripe element (PESE) is also repressed by Runt, but in this case, Runt prevents PESE-dependent Pol II recruitment and preinitiation complex (PIC) assembly. PESE is also repressed by Even-skipped (Eve), but, of interest, this repression involves regulation of P-TEFb recruitment and promoter-proximal Pol II pausing. These results demonstrate that the mode of slp1 repression by Runt is enhancer specific, whereas the mode of repression of the slp1 PESE enhancer is transcription factor specific. We propose a model based on these differential regulatory interactions that accounts for the nonadditive interactions between the PESE and DESE enhancers during Drosophila segmentation.
INTRODUCTION
The differential regulation of gene transcription is critically important for the development of multicellular organisms. The cycle of events that characterize transcription of protein-coding genes by RNA polymerase II (Pol II) can be divided into three phases: initiation, when Pol II is recruited to form a preinitiation complex (PIC) at the promoter and begins RNA synthesis; elongation, during which Pol II is modified into an elongating form and escapes the promoter; and termination, when both Pol II and the nascent RNA transcript are released from the DNA template. Each phase of this multistep process is subject to regulation (Core and Lis, 2008). Transcription regulation in eukaryotes involves interactions between sequence-specific DNA-binding transcription factors and cis-regulatory DNA elements referred to as enhancers, which can be located many kilobases upstream or downstream of the transcription start site (TSS). Although recent advances in genome-wide chromatin immunoprecipitation (ChIP)-chip analysis and the ENCODE and modENCODE projects have resulted in the identification of many enhancer elements (Roy et al., 2010; Negre et al., 2011), more studies are required to understand how these elements influence molecular events that occur at the promoter, in particular in animal cells, in which genes frequently contain multiple enhancers.
Many of the DNA-binding transcription factors that regulate gene expression during development can act either as activators or repressors, depending on the architecture of binding sites in the enhancer, the presence of other DNA-binding proteins, and other environmental cues (Umayahara et al., 1994; Aronson et al., 1997; Dubnicoff et al., 1997; Porter et al., 1997; Kramer et al., 1999; Cheung et al., 2005). Although the mechanism of activation and repression by different context-dependent regulators has been studied in several systems (Javed et al., 2000; Peng and Jahroudi, 2002; Seufert et al., 2005; Sakabe et al., 2012), it is difficult to completely define or carefully control the in vivo cellular context. Studies of the Drosophila segmentation pathway have generated a rich framework that provides advantages for investigating the properties of context-dependent transcription factors. For example, the dual-regulatory properties of Runt, the founding member of the Runx family of transcriptional regulators, are exemplified by the parasegment-specific effects of Runt on engrailed (en), wingless (wg), and slp1 (Manoukian and Krause, 1993; Tracey et al., 2000; Swantek and Gergen, 2004).
The 14-stripe slp1 expression pattern in the late blastoderm embryo is generated in response to combinatorial regulation by Runt and three other pair-rule transcription factors: the Zn-finger transcription factor Odd-paired (Opa) and the homeodomain proteins Eve and Fushi tarazu (Ftz; Swantek and Gergen, 2004). The 14-stripe pattern consists of seven repetitive units, each containing four different cellular contexts for slp1 transcription: type I cells are the two cells located in the anterior half of odd-numbered parasegments that do not express slp1 (Figure 1A); type II cells comprise the posterior half of the odd-numbered parasegments and express the odd-numbered slp1 stripes; type III cells comprise the anterior half of the even-numbered parasegments and do not express slp1; and type IV cells comprise the posterior half of the even-numbered parasegments and express the even-numbered slp1 stripes. Different factors are responsible for slp1 regulation in each of these four contexts. Eve represses slp1 in type I cells, whereas repression in type III cells requires both Runt and Ftz. Expression of slp1 in type II cells requires Runt in combination with Opa. Expression in type IV cells also depends on Opa, in this case without Runt but with a contribution from an as- yet-unidentified Factor X (Swantek and Gergen, 2004). Of importance, slp1 responds to these four transcription factors in all somatic nuclei of late blastoderm–stage embryos, indicating that preexisting, spatially regulated epigenetic modifications in chromatin are not critical to slp1 regulation during this stage of development.
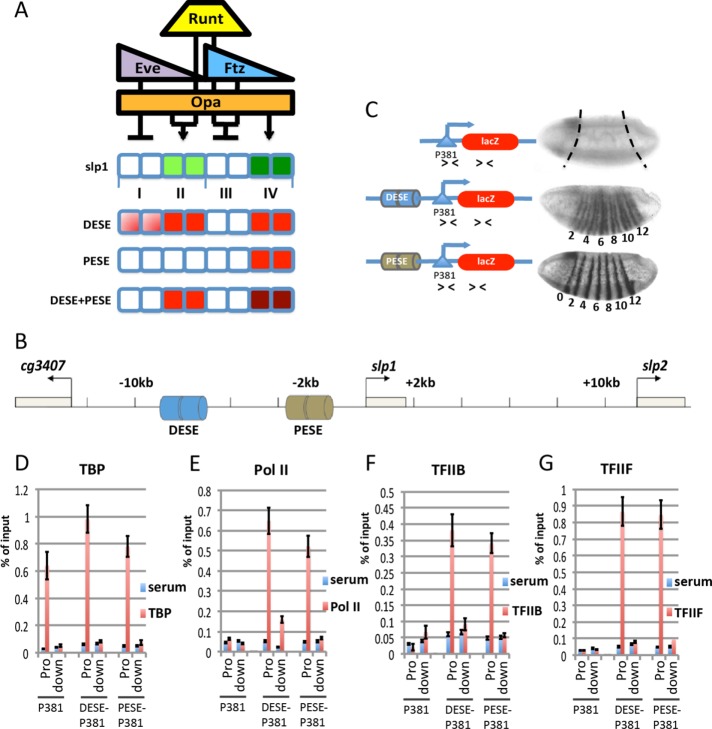
FIGURE 1:
DESE and PESE stimulate preinitiation complex formation on the slp1 promoter. (A) Rules for slp1 regulation by Runt, Eve, Ftz, and Opa for an eight-cell unit spanning two parasegments along the anterior–posterior axis. The expression domains of these regulators are indicated above cells that express (green) or repress (white) slp1 in four cell types, labeled I–IV, respectively. The expression patterns of reporter genes containing different slp1 enhancers are depicted below, with lacZ-expressing cells shaded in red. (B) The slp1 locus flanked by the cg3407 and slp2 genes, showing location of the DESE and PESE enhancers. (C) Expression of slp1[p381]lacZ (P381), slp1[8765/p381]lacZ (DESE-P381), or slp1[3125/p381]lacZ (PESE-P381) transgenes in stage 6 Drosophila embryos as visualized by in situ hybridization. Embryos are oriented anterior to the left, dorsal side up. Numbers beneath the embryos indicate even-numbered stripes. The schematic representations of these reporter constructs show the location of primers (arrowheads) used for qPCR to specifically detect the reporter gene promoter (Pro) and downstream (down) lacZ gene in ChIP assays. (D–G) Results of ChIP assays with control serum (blue bars) and antibodies specific for TBP, Pol II (8WG16), TFIIB, and TFIIF (red bars) using chromatin from embryos homozygous for the reporter genes shown in C.
Two distinct cis-acting DNA elements from the slp1 locus that drive expression of stripes in the blastoderm embryo have been identified, and extensive genetic experiments have revealed how these elements respond to manipulations in the activity of the pair-rule transcription factors (Prazak et al., 2010). The proximal early stripe element (PESE) enhancer, located between 3.1 and 2.5 kb upstream of the slp1 TSS, drives expression in type IV cells, corresponding to the even-numbered slp1 stripes (Figure 1, B and C). The distal early stripe element (DESE) enhancer, located between 8.1 and 7.2 kb upstream of the TSS (Figure 1B), drives expression in cells corresponding to both the odd- and even-numbered slp1 stripes (type II and type IV cells) with stronger-than-normal expression of the odd stripes in cell type II. Of note, DESE also drives expression in type I cells, in which Eve normally represses slp1 (Figure 1, A and C). The inappropriate expression of the DESE-lacZ reporter gene in type I cells is due to the insensitivity of DESE to repression by Eve (Prazak et al., 2010). A striking finding from this prior work is that a composite reporter construct containing both the DESE and PESE enhancers recapitulates the wild-type slp1 pattern. Critical to understanding this nonadditive interaction is determining how PESE prevents inappropriate activity of DESE in Eve-expressing type I cells. Of importance, these results provide a platform for using genetic manipulations to investigate the in vivo mechanisms by which DESE and PESE mediate activation and repression in response to these different pair-rule transcription factors.
Here we combine genetic approaches with ChIP assays using chromatin from carefully staged embryos containing different slp1 reporter genes to investigate the molecular mechanisms by which DESE and PESE regulate transcription in response to the pair-rule transcription factors in these different cellular contexts. We find that Runt represses DESE and PESE by two different mechanisms. Runt and Ftz repress DESE by antagonizing P-TEFb recruitment and preventing release of promoter-proximal paused Pol II. In contrast, Runt prevents PESE-dependent recruitment of Pol II and other PIC components. PESE-dependent transcription is also repressed by Eve. However, in this case, the repression is due to antagonism of P-TEFb recruitment and a block to release of promoter-proximal paused Pol II. On the basis of these findings, we propose a model for how these two enhancers contribute to regulating transcription of the endogenous slp1 gene in these four different cellular contexts. This model strongly suggests that regulation of Pol II pausing has an unappreciated and potentially widespread role in restricting gene expression in developmental systems.
RESULTS
DESE and PESE facilitate PIC assembly on the slp1 promoter
Studies on slp1 cis-regulatory architecture have used both conventional P-element and ΦC31-mediated transgenic approaches with different basal promoter segments (Prazak et al., 2010; Fujioka and Jaynes, 2012). To better compare results of ChIP assays done with different reporter genes, we used ΦC31-mediated transgenesis to integrate different constructs into the same P{CaryP}attP2 landing site. The backbone construct for these experiments was a lacZ reporter with a promoter-containing DNA segment that spans from 260 base pairs upstream to 121 base pairs downstream of the slp1 TSS. A reporter gene containing only this region, slp[p381]lacZ, is expressed at low levels in a dorsal anterior patch of cells, with no expression throughout the presumptive segmented region of the embryo (Figure 1C). This observation agrees with previous findings (Prazak et al., 2010; Fujioka and Jaynes, 2012) and indicates that this promoter-containing DNA segment does not by itself respond to the pair-rule gene regulatory circuitry.
DESE was originally identified within a DNA segment extending from 8.7 to 6.5 kb upstream of the slp1 TSS. A reporter with this DNA segment, slp1[8765/p381]lacZ, generated the characteristic DESE-driven pattern with strong expression of both odd- and even-numbered stripes in type II and type IV cells, respectively, along with inappropriate expression in type I cells anterior to the odd stripes (Figure 1C). Similarly, inclusion of a PESE-containing segment that extends from 3.1 to 2.5 kb upstream of the TSS in the slp1[3125/p381]lacZ reporter resulted in expression of only the even-numbered stripes in type IV cells (Figure 1C). These results confirm observations made with other basal promoter segments and provide a starting point for using ChIP assays to investigate the effects of these two enhancer elements on the association of different components of the transcriptional machinery with the slp1 promoter.
We used chromatin from carefully staged collections of Drosophila embryos to investigate the effects of DESE and PESE on association of Pol II and other general transcription factors with the p381 promoter. TBP, the TATA-binding component of TFIID, was associated with the promoter region of all three reporters (Figure 1D). The specificity of this association is confirmed by the lack of a ChIP signal, using primers from the downstream lacZ gene (Figure 1D). In contrast to the results with TBP, there was little association of Pol II, TFIIB, or TFIIF with the promoter region of the slp1[p381]lacZ reporter (Figure 1, E–G). Of importance, inclusion of either DESE or PESE increased association of all three factors with the promoter. As expected, the TFIIB and TFIIF signals were specific for the promoter regions (Figure 1, F and G), whereas a modest increase in association of Pol II with the lacZ structural gene is detected for the more widely expressed DESE-containing reporter gene (Figure 1E). On the basis of these results, we conclude that p381 is sufficient for recruiting TBP and that DESE and PESE act to stimulate the recruitment and/or stabilize the association of Pol II and other PIC components with the slp1 promoter.
Runt and Ftz repress DESE by preventing release of promoter-proximal paused Pol II
Ectopic coexpression of Runt and Ftz using the NGT maternal GAL4 expression system (Tracey et al., 2000) results in uniform slp1 repression in all somatic cells of the late blastoderm–stage embryo (Swantek and Gergen, 2004). This approach was previously used to demonstrate that slp1 repression by Runt and Ftz occurs downstream of Pol II recruitment and the initiation of transcription and involves preventing release of promoter-proximal paused Pol II into active elongation (Wang et al., 2007). The DESE-containing slp1[8765/p381]lacZ reporter recapitulated the repression of slp1 by Runt and Ftz (Figure 2A). ChIP assays showed no difference in association of either TBP (Figure 2B) or Pol II (Figure 2C) with the reporter gene promoter in Runt and Ftz–coexpressing (RF) embryos, in which the reporter gene is repressed, versus the ChIP signal obtained in wild-type (WT) embryos, which express the reporter in multiple cell types. Repression by Runt and Ftz did reduce the more modest ChIP signal for Pol II association with the lacZ structural gene that is observed in WT embryos to background levels (Figure 2C). Of importance, repression by Runt and Ftz also increased the ChIP signals on the slp1[8765/p381]lacZ promoter for Pol II containing the phospho–Ser-5 (pSer5) modification on the C-terminal domain (CTD) and for a subunit of the negative elongation factor NELF (Figure 2, D and E). The CTD pSer5 modification is a signature of transcription initiation, and association with NELF is a hallmark of Pol II pausing (Saunders et al., 2006; Hirose and Ohkuma, 2007; Lee et al., 2008). These results mirror those obtained previously for slp1 and indicate that Runt and Ftz repress DESE-driven expression by preventing release of promoter-proximal paused Pol II.
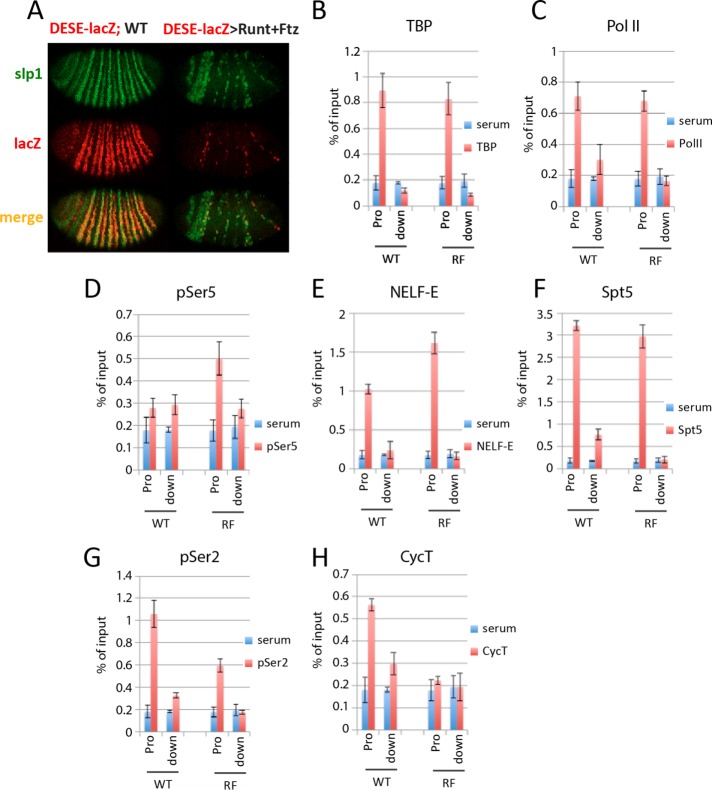
FIGURE 2:
Runt and Ftz repress DESE by blocking paused Pol II release. (A) In situ hybridization showing slp1 (green) and slp1[8765/p381]lacZ (DESE-lacZ; red) expression in a wild-type (WT) embryo and in response to NGT-driven coexpression of Runt and Ftz. The WT (left) embryo was from a cross between homozygous NGT[40]; slp1[8765/p381]lacZ NGT[A] females and WT males. The embryo with ectopic expression of Runt and Ftz (right) was from a cross between females of the same genotype with homozygous UAS-runt[232]; UAS-ftz[263] males. (B–H) Results of ChIP assays with control serum (blue bars) and antibodies against (red bars) TBP (B), Pol II antibody 8WG16 (C), Pol II antibody H14 (D), NELF-E (E), Spt5 (F), Pol II antibody ab5095 (G), and CycT (H), using primers to detect the promoter and downstream lacZ gene of the slp1[8765/p381]lacZ reporter. Chromatin from WT and Runt plus Ftz–repressed (RF) embryos was obtained from the same crosses as in A.
DSIF and P-TEFb are two additional protein complexes involved in promoter-proximal pausing. Similar to NELF, DSIF is associated with paused Pol II and is believed to inhibit elongation (Saunders et al., 2006). However, unlike NELF, DSIF remains associated with elongating complexes that have been released from the promoter (Andrulis et al., 2000). Association of the Spt5 subunit of DSIF with the slp1[8765/p381]lacZ promoter was not affected in RF embryos, although repression did result in reduced Spt5 association with the lacZ structural gene (Figure 2F). P-TEFb, a protein kinase complex comprising Cdk9 and cyclin T (CycT), is responsible for phosphorylating Ser-2 residues within the Pol II CTD, a modification that releases polymerase from the negative effect of NELF and DSIF and is found on elongating Pol II (Lis et al., 2000; Hirose and Ohkuma, 2007). Repression by Runt and Ftz reduced association of both pSer-2–modified Pol II and CycT with the slp1[8765/p381]lacZ promoter (Figure 2, G and H). These results provide additional evidence that DESE-mediated repression by Runt and Ftz involves preventing release of promoter-proximal paused Pol II and further suggest that this is due to inhibition of P-TEFb recruitment.
Activation by Runt is associated with increased P-TEFb recruitment
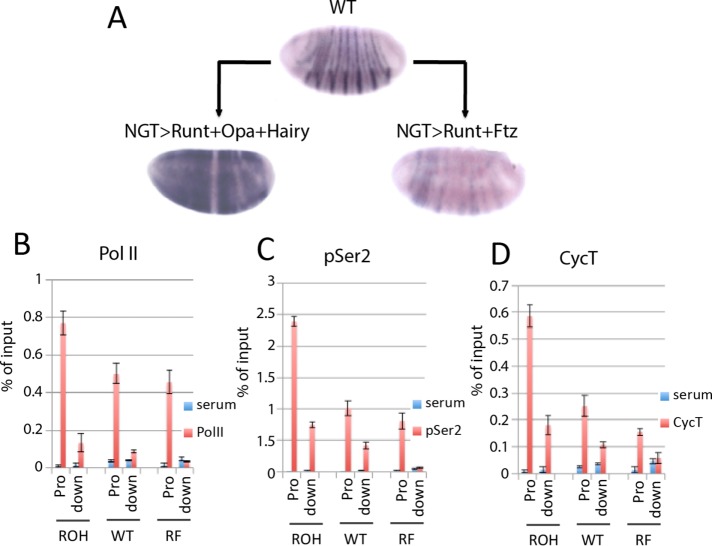
P-TEFb recruitment is a well-documented rate-limiting step for transcription (Bieniasz et al., 1999; Majello et al., 1999; Lis et al., 2000; Chao and Price, 2001). DESE also mediates Runt-dependent activation and drives expression in all somatic blastoderm cells that have Runt and Opa and do not express Ftz (Prazak et al., 2010), a regulatory response that faithfully recapitulates the response of the endogenous slp1 gene to this specific combination of these three pair-rule transcription factors. We wondered whether Runt-dependent activation of DESE and, by extension, the activation of the endogenous slp1 gene involve P-TEFb recruitment. To investigate this, we used the NGT system to generate embryos that in addition to Runt and Opa also coexpress the pair-rule transcription factor Hairy. One function of hairy is to repress ftz (Howard and Ingham, 1986; Ish-Horowicz and Pinchin, 1987; Tsai and Gergen, 1995), which in turn should allow for activation of slp1 by Runt and Opa in cells in which Ftz is repressed, that is, in nearly all cells of these ROH embryos (Figure 3A). ChIP assays revealed increased Pol II association with the slp1 promoter in slp1-expressing ROH embryos compared with both WT and RF embryos (Figure 3B). Of importance, the ROH embryos also showed increased ChIP signals relative to those from WT and RF embryos for pSer-2 (Figure 3C) and CycT (Figure 3D). These results provide strong evidence that the effect of Ftz on Runt’s dual role in slp1 regulation involves differential recruitment of P-TEFb.
FIGURE 3:
P-TEFb recruitment is central to Runt-dependent slp1 regulation. (A) In situ hybridization showing slp1 mRNA expression in WT embryos compared with those with NGT-driven coexpression of Runt, Opa, and Hairy (ROH) or Runt and Ftz (RF). ROH embryos were produced by crossing homozygous NGT[40]; NGT[A] females to homozygous UAS-runt[232] UAS-hairy[211]; UAS-opa[D10] males. RF embryos were produced by crossing homozygous NGT[40]; NGT[A] females to homozygous UAS-runt[232]; UAS-ftz[263] males. (B–D) Results of ChIP assays with control serum (blue bars) and (red bars) Pol II antibody 8WG16 (B), Pol II antibody ab5095 (C), and antisera against CycT (D), using chromatin from ROH, WT, and RF embryos as indicated. ChIP signal detected at the slp1 promoter (Pro) and with a region of the slp1 structural gene 700 base pairs downstream (down) of the TSS.
Runt-dependent repression of PESE occurs upstream of transcription initiation
Reporter genes containing the slp1 PESE enhancer are also repressed by Runt (Prazak et al., 2010). Thus, as expected, the PESE-containing slp1[3125/p381]lacZ reporter is repressed by coexpression of Runt and Ftz (Figure 4A). However, ChIP assays revealed that this repression involves a different mechanism because NGT-driven coexpression of Runt and Ftz reduced association of TBP and Pol II with the promoter region of the slp1[3125/p381]lacZ reporter gene (Figure 4, B and C). The reduction of promoter-associated Pol II suggests that assembly of a PESE-dependent PIC is disrupted in these embryos. Consistent with this, promoter association of TFIIB and TFIIF was reduced to background levels, and association of both the pSer-5– and pSer-2–modified forms of Pol II with this PESE-containing reporter is reduced in response to this repression (Figure 4, D–G). It is notable that TBP association with the reporter gene promoter was reduced in the repressed RF embryos because a reporter gene lacking either PESE or DESE and containing only the p381 slp1 basal promoter segment was sufficient for TBP recruitment (Figure 1D). Although further studies are needed to understand the mechanism by which Runt represses PESE, we conclude that this occurs upstream of transcription initiation and does not involve regulation of Pol II pausing.
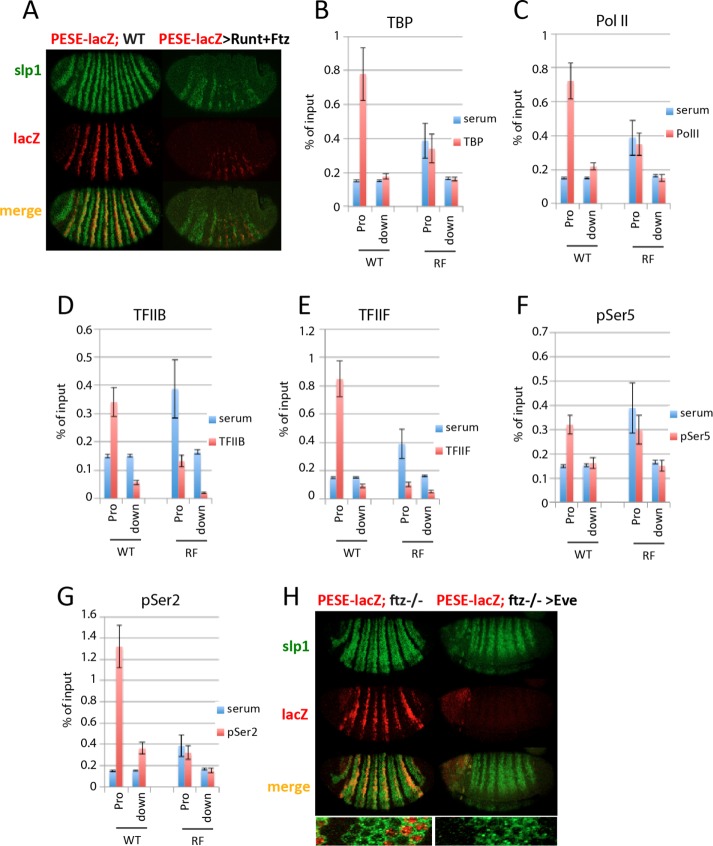
FIGURE 4:
Runt represses PESE by a different mechanism. (A) In situ hybridization showing slp1 (green) and slp1[3125/p381]lacZ (PESE-lacZ; red) expression in WT and in response to NGT-driven coexpression of Runt and Ftz. The WT embryo (left) was from a cross between homozygous NGT[40]; slp1[3125/p381]lacZ females and WT males. The Runt plus Ftz embryo (right) was from a cross between similar females and homozygous UAS-runt[232]; UAS-ftz[263] males. (B–G) Results of ChIP assays with control serum (blue bars) and antibodies against (red bars) TBP (B), Pol II antibody 8WG16 (C), TFIIB (D), TFIIF (E), Pol II antibody H14 (F), and Pol II antibody ab5095 (G) using chromatin from WT and slp1-repressed embryos. WT embryos were from a stock homozygous for the reporter gene. RF embryos were from the same cross as in A. It is notable that the reporter gene promoter region consistently gave higher background signals in RF embryos than in WT embryos. One explanation is that the solubility of chromatin containing the slp1[3125/p381]lacZ promoter is affected in RF embryos. (H) In situ hybridization showing slp1 (green) and slp1[3918/p126]lacZatt (PESE-lacZ; red) expression in ftz mutants in the absence (left) and presence (right) of ectopic Eve. Inset, merged patterns for a region containing one six-cell-wide slp1 stripe. These embryos are from a cross between NGT[40]; slp1[3918/p126]lacZatt ftz[11]/TM3 females and males heterozygous for the second chromosome UAS-eve[12] transgene and the slp1[3918/p126]lacZatt ftz[11] third chromosome. In this cross, one-fourth of the progeny are mutant for ftz and can be unambiguously identified based on the derepression of the endogenous slp1 gene. These ftz mutant embryos are also homozygous for the PESE-lacZ reporter. Half of the progeny from this cross express Eve from the UAS-eve[12] transgene. Derepression of slp1 as shown in H was observed in 25% of similarly staged embryos from this cross; half of these embryos expressed the even-numbered PESE-lacZ stripes, whereas the other half failed to express lacZ.
Runt and Ftz are both required to repress endogenous slp1 in type III cells, as expression is derepressed in embryos that are mutant for either factor (Prazak et al., 2010). Of interest, the PESE-containing slp1[3918]lacZ reporter shows little evidence of derepression in ftz mutants (Figure 4H). This observation suggests that Ftz is not required for the Runt-dependent repression of PESE in these cells. To further investigate whether PESE contributes to the Ftz-dependent repression of endogenous slp1 in type III cells, we took advantage of the differential sensitivity of the DESE and PESE enhancers to repression by Eve. NGT-driven Eve expression had little effect on the derepression of slp1 observed in ftz mutants, whereas the PESE-containing slp1[3918]lacZ reporter was fully repressed in these same embryos (Figure 4H). These results strongly suggest that DESE is responsible for the slp1 expression observed in presumptive type III cells in ftz mutant embryos and provide a second indication that the mechanism of repression by Runt is different for the DESE and PESE enhancers. Runt-dependent repression of DESE requires Ftz and involves preventing release of promoter-proximal paused Pol II. In contrast, Runt-dependent repression of PESE does not require Ftz and is due to a failure to recruit Pol II and other PIC components to the promoter.
Eve represses PESE by preventing release of promoter-proximal paused Pol II
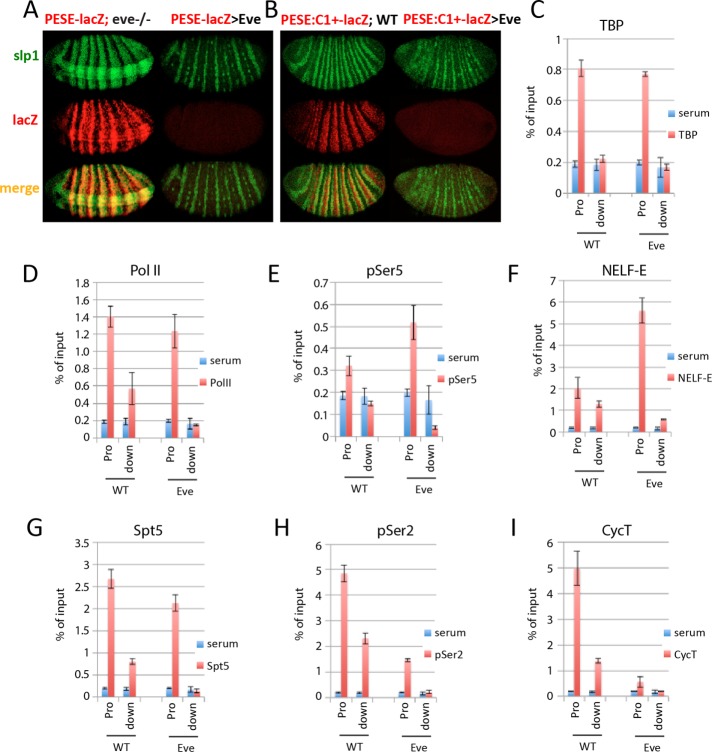
Eve is responsible for slp1 repression in type I cells (Swantek and Gergen, 2004). The PESE-containing slp1[3125/p381]lacZ reporter faithfully recapitulated the derepression observed for slp1 in presumptive type I cells in response to a transient loss of eve and also recapitulated the loss of expression of the even-numbered stripes in presumptive type IV cells in response to ectopic Eve (Figure 5A). As shown earlier, this reporter is repressed by Runt. Therefore, to investigate the molecular basis for Eve-dependent repression independent of the effects of Runt, we used a PESE construct deleted for regions required for repression by Runt. This reporter, slp1[PESE:C1+]lacZatt, fails to be repressed in Runt-expressing type III cells in wild-type embryos, is uniformly expressed throughout the presumptive segmented region of the embryo in response to the transient elimination of eve (Prazak et al., 2010), and is also repressed in all of these cells in response to ectopic Eve (Figure 5B). ChIP assays revealed that repression by Eve does not affect association of either TBP or Pol II with the reporter gene promoter (Figure 5, C and D). These results are similar to those obtained for DESE-mediated repression by Runt and Ftz, suggesting that Eve represses this reporter by preventing release of promoter-proximal paused Pol II. In support of this, repression by Eve increased association of pSer-5–modified Pol II and NELF-E with the slp1[PESE:C1+]lacZatt promoter (Figure 5, E and F). Repression by Eve had little effect on association of the Spt5 subunit of DSIF with the promoter region and specifically reduced association of this elongation factor with the downstream lacZ gene (Figure 5G). Finally, ChIP signals for both the pSer-2–modified form of Pol II and the CycT subunit of P-TEFb were significantly lower at the reporter gene promoter and reduced to background levels within the downstream lacZ gene in response to ectopic Eve (Figure 5, H and I). On the basis of these results, we conclude that Eve represses PESE-dependent transcription by preventing release of promoter-proximal paused Pol II and propose that this is due to inhibition of P-TEFb recruitment.
FIGURE 5:
Eve represses PESE by blocking paused Pol II release. (A) Fluorescence in situ hybridization showing slp1 (green) and the slp1[3125/p381]lacZ (PESE-lacZ; red) expression in embryos mutant for eve (left) and in response to NGT-driven Eve (right). The eve mutant embryos were collected from a cross between flies doubly heterozygous for the temperature-sensitive eve[1] mutation and the reporter gene. Transient elimination of eve was accomplished by collecting embryos for 2 h at 25°C, allowing them to develop an additional 4 h at 18°C, and then shifting to 30°C for 20 min immediately before fixation and processing for in situ hybridization. The response of the reporter gene to ectopic Eve was examined in embryos from a cross between homozygous NGT[40]; slp1[3125/p381]lacZ females and homozygous UAS-eve[12] males. The ectopic Eve in these embryos repressed the even-numbered slp1 stripes and also eliminated expression of this PESE-lacZ reporter in presumptive type IV cells but did not prevent expression of the odd-numbered slp1 stripes in type II cells. (B) Fluorescence in situ hybridization shows slp1 (green) and the slp1[PESE:C1+]lacZatt (PESE:C1+-lacZ; red) expression in WT embryos (left) and in response to ectopic Eve (right). These embryos were collected from a cross between homozygous NGT[40]; slp1[PESE:C1+]lacZatt females and either WT or homozygous UAS-eve[12] males, respectively. The insensitivity of this reporter to repression by Runt results in expanded four-cell-wide stripes in WT embryos due to derepression in type III cells. As observed in A, ectopic Eve specifically repressed the even-numbered slp1 stripes, as well as all expression from the PESE:C1+-lacZ reporter. (C–I) Results of ChIP assays with control serum (blue bars) and antibodies against (red bars) TBP (C), Pol II antibody 8WG16 (D) Pol II antibody H14 (E), NELF-E (F), Spt5 (G), Pol II antibody ab5095 (H), and CycT (I) using primers to detect the promoter and downstream lacZ gene of the PESE:C1+-lacZ reporter. Chromatin from WT and Eve-repressed (Eve) embryos was obtained from the same crosses as in B.
DISCUSSION
The studies presented here take advantage of two key attributes of the slp1 gene as a model for investigating the mechanisms of transcriptional regulation by Runt and other pair-rule transcription factors during Drosophila segmentation. The circuitry for the initial regulation of slp1 is relatively simple and involves only the pair-rule transcription factors Runt, Eve, Ftz, and Opa. This is in contrast to the segment-polarity genes en and wg, which also respond to regulatory inputs from Odd-skipped and Paired, neither of which is involved in the initial regulation of slp1. This simple combinatorial code allows for genetic manipulations that essentially convert the blastoderm embryo into an in vivo test tube for studies on transcription regulation using molecular techniques such as ChIP. A second key attribute of slp1 is the identification of two distinct cis-regulatory DNA elements that together faithfully recapitulate the regulation of slp1 in response to the pair-rule transcription factors. The focus of the experiments presented here has been to investigate the mechanism of regulation of the slp1 DESE and PESE enhancers by Runt and these other pair-rule transcription factors.
The results of these experiments provide new insights into the role of repression by these pair-rule transcription factors in establishing the initial metameric slp1 expression pattern. The PESE enhancer, which normally drives expression in the type IV cells that express the even-numbered slp1 stripes, is repressed by both Runt and Eve, but, of importance, these two transcription factors repress this enhancer by two different mechanisms. Runt prevents PESE-dependent recruitment of Pol II and PIC assembly at the promoter, whereas Eve represses PESE by preventing release of promoter-proximal paused Pol II. Runt also represses DESE, but this is by a mechanism distinct from that used by Runt to repress PESE. Runt-dependent repression of DESE requires Ftz and, similar to Eve-dependent repression of PESE, involves preventing the release of promoter-proximal paused Pol II.
Regulation of paused Pol II release by Eve, Ftz, and Runt
Regulating the release of promoter-proximal paused Pol II has emerged as a widespread phenomenon, especially in developmental systems (Guenther et al., 2007; Muse et al., 2007; Wang et al., 2007; Zeitlinger et al., 2007; Core and Lis, 2008; Gilmour, 2009; Chiba et al., 2010). Our results extend previous findings that Runt and Ftz repress slp1 during Drosophila segmentation by preventing release of promoter-proximal paused Pol II (Wang et al., 2007) and further indicate that this repression is mediated by the slp1 DESE enhancer. Of interest, we found that expression driven by the slp1 PESE enhancer can also be repressed by preventing release of promoter-proximal paused Pol II, but in this case, the repression is due to Eve. A shared feature in the regulated release of paused Pol II by these pair-rule transcription factors is the central role of P-TEFb recruitment. P-TEFb is one of three protein complexes associated with promoter-proximal pausing and is the only factor known to convert paused Pol II complexes into productive elongation (Price, 2008). Although it is possible that differential recruitment of negative factors such as DSIF and NELF could contribute to this regulation, it is questionable whether this occurs under normal cellular conditions (Price, 2008). We found that Runt-dependent repression of DESE correlates with reduced association of the CycT subunit of P-TEFb with the promoter and a corresponding decrease of pSer-2–modified Pol II (Figure 2, G and H). Runx1-dependent repression of CD4 also involves regulation of Pol II pausing (Jiang et al., 2005), and targeting Runx1 to either the promoter or the CD4 silencer interferes with transcription in a manner that appears to involve direct interactions with the CycT subunit of P-TEFb. These observations suggest that antagonism of P-TEFb activity is a conserved property of the Runx proteins. However, DESE also mediates Runt-dependent slp1 activation (Prazak et al., 2010), and this involves increased association of pSer-2–modified Pol II and CycT with the promoter (Figure 3, C and D). The key difference between Runt-dependent repression and activation of DESE is the presence or absence of Ftz (Swantek and Gergen, 2004; Prazak et al., 2010). Thus Ftz is implicated as directly or indirectly antagonizing recruitment of CycT, and, by extension, P-TEFb, to the slp1 promoter in Runt-expressing cells. Eve-dependent repression of PESE also results in reduced promoter association of CycT and pSer-2–modified Pol II (Figure 5, H and I). Eve interacts with TBP, which has been interpreted as evidence that Eve may prevent PIC formation (Han and Manley, 1993; McKay et al., 1999). Alternatively, this could be a strategy to augment interactions between Eve-associated PESE and the slp1 promoter. In any event, with these results, the slp1 enhancers now provide a useful platform for investigating the in vivo regulation of P-TEFb recruitment by these different transcription factors.
Different modes of repression by Runt
It is interesting to consider the different modes of slp1 repression in light of previous findings on Runt’s properties as a transcriptional regulator. Runt interacts with the corepressor protein Groucho via a conserved C-terminal VWRPY motif (Aronson et al., 1997). This interaction is important for maintaining the Runt-dependent repression of en, a process that is also sensitive to the dosage of the corepressors Rpd3 and dCtBP (Wheeler et al., 2002). Of interest, the VWRPY motif has no role in slp1 repression but instead appears to contribute to slp1 activation (Walrad et al., 2010). Studies on other transcription factors in the early Drosophila embryo identified two modes of repression. Short-range repressors, typified by the gap gene transcription factors Giant and Knirps, are believed to interfere with the function of nearby bound activators, whereas long-range repressors, typified by the pair-rule transcription factor Hairy, have the ability to act over distances of several hundred base pairs through chromatin modifications (Hewitt et al., 1999; Strunk et al., 2001; Li and Arnosti, 2011). The minimal 272–base pairPESE:C1+ element contains the 155–base pair PESE:C1 region required for PESE-dependent activation plus distal and proximal extensions of 44 and 73 base pairs, respectively (Prazak et al., 2010). The observation that the PESE:C1+ element is not sensitive to repression by Runt indicates that the regions of PESE involved in mediating repression by Runt are not directly adjacent to sites within the smaller C1 interval that are required for activation. This observation is consistent with the idea that Runt may repress PESE by a long-range mechanism. In contrast, the PESE:C1+ element is fully capable of mediating repression by Eve, suggesting that this repression may involve a shorter-range mechanism. It will be of great interest to determine whether Runt and Eve both interact directly with the PESE enhancer and, if they do, to map the relative location of their binding sites relative to the sites required for activation. Similarly, understanding the mechanistic differences in the mode of Runt-dependent repression of PESE and DESE requires determining whether Runt interacts directly with both enhancers and, if so, defining the location of the Runt sites relative to the binding sites of factors involved in their activation.
A leading candidate for a direct activator of both PESE and DESE is the Zn-finger transcription factor Opa. The loss of the DESE-dependent odd-numbered slp1 stripes in presumptive type III cells in opa mutant embryos provides evidence that Opa is required for activation of DESE. The even-numbered slp1 stripes are also greatly reduced in opa mutants (Swantek and Gergen, 2004). Both DESE and PESE can drive expression in the type IV cells that express these stripes. It will be interesting to determine whether it is DESE or PESE that is responsible for the residual expression of slp1 in presumptive type IV cells in opa mutant embryos. It will also be important to identify the unknown Factor X responsible for this expression. Indeed, a complete understanding of the factors involved in activating these slp1 enhancers and the architecture of the binding sites that contribute to their activation and repression should indicate whether the differing modes of slp1 repression can be properly categorized as involving either the short-range or long-range mechanisms and provide great insights into the context-dependent activities of the Runt transcription factor.
Context-dependent contributions of the slp1 early stripe elements
One of the most interesting aspects of the two slp1 early stripe elements is their combined ability to faithfully integrate regulatory inputs from the pair-rule transcription factors in a nonadditive manner not predicted by the autonomous properties of each enhancer. The results presented here provide information on the potential contributions of these two enhancers to slp1 regulation in each of the four different cellular contexts comprising the segmented region of the blastoderm embryo. On the basis of the finding that Runt prevents PESE-dependent recruitment of Pol II, we propose that only the DESE enhancer is capable of driving slp1 transcription in Runt-expressing cells. DESE is capable of activating transcription in the type II cells that do not express Ftz, resulting in expression of the odd-numbered slp1 stripes (Figure 6C). In contrast, the presence of both Ftz and Runt in type III cells results in the DESE-dependent antagonism of P-TEFb recruitment and a failure of paused Pol II release from the slp1 promoter (Figure 6D). Indeed, the finding that the slp1 expression observed in presumptive type III cells in ftz mutants is insensitive to repression by Eve (Figure 4H) supports the proposal that DESE and not PESE is primarily responsible for regulating expression of the endogenous slp1 gene in these Runt-expressing cells. Finally, the results of ChIP experiments demonstrating that the block to promoter-proximal paused Pol II release observed for slp1 in all somatic blastoderm nuclei in response to NGT-driven Runt and Ftz is emulated by a DESE-lacZ (Figure 2) but not by a PESE-lacZ (Figure 4) reporter provide strong evidence that slp1 repression by Runt and Ftz in type III cells is mediated by DESE.
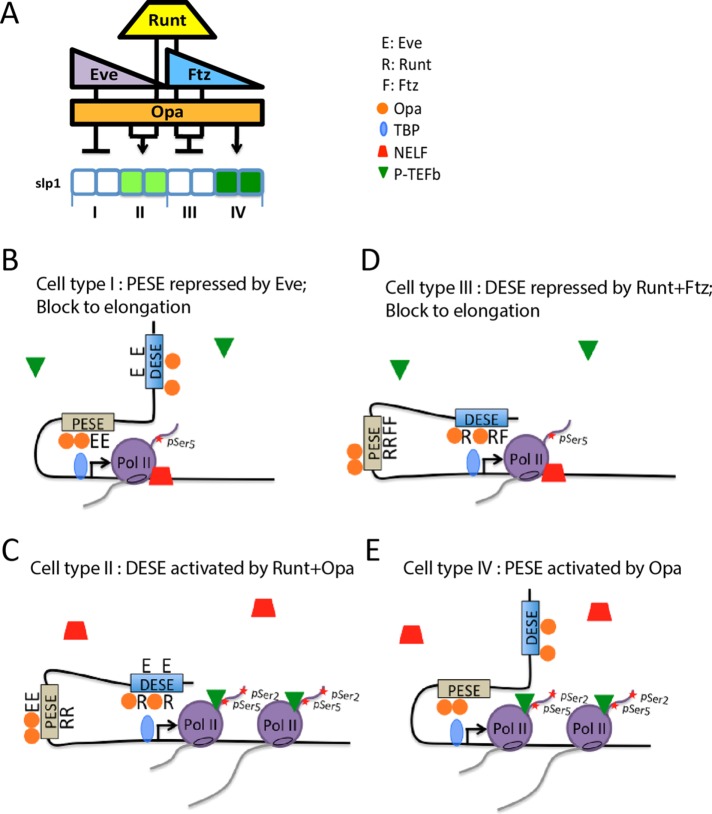
FIGURE 6:
Context-dependent regulation of slp1 transcription. (A) Schematic summarizing slp1 regulation in four different cellular contexts as also depicted in Figure 1A. The key to the right shows symbols representing the different pair-rule transcription factors, TBP, NELF, and P-TEFb in cartoons of proposed enhancer-promoter interactions in these different contexts. (B) In type I cells, PESE interacts with the promoter to recruit TBP, Pol II, and other PIC components to the promoter, and transcription is initiated. However, Eve inhibits P-TEFb recruitment, thereby blocking Pol II release and resulting in a NELF-associated paused Pol II complex. (C) In type II cells, Runt prevents PESE from interacting with the slp1 promoter. Instead, DESE mediates activation by Runt and Opa by increasing P-TEFb recruitment, pSer-2 modification of the Pol II CTD, and a release into productive transcription elongation that results in production of the odd-numbered slp1 stripes. (D) DESE also interacts with the promoter in type III cells, but the combination of Runt and Ftz inhibits P-TEFb recruitment, resulting in a paused Pol II complex and slp1 repression. (E) In type IV cells, the absence of Runt allows PESE to interact with the promoter, recruit Pol II, and promote transcription initiation. The absence of Eve allows P-TEFb recruitment, CTD Ser-2 phosphorylation, and Pol II release into productive transcription elongation, which results in production of the even-numbered stripes of slp1 expression.
It is perhaps more interesting to consider the relative contributions of PESE and DESE to slp1 regulation in the type I and type IV cells that do not express Runt. We consider first cell type I, in which Eve is responsible for slp1 repression (Swantek and Gergen, 2004). This repression should be mediated by PESE because DESE is insensitive to repression by Eve (Prazak et al., 2010). Indeed, key to explaining the nonadditive interaction between PESE and DESE is to understand how the presence of PESE prevents DESE-dependent expression of slp1 in these type I cells. We propose that Eve-dependent repression of PESE involves interactions between PESE and the slp1 promoter that not only prevent release of promoter-proximal paused Pol II but also interfere with the ability of the DESE enhancer to drive transcription from the slp1 promoter (Figure 6B). Prevention of promoter access by steric hindrance is a straightforward explanation for this proposed dominant interfering effect of PESE on DESE-dependent transcription.
It is interesting to further consider which enhancer is responsible for the even-numbered slp1 stripes in cell type IV (Figure 6E). When tested as autonomous elements, both DESE and PESE drive expression in these cells (Prazak et al., 2010). Ectopic Eve specifically represses the even-numbered stripes of slp1 (Swantek and Gergen, 2004) and of a composite reporter containing both the DESE and PESE enhancers but has no effect on the activity of DESE when it is tested as an autonomous enhancer (Prazak et al., 2010). Thus Eve and PESE can also cooperate to dominantly interfere with the activity of DESE in these presumptive type IV cells. Eve-dependent repression of PESE in these cells should result in a promoter-proximal paused Pol II complex that prevents DESE-dependent expression, just as proposed for cell type I. Although it is possible that DESE normally contributes to slp1 expression in type IV cells, this should occur under dynamic conditions under which both enhancers share access to the promoter, thereby allowing PESE to establish repression in response to the ectopic expression of Eve.
The mechanism we proposed to account for the nonadditive interactions between these two slp1 enhancers may also explain other nonadditive interactions, such as the ability of a distal “shadow enhancer” to block the action of a proximal enhancer on the snail promoter (Dunipace et al., 2011). Individual enhancers frequently drive expression in cells that do not express the endogenous gene (Pfeiffer et al., 2008), and in silico attempts to model integration of cis-regulatory information at a promoter strongly suggest that there are nonautonomous mechanisms for silencing unwanted activation (Kim et al., 2013; Samee and Sinha, 2014). Dominant repression by an enhancer-dependent block to release of a promoter-proximal paused Pol II complex provides a potentially widespread route for silencing such spurious transcription. Regulation of transcription elongation was initially believed to facilitate rapid changes in gene expression (Lis, 1998), although more recent work indicates other roles in contributing to the fidelity and/or synchronicity of promoter activity (Boettiger and Levine, 2009; Lagha et al., 2013). Control of promoter-proximal pausing is clearly critical for integrating regulatory inputs from Runt and other pair-rule transcription factors during Drosophila segmentation. The interplay between this mode of regulation and other, more direct modes of regulating enhancer-promoter interactions is extremely likely to be of broad importance for the regulation of gene expression in developmental systems.
MATERIALS AND METHODS
Transgenic lacZ reporters and Drosophila strains
The slp1[3918]lacZatt and slp1[PESE:C1+]lacZatt reporter lines generated by ΦC31-mediated transgenesis and integrated into the P{CaryP}attP2 docking site used for other reporter genes in this study were described previously (Prazak et al., 2010). These constructs contain slp1 basal promoter sequences spanning from −72 to +57 base pairs. Although this promoter region supports DESE-driven expression, more robust activity is observed with a promoter segment that extends from 260 base pairs upstream to 121 base pairs downstream of the TSS. Thus reporter constructs used to assess the effects of DESE and PESE on association of different factors with the promoter contained this larger slp1:p381 segment. This extended promoter, obtained by PCR amplification with flanking XhoI and KpnI sites, was used to replace the shorter promoter segment contained in pC:slp1-link-lacZatt. The DESE-containing slp1[8765/p381]lacZ reporter contains DNA extending from 8710 to 6506 base pairs upstream of the slp1 TSS inserted as a NotI–StuI fragment upstream of the p381 segment. The PESE-containing slp1[3125/p381]lacZ reporter contains DNA extending from 3140 to 2519 base pairs upstream of the TSS inserted as an SpeI fragment. Transformants containing the slp1[8765/p381]lacZ transgene were generated as described (Prazak et al., 2010). Transformants containing the slp1[p381]lacZ and slp1[3125/p381]lacZ transgenes were generated by BestGene. Homozygous stocks were generated through crosses with third chromosome balancers.
Drosophila stocks
The second chromosome–linked P{GAL4-nos.NGT}40 (NGT[40]) and third chromosome–linked P{GAL4-nos.NGT}A (NGT[A]) maternal GAL4-driver lines, as well as the compound stock homozygous for both transgenes, have been described (Tracey et al., 2000; Wheeler et al., 2002). Standard crossing schemes were used to generate stocks homozygous for the second chromosome NGT[40] maternal GAL4-driver and different lacZ reporters inserted into the third chromosome P{CaryP}attP2 docking site. Similarly, a recombinant third chromosome containing both the NGT[A] GAL4-driver and slp1[8765]p381-lacZ reporter was used to create a compound stock homozygous for both these transgenes and also homozygous for NGT[40]. The temperature-sensitive eve[1] mutation and the ftz[11] mutation were obtained from the Bloomington Drosophila Stock Center. The second chromosome–linked UAS-eve[12] and UAS-runt[232] transgenes and third chromosome–linked UAS-ftz[263] and UAS-opa[10] transgenes, as well as the compound UAS-runt[232]; UAS-ftz[263] stock, have been described (Prazak et al., 2010). A transgene that allows for GAL4-driven expression of Hairy was obtained from D. Ish-Horowicz (Imperial Cancer Research Fund, London, United Kingdom) and used to generate a recombinant UAS-runt[232] UAS-hairy[211] second chromosome, which was then combined in a compound stock with the third chromosome–linked UAS-opa[10]. Flies from either the y w[67c23] or y w[67c23]; P{CaryP}attP[2] strain were used for crosses described as being done with wild-type males.
In situ hybridization
Immunohistochemical and fluorescence in situ hybridization was done as described (Prazak et al., 2010). Antisense RNA probes were produced using T7 or T3 RNA polymerase and digoxigenin-UTP (Roche) or fluorescein isothiocyanate (FITC)–UTP (Roche). Antibodies for fluorescent in situ mouse anti-DIG, rabbit anti-FITC, goat anti-mouse Alexa Fluor 555, donkey anti-rabbit Alexa Fluor 647, and donkey anti-goat Alexa Fluor 555 were obtained from Molecular Probes. Confocal images were acquired on a Leica TCS SP5 Microscope system.
ChIP assays
ChIP assays were performed as described (Wang et al., 2007) using chromatin prepared from 25 mg of pooled collections of 3- to 4-h embryos (∼2500 embryos) with the following antisera: mouse monoclonal antibodies 8WG16 (recognizes Pol II CTD lacking pSer-2; Millipore) and H14 (recognizes pSer-5–modified CTD; Millipore; Jones et al., 2004), rabbit ab5095 (recognizes pSer-2–modified CTD; Abcam); rabbit anti-dTBP, anti-dTFIIB, and anti-dTFIIF30 (RAP30; Lebedeva et al., 2005), rabbit anti-dSpt5 and anti-NELF-E (Wu et al., 2003), and rabbit anti-CycT (Hanyu-Nakamura et al., 2008). Quantitative PCR (qPCR) was used to measure enrichment of different DNA segments in the immunoprecipitates. This enrichment is reported as a percentage of input DNA, with error bars representing the mean ± SD from three independent immunoprecipitation experiments. Background precipitation was determined using rabbit serum (Sigma-Aldrich). ChIP signals on the hsp70a promoter were used to normalize results obtained from replicates of the same experiment or with chromatin preparations from different genotypes for each antibody and chromatin preparation as described previously (Wang et al., 2007). Primer sequences used for qPCR are available upon request.
Acknowledgments
Technical assistance from Liujing Xing, Chih-Li Lin, and Shabbir Alam and advice from Xiaoling Wang, Lisa Prazak, Kimberly Bell, and Mike Higgins contributed to this work. The manuscript benefitted from comments of Deniz Erezyilmaz, Ben Martin, and Xiaoling Wang. We thank David Gilmour, David Ish-Horowicz, James Kadonaga, Akira Nakamura, and John Reinitz for providing antibodies and Drosophila stocks. This work was supported by Award MCB 0721430 from the National Science Foundation and Award R01GM094401 from the National Institute of General Medical Sciences.
Abbreviations used:
- Cdk9
cyclin-dependent kinase 9
- ChIP
chromatin immuno-precipitation
- CTD
C-terminal domain
- CycT
cyclin T
- dCtBP
Drosophila C-terminal binding protein
- DESE
distal early stripe element
- DSIF
DRB sensitivity inducing factor
- en
engrailed
- Eve
Even-skipped
- Ftz
Fushi tarazu
- NELF
negative elongation factor
- Opa
Odd-paired
- PESE
proximal early stripe element
- PIC
preinitiation complex
- Pol II
RNA polymerase II
- pSer2
phospho-serine-2
- pSer5
phospho-serine-5
- P-TEFb
positive transcription elongation factor b
- Rpd3
reduced potassium dependency 3
- slp1
sloppy paired 1
- TBP
TATA-binding protein
- TFIIB
transcription factor II B
- TFIID
transcription factor II D
- TFIIF
transcription factor II F
- TSS
transcription start site
- wg
wingless.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-09-0630) on January 11, 2017.
REFERENCES
- Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson BD, Fisher AL, Blechman K, Caudy M, Gergen JP. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz PD, Grdina TA, Bogerd HP, Cullen BR. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc Natl Acad Sci USA. 1999;96:7791–7796. doi: 10.1073/pnas.96.14.7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem. 2001;276:31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- Cheung E, Acevedo ML, Cole PA, Kraus WL. Altered pharmacology and distinct coactivator usage for estrogen receptor-dependent transcription through activating protein-1. Proc Natl Acad Sci USA. 2005;102:559–564. doi: 10.1073/pnas.0407113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba K, Yamamoto J, Yamaguchi Y, Handa H. Promoter-proximal pausing and its release: molecular mechanisms and physiological functions. Exp Cell Res. 2010;316:2723–2730. doi: 10.1016/j.yexcr.2010.05.036. [DOI] [PubMed] [Google Scholar]
- Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnicoff T, Valentine SA, Chen G, Shi T, Lengyel JA, Paroush Z, Courey AJ. Conversion of dorsal from an activator to a repressor by the global corepressor Groucho. Genes Dev. 1997;11:2952–2957. doi: 10.1101/gad.11.22.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunipace L, Ozdemir A, Stathopoulos A. Complex interactions between cis-regulatory modules in native conformation are critical for Drosophila snail expression. Development. 2011;138:4075–4084. doi: 10.1242/dev.069146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Jaynes JB. Regulation of a duplicated locus: Drosophila sloppy paired is replete with functionally overlapping enhancers. Dev Biol. 2012;362:309–319. doi: 10.1016/j.ydbio.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS. Promoter proximal pausing on genes in metazoans. Chromosoma. 2009;118:1–10. doi: 10.1007/s00412-008-0182-4. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Manley JL. Transcriptional repression by the Drosophila even-skipped protein: definition of a minimal repression domain. Genes Dev. 1993;7:491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature. 2008;451:730–733. doi: 10.1038/nature06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt GF, Strunk BS, Margulies C, Priputin T, Wang XD, Amey R, Pabst BA, Kosman D, Reinitz J, Arnosti DN. Transcriptional repression by the Drosophila giant protein: cis element positioning provides an alternative means of interpreting an effector gradient. Development. 1999;126:1201–1210. doi: 10.1242/dev.126.6.1201. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Ohkuma Y. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J Biochem. 2007;141:601–608. doi: 10.1093/jb/mvm090. [DOI] [PubMed] [Google Scholar]
- Howard K, Ingham P. Regulatory interactions between the segmentation genes fushi tarazu, hairy, and engrailed in the Drosophila blastoderm. Cell. 1986;44:949–957. doi: 10.1016/0092-8674(86)90018-8. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D, Pinchin SM. Pattern abnormalities induced by ectopic expression of the Drosophila gene hairy are associated with repression of ftz transcription. Cell. 1987;51:405–415. doi: 10.1016/0092-8674(87)90636-2. [DOI] [PubMed] [Google Scholar]
- Javed A, Guo B, Hiebert S, Choi JY, Green J, Zhao SC, Osborne MA, Stifani S, Stein JL, Lian JB, et al. Groucho/TLE/R-esp proteins associate with the nuclear matrix and repress RUNX (CBF(alpha)/AML/PEBP2(alpha)) dependent activation of tissue-specific gene transcription. J Cell Sci. 2000;113:2221–2231. doi: 10.1242/jcs.113.12.2221. [DOI] [PubMed] [Google Scholar]
- Jiang H, Zhang F, Kurosu T, Peterlin BM. Runx1 binds positive transcription elongation factor b and represses transcriptional elongation by RNA polymerase II: possible mechanism of CD4 silencing. Mol Cell Biol. 2005;25:10675–10683. doi: 10.1128/MCB.25.24.10675-10683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Phatnani HP, Haystead TA, MacDonald JA, Alam SM, Greenleaf AL. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J Biol Chem. 2004;279:24957–24964. doi: 10.1074/jbc.M402218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AR, Martinez C, Ionides J, Ramos AF, Ludwig MZ, Ogawa N, Sharp DH, Reinitz J. Rearrangements of 2.5 kilobases of noncoding DNA from the Drosophila even-skipped locus define predictive rules of genomic cis-regulatory logic. PLoS Genet. 2013;9:e1003243. doi: 10.1371/journal.pgen.1003243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer SG, Jinks TM, Schedl P, Gergen JP. Direct activation of Sex-lethal transcription by the Drosophila runt protein. Development. 1999;126:191–200. doi: 10.1242/dev.126.1.191. [DOI] [PubMed] [Google Scholar]
- Lagha M, Bothma JP, Esposito E, Ng S, Stefanik L, Tsui C, Johnston J, Chen K, Gilmour DS, Zeitlinger J, et al. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell. 2013;153:976–987. doi: 10.1016/j.cell.2013.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva LA, Nabirochkina EN, Kurshakova MM, Robert F, Krasnov AN, Evgen’ev MB, Kadonaga JT, Georgieva SG, Tora L. Occupancy of the Drosophila hsp70 promoter by a subset of basal transcription factors diminishes upon transcriptional activation. Proc Natl Acad Sci USA. 2005;102:18087–18092. doi: 10.1073/pnas.0509063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LM, Arnosti DN. Long- and short-range transcriptional repressors induce distinct chromatin states on repressed genes. Curr Biol. 2011;21:406–412. doi: 10.1016/j.cub.2011.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- Majello B, Napolitano G, Giordano A, Lania L. Transcriptional regulation by targeted recruitment of cyclin-dependent CDK9 kinase in vivo. Oncogene. 1999;18:4598–4605. doi: 10.1038/sj.onc.1202822. [DOI] [PubMed] [Google Scholar]
- Manoukian AS, Krause HM. Control of segmental asymmetry in Drosophila embryos. Development. 1993;118:785–796. doi: 10.1242/dev.118.3.785. [DOI] [PubMed] [Google Scholar]
- McKay LM, Carpenter B, Roberts SG. Evolutionary conserved mechanism of transcriptional repression by even-skipped. Nucleic Acids Res. 1999;27:3064–3070. doi: 10.1093/nar/27.15.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Jahroudi N. The NFY transcription factor functions as a repressor and activator of the von Willebrand factor promoter. Blood. 2002;99:2408–2417. doi: 10.1182/blood.v99.7.2408. [DOI] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter W, Saville B, Hoivik D, Safe S. Functional synergy between the transcription factor Sp1 and the estrogen receptor. Mol Endocrinol. 1997;11:1569–1580. doi: 10.1210/mend.11.11.9916. [DOI] [PubMed] [Google Scholar]
- Prazak L, Fujioka M, Gergen JP. Non-additive interactions involving two distinct elements mediate sloppy-paired regulation by pair-rule transcription factors. Dev Biol. 2010;344:1048–1059. doi: 10.1016/j.ydbio.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DH. Poised polymerases: on your mark...get set...go! Mol Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe NJ, Aneas I, Shen T, Shokri L, Park SY, Bulyk ML, Evans SM, Nobrega MA. Dual transcriptional activator and repressor roles of TBX20 regulate adult cardiac structure and function. Hum Mol Genet. 2012;21:2194–2204. doi: 10.1093/hmg/dds034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samee MA, Sinha S. Quantitative modeling of a gene’s expression from its intergenic sequence. PLoS Comput Biol. 2014;10:e1003467. doi: 10.1371/journal.pcbi.1003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Seufert DW, Prescott NL, El-Hodiri HM. Xenopus aristaless-related homeobox (xARX) gene product functions as both a transcriptional activator and repressor in forebrain development. Dev Dyn. 2005;232:313–324. doi: 10.1002/dvdy.20234. [DOI] [PubMed] [Google Scholar]
- Strunk B, Struffi P, Wright K, Pabst B, Thomas J, Qin L, Arnosti DN. Role of CtBP in transcriptional repression by the Drosophila giant protein. Dev Biol. 2001;239:229–240. doi: 10.1006/dbio.2001.0454. [DOI] [PubMed] [Google Scholar]
- Swantek D, Gergen JP. Ftz modulates Runt-dependent activation and repression of segment-polarity gene transcription. Development. 2004;131:2281–2290. doi: 10.1242/dev.01109. [DOI] [PubMed] [Google Scholar]
- Tracey WD, Jr, Ning X, Klingler M, Kramer SG, Gergen JP. Quantitative analysis of gene function in the Drosophila embryo. Genetics. 2000;154:273–284. doi: 10.1093/genetics/154.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C, Gergen P. Pair-rule expression of the Drosophila fushi tarazu gene: a nuclear receptor response element mediates the opposing regulatory effects of runt and hairy. Development. 1995;121:453–462. doi: 10.1242/dev.121.2.453. [DOI] [PubMed] [Google Scholar]
- Umayahara Y, Kawamori R, Watada H, Imano E, Iwama N, Morishima T, Yamasaki Y, Kajimoto Y, Kamada T. Estrogen regulation of the insulin-like growth factor I gene transcription involves an AP-1 enhancer. J Biol Chem. 1994;269:16433–16442. [PubMed] [Google Scholar]
- Walrad PB, Hang S, Joseph GS, Salas J, Gergen JP. Distinct contributions of conserved modules to Runt transcription factor activity. Mol Biol Cell. 2010;21:2315–2326. doi: 10.1091/mbc.E09-11-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lee C, Gilmour DS, Gergen JP. Transcription elongation controls cell fate specification in the Drosophila embryo. Genes Dev. 2007;21:1031–1036. doi: 10.1101/gad.1521207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JC, VanderZwan C, Xu X, Swantek D, Tracey WD, Gergen JP. Distinct in vivo requirements for establishment versus maintenance of transcriptional repression. Nat Genet. 2002;32:206–210. doi: 10.1038/ng942. [DOI] [PubMed] [Google Scholar]
- Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]