Abstract
Cancer is intrinsically complex, comprising both heterogeneous cellular compositions and microenvironmental cues. During the various stages of cancer initiation, development, and metastasis, cell–cell interactions (involving vascular and immune cells besides cancerous cells) as well as cell-extracellular matrix (ECM) interactions (e.g., alteration in stiffness and composition of the surrounding matrix) play major roles. Conventional cancer models both two- and three-dimensional (2D and 3D) present numerous limitations as they lack good vascularization and cannot mimic the complexity of tumors, thereby restricting their use as biomimetic models for applications such as drug screening and fundamental cancer biology studies. Bioprinting as an emerging biofabrication platform enables the creation of high-resolution 3D structures and has been extensively used in the past decade to model multiple organs and diseases. More recently, this versatile technique has further found its application in studying cancer genesis, growth, metastasis, and drug responses through creation of accurate models that recreate the complexity of the cancer microenvironment. In this review we will focus first on cancer biology and limitations with current cancer models. We then detail the current bioprinting strategies including the selection of bioinks for capturing the properties of the tumor matrices, after which we discuss bioprinting of vascular structures that are critical toward construction of complex 3D cancer organoids. We finally conclude with current literature on bioprinted cancer models and propose future perspectives.
Keywords: cancer biology, cancer model, bioprinting, vascularization, drug screening
Graphical Abstract
INTRODUCTION
Cancer is one of the major causes of morbidity and mortality, accounting for around 14 million new cases and 8.2 million deaths in 2012 worldwide.1 It is further expected that the annual cancer incidences will rise from 14 million in 2012 to 22 million within the next 20 years,1 leading to significantly increased healthcare costs and the great need to better understand cancer to improve therapy. The most prevalent types of cancer (e.g., breast, colon, stomach, liver, and lung) share common features, each with a unique microenvironment and a hypoxic core surrounded by a dense tissue.2–4 Cancer cells are otherwise healthy cells that upon acquiring genetic mutations develop capabilities specific for cancer such as displaying invasive behavior, resisting cell death, evading growth suppression, experiencing uncontrolled replication, showing sustained growth, and triggering abnormal angiogenesis.5 This last capability constitutes a limiting step for tumors to be able to fully develop and evade hypoxia (only cells within a 200 µm range from a blood vessels experience sufficient oxygen levels6). The cancer microenvironment is highly complex (Figure 1). For example, a vast majority of cancers originate from mutated epithelial cells (developing carcinomas).5,7,8 The tumor then grows as it interacts with its microenvironment, ranging from the surrounding stroma composed of both cells (mainly endothelial cells and fibroblasts) and the extracellular matrix (ECM). The tumor cells modulate their proliferation and migration by producing pro-angiogenic factors (e.g., vascular endothelial growth factor, VEGF) and promoting their interactions with the stromal cells and the surrounding inflammatory cells (macrophages, neutrophils, and mast cells).9
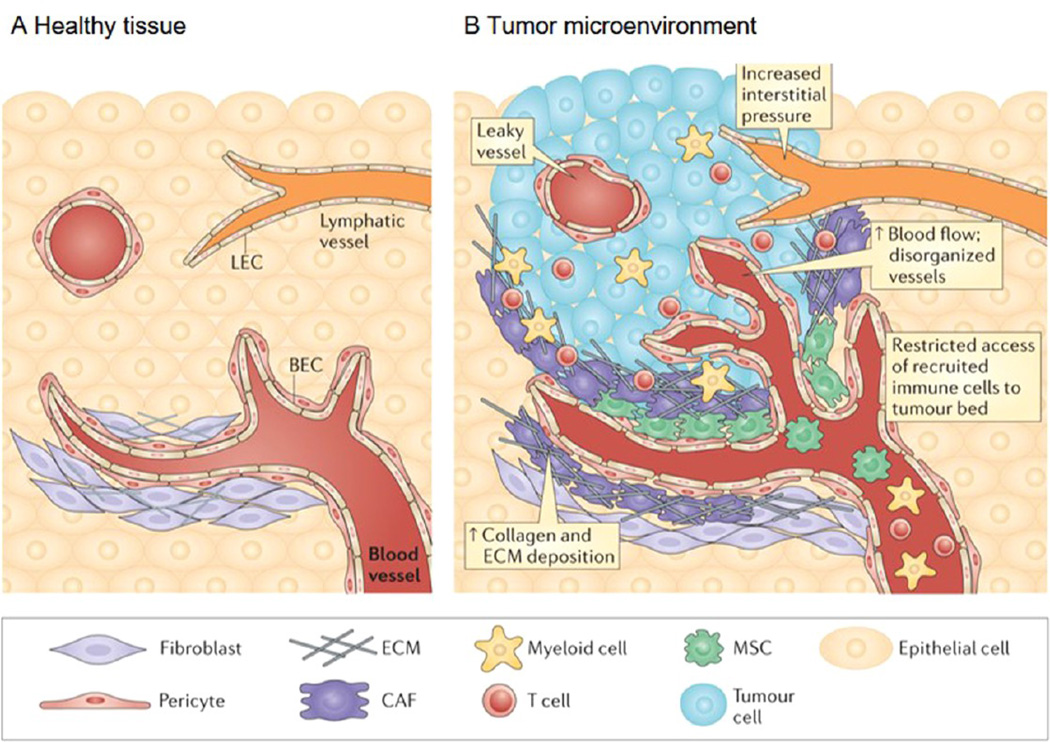
Figure 1.
Healthy tissue and the tumor microenvironment. (A) Stromal cells present in interstitial spaces surrounding the parenchyma of various organs promote tissue integrity by providing growth factors and structural support. Blood endothelial cells (BECs) and pericytes maintain the integrity of blood vessels and ensure the supply of oxygen and other nutrients to the tissue. Lymphatic vessels composed of lymphatic endothelial cells (LECs) drain interstitial fluid. Fibroblasts are constantly remodeling the ECM to cope with mechanical stress within connective tissue. (B) Neoplastic transformation is often accompanied by the formation of a tumor bed and profound alterations in the surrounding connective tissue and stroma, a process that culminates in the establishment of a pathological tumor microenvironment. An imbalance between pro- and antiangiogenic factors results in the formation of aberrant vasculature, characterized by numerous leaky blood vessels. Increased interstitial pressure and inadequate drainage by the lymphatic vessels is also observed. The increased hydrostatic load, along with tumor-secreted molecules (not shown), induces the recruitment of circulating mesenchymal stem cells (MSCs), the activation of cancer-associated fibroblasts (CAFs) and a marked accumulation of ECM. Finally, various chemokines and cytokines in the tumor microenvironment attract activated T cells and myeloid cells to the tumor lesion, but tortuous blood vessels and dense ECM often hinder their access to the tumor nest. Although the makeup of the cellular and extracellular milieu can differ between tumor types and stages of growth, it is becoming clear that changes in the cellular architecture of the tumor microenvironment can influence tumor growth, metastasis, and drug resistance. Adapted with permission from ref 10. Copyright 2015 Nature Publishing Group.
The tumor microenvironment, including both chemical cues (growth factors and cytokines) and biophysical cues (interstitial pressure and matrix mechanics), is extremely complex and plays a major role in the progression and metastasis of the tumor as it evolves over time.9,10 This microenvironment is highly dynamic with distinctive key features (e.g., cellular and ECM compositions, matrix stiffness, and degree of vascularization) present at each of the different stage of the disease.11,12 Therefore, it is of primary necessity to be able to decompose all the key elements interacting with the cancer cells and then understand how, why, and when they become implicated in the tumor growth and migration. In addition, there is a high variability between the cancer types (i.e., intertumor heterogeneity) as well as within each tumor (i.e., intratumor heterogeneity).13–16 This heterogeneity leads to an extremely high complexity and variability requiring personalized care for the patient.17 To fully grasp the complexity of the origins of cancer, its development, metastasis, and interactions between the various key players in the tumor microenvironment, as well as screening of various anticancer drugs, it has been increasingly realized that in vitro engineered human cancer models are strongly desired, as the conventional animal-based xenograft cancer models do not necessarily recapitulate the human physiology and drug responses.18–20 To address these challenges, three-dimensional (3D) cancer models are anticipated to precisely mimic the in vivo tumor microenvironment in human patients by recapitulating the proper tumor cell/ matrix composition and properties that match both the type and stage of the disease, and therefore provide accurate mechanistic studies as well as a tool for personalized anticancer therapeutics screening.
Here we review the conventional methods to generate in vitro 3D cancer models through tissue engineering approaches. We further state the limitations of these methods and introduce the 3D bioprinting technologies that promise to produce complex tissue constructs at high spatial precision and compositional accuracy. We finally discuss the application of 3D bioprinting to fabrication of biomimetic cancer models and envision future perspectives.
CONVENTIONAL APPROACHES FOR CONSTRUCTION OF IN VITRO 3D CANCER MODELS
State-of-the-art in vitro cancer models range from two-dimensional (2D) monolayer to 3D (co)cultures using conventional tissue engineering approaches (Figure 2). Planar cancer models have a long history, as early as the 1950s (Figure 2A i);21,22 however, it was quickly recognized that these models exhibit strong discrepancies specifically drug responses when compared to the in vivo counterparts. The differences arise from the fact that these 2D models lack the functional 3D tumor microenvironment leading to insufficient cell–cell and cell-ECM interactions.23–26 Moreover, during the culture process on plastic surfaces cells tend to be selected for their phenotypes that promote their adhesion to stiffer substrates.27 As a result, the applications and efficacy of 2D cancer models have been limited.
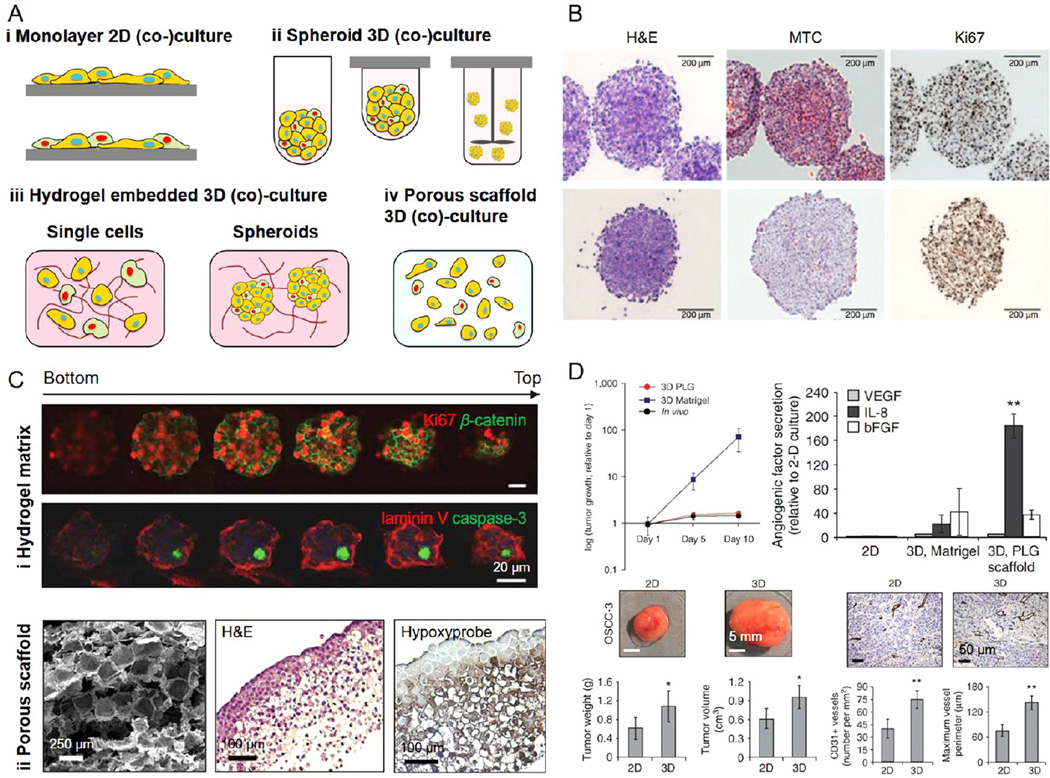
Figure 2. Conventional cancer models.
(A) Schematic illustration of cancer models based on (i) 2D monolayer (co)cultures, (ii) spheroid 3D (co)cultures derived from microwells, hanging drops, and spinner flask methods, (iii) hydrogel-embedded 3D (co)cultures, and (iv) porous scaffold-enabled 3D (co)cultures. Adapted with permission from ref 28. Copyright 2014 Wiley–VCH. (B) Multicellular tumor spheroids composed of A375 human melanoma cells were characterized by hematoxylin and eosin (H&E), Mason’s trichrome, and Ki67 staining. Adapted with permission from ref 31. Copyright 2014 Nature Publishing Group. (C) (i) Matrigel-embedded 3D OSCC-3 human oral cancer model stained for Ki67 (red) and β-catenin (green), or laminin V (red) and caspase-3 (green); and (ii) PLG porous scaffold-based 3D OSCC-3 human oral cancer model stained by H&E and hydroxyprobe. (D) Comparisons of tumor growth, angiogenic factor secretion, and vascularization in 3D Matrigel-embedded model, PLG scaffold-based model, and in vivo. Adapted with permission from ref 38. Copyright 2007 Nature Publishing Group.
In an attempt to mitigate these drawbacks that 2D monolayer cultures present, in vitro 3D cancer models have been subsequently developed mainly based on spheroid (co)cultures and those based on the utilization of 3D matrices (Figure 2A ii-iv).28 Aggregates of tumor cells, usually referred to as multicellular tumor spheroids, can be generated by seeding the cells onto a nonadherent surface such as agarose and polydimethylsiloxane (PDMS).28,29 When these substrates are processed to possess confined features of microwells, uniform spheroids can form inside these microwells that prevent the cells from adhering to the walls and hence self-aggregate.29 Other methods for spheroid generation include inducing aggregation of tumor cells at the bottom of “hanging drops”30,31 and through the use of a constantly stirred spinning flask.32 The cells that aggregate in spheroids tend to display similar features as their in vivo counterparts (Figure 2B), where a necrotic core will form at the center of the spheroid upon the growth of the aggregate will start because of the lack of nutrients and oxygen.33 The multicellular tumor spheroids with improved functionality have been used for studies of fundamental cancer biology33 as well as drug screening and validation.34–36 Although more advanced compared to the 2D cultures, the 3D models based on multicellular tumor spheroids still lack the major ECM component of the tumor microenvironment.9,37
To this end, 3D matrices mimicking the cancer ECM, either based on hydrogels (Figure 2A iii) or porous scaffolds (Figure 2A iv), have been combined with tumor cells to construct more biomimetic cancer models.38–42 Multiple types of matrices have been used for this purpose. The biomaterials for matrix reconstitution can be derived from natural polymers such as collagen,43 fibrin,44 and Matrigel45,46 to form a variety of hydrogels (Figure 2C i). In particular, the latter is commonly used as matrix in the context of cancer modeling as it is harvested from Engelbreth–Holm–Swarm (EHS) mouse sarcoma and thus intrinsically features important properties of the tumor ECM.46 Indeed, the use of these hydrogel matrices with precisely controllable stiffness has shown promising results in recapitulating the in vivo tumor behaviors.27 On the other hand, the 3D scaffolds can also be made from synthetic polymers that present better control over their chemical and physical properties including polyethylene glycol (PEG), poly(lactic acid) (PLA), poly(glycolic acid) (PGA), and their copolymer poly(lactic-co-glycolic acid) (PLG), among others.47,48 These scaffolds, usually processed to be porous, can be populated with tumor cells to fabricate cancer models possessing hypoxic cores (Figure 2C ii).38 The 3D cancer models generated using porous scaffolds have also been shown to indicate enhanced performance (e.g., proliferation, growth factor and cytokine secretion, and vascularization) than the 2D monolayer cultures and 3D tissues embedded in Matrigel both in vitro and after in vivo implantation (Figure 2D).38
BIOPRINTING AS AN INNOVATIVE TECHNOLOGY TO BUILD BIOMIMETIC CANCER MODELS
Major challenges associated with current 3D in vitro models include their oversimplified structures and limited vascularization potential. For example, the multicellular tumor spheroids or 3D scaffold-based tumors will be size-limited due to the lack of vascularization and thereby will likely faithfully model the genesis of tumors but not the later stages of their development. Moreover, most of those models lack well organized spatial distribution of tumor cells and ECM compositions. Recently a strong interest toward the utilization of 3D bioprinting technologies has arisen to solve these issues with an aim of constructing more biomimetic 3D in vitro cancer models.49
Bioprinting Techniques
The general term of bioprinting refers to multiple strategies that are capable of dispensing biological components such as biomaterials (i.e., the bioinks) and cells in a spatially defined manner.50,51 Although 3D printing was developed decades ago, only until recently was this versatile technology adapted to the field of biomedicine in fabricating sophisticated biological structures through the use of biocompatible materials.52 The common strategies of bioprinting include inkjet printing,53,54 extrusion-based printing,55,56 laser-assisted printing,57,58 and stereolithography.59–61
Inkjet bioprinting is based on a well-established drop-by-drop deposition mechanism enabled by thermal or piezoelectric actuation (Figure 3A).50,51 In a typical setup the cartridge is filled up with the (cell-laden) bioink, and the droplet ejection through the printhead synchronized with a motorized stage driven by a computer program lead to deposition of the droplets at specific locations to build the 3D architecture. Inkjet bioprinting features relatively low cost, fast speed, and high viability of cells in the deposited structures.62,63 In comparison, the extrusion-based bioprinting relies on propelling the bioink through the nozzle using pneumatic or mechanical pressure (Figure 3B),50,51 which affords low cost and medium-to-fast deposition speed. In the case of laser-assisted bioprinting or laser-induced forward transfer (LIFT) it requires a donor layer that responds to laser stimulation by absorbing the focused energy and local generation of high-pressure bubble that pushes a droplet of the donor bioink onto the collecting substrate (Figure 3C).50,51 However, this method suffers from high cost and bulky instrumentation, as well as limited choice over the bioinks. Another versatile 3D bioprinting technique capable of patterning cell-laden hydrogels has been adopted from the stereolithography that was conventionally used for materials fabrication. In this strategy, a digital mask generated by an array of programmed mirrors is projected to a bioink reservoir to selectively photo-cross-link patterns of interest in a layer-by-layer manner; along with the vertical movement of the stage 3D objects containing prescribed architecture can be produced at high fidelity (Figure 3D).59–61,64,65 Stereolithography has multiple advantages including high resolution (up to a few micrometers) and a fast bioprinting speed.
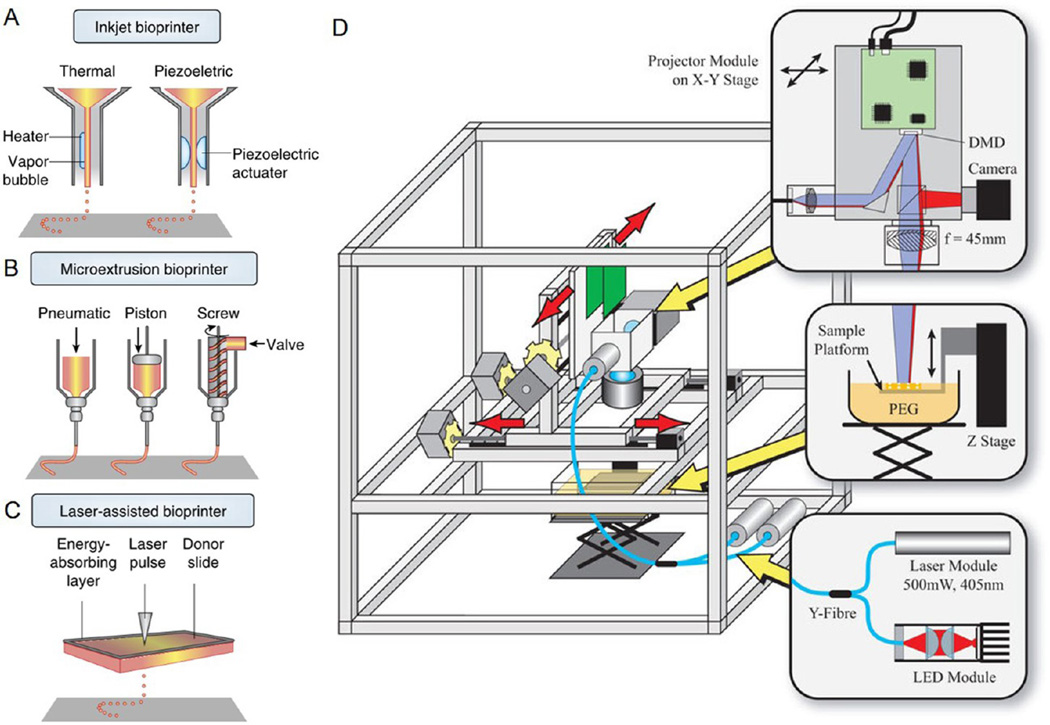
Figure 3. Common bioprinting modalities.
(A) Thermal inkjet printers electrically heat the printhead to produce air-pressure pulses that force droplets from the nozzle, whereas piezoelectric printers utilize pulses formed by piezoelectric actuation for droplet ejection. (B) Microextrusion bioprinters use pneumatic or mechanical (piston or screw) dispensing systems to extrude continuous bioinks. (C) Laser-assisted bioprinters use laser focused on an absorbing substrate to generate localized pressures that propel cell-containing bioink onto a collector substrate. Adapted with permission from ref 51. Copyright 2014 Nature Publishing Group. (D) Schematic of the stereolithography-based 3D bioprinter. A frame is used to support the custom precision translation stages and projector system; light from a UV laser then illuminates a digital micromirror device (DMD) projector; a lens is adopted to project the image of the DMD pattern onto the bioink for layer-by-layer cross-linking, which along with vertical movement of the stage generates 3D bioprinted structures. Adapted with permission from ref 65. Copyright 2015 Nature Publishing Group.
All these bioprinting methods enable a highly improved control of cell distribution within the 3D space compared to conventional approaches. Moreover, they permit the creation of large-scale constructs possessing well-organized architecture and arbitrary shapes, hence providing excellent flexibility and ability to reproduce the complex cancer microenvironment that mimic that of their counterparts in vivo.
Designing the Bioinks
The composition of the tumor ECM is highly complex, consisting of a wide variety of extracellular proteins and molecules that constitute the tumor microenvironment together with tumor and stromal cells.9,12,23,26 Studies have also revealed that the composition of the tumor matrix is unique for tumor type and stage and exhibits strong heterogeneity between patients, which is critical in maintaining the phenotype of the tumor in vitro.66 In addition, the matrix stiffness plays a nontrivial role in regulating tumor behaviors including their initiation, progression, and metastasis.67–71 Therefore, the bioinks used for fabricating the tumor models should be carefully selected or designed to satisfy the needs of cancer bioprinting.
The bioinks used for bioprinting, typically in the form of hydrogels, should possess specific characteristics including printability, cross-linking mechanisms, and biocompatibility.50–52,72–75 Although immediate printability of the bio-printed tissue constructs may depend on the physical properties (e.g., viscosity) of the bioink itself, subsequent stabilization of the structures relies on additional cross-linking steps.52,76 In general, two types of cross-linking strategies can be applied, that based on physical cross-linking through ionic interactions such as in the case of alginate (gels by Ca2+ ions),77,78 and those hydrogel precursors able to form covalent bonds such as methacrylated forms of the polymers that are photocross-linkable.79–82 The mechanics of the resulting hydrogel matrices post-bioprinting can further be finely tuned within a wide range covering the entire spectrum of tissue stiffness and elasticity by changing the concentration of the polymers making up the bioink or cross-linking density.50,83,84
Existing bioinks have been derived from myriad classes of biomaterials including those of both synthetic (e.g., PEG and its derivatives, pluronics) and natural (e.g., collagen, hyaluronic acid, fibrin, alginate, gelatin) origins.50–52,85,86 The major advantage of natural biomaterials lies in their excellent bioactivity such as biocompatibility and presence of intrinsic cell-adhesion ligands (e.g., RGD); however, the usually weak mechanical properties and limited freedom of customization have been their consistent drawbacks.52,87 On the contrary, the properties of synthetic biomaterials can be easily tuned over a large range, which include their mechanical properties, degradation, and bioactivity.47,88,89 It is expected that by rational combination of the natural and synthetic biomaterials for use as the bioink optimized for each cancer type, maximal recapitulation of the viability, biological behaviors, and functionality can be achieved for the bioprinted cancer tissues.
More recently, bioinks derived from decellularized ECMs (dECMs) have attracted great attention, which are formed by rehydrating powdered tissue-specific dECMs obtained following the standard decellularization procedures.90–93 One unique advantage of dECM-based bioinks compared to other artificial bioink formulations lies in the possibility to apply biomaterials and biochemical cues from the target tissue in the bioprinting process, therefore achieving well-matched compositional complexity between the bioprinted structures and their native counterparts in the body.91 Although this concept has not yet been applied to cancer bioprinting, we anticipate that when tumor-derived dECM bioinks are used in a tumor-specific manner the performance of the resulting bioprinted tumor tissues will be enhanced due to the ability to maintain the biochemical cues of the microenvironment. In combination with well-defined synthetic biomaterials, the biophysical parameters may be simultaneously incorporated to maximally match the properties of the in vivo tumor niches.
Bioprinting the Vasculature
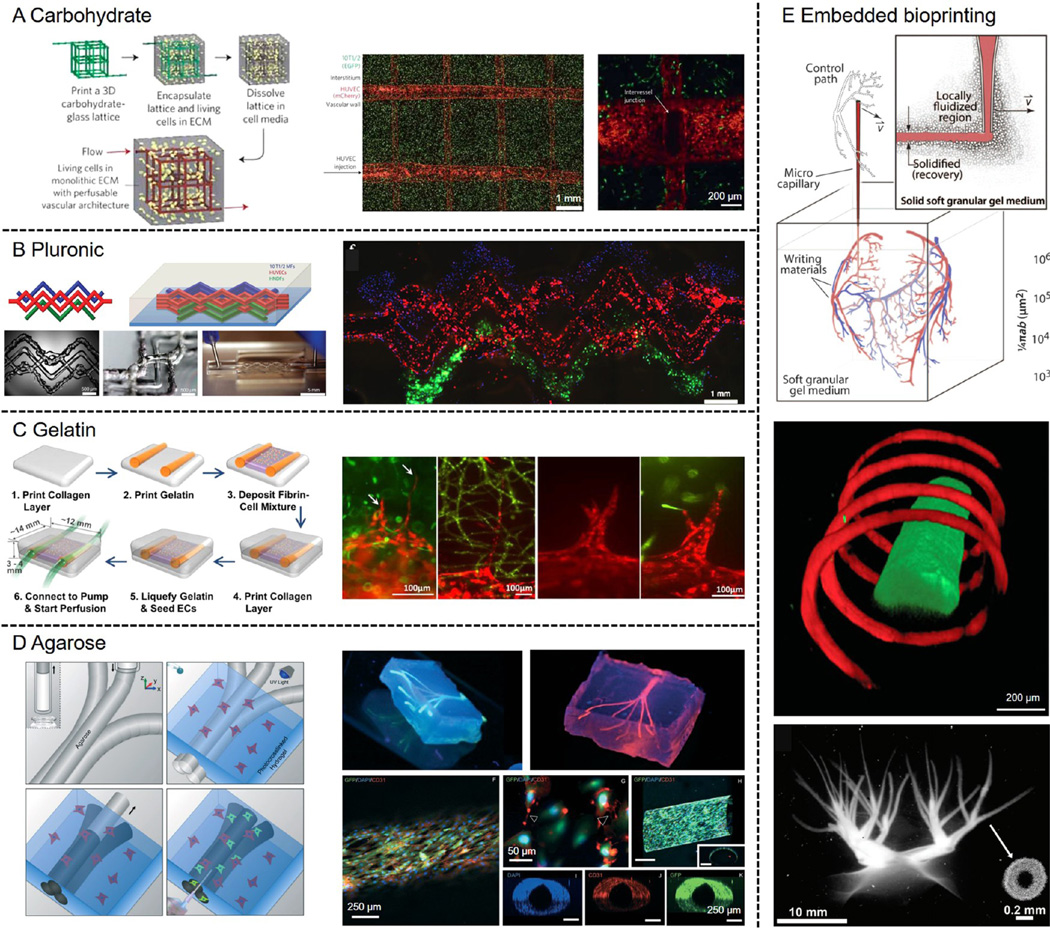
Extensive vascularization is a critical hallmark of cancer and thus a critical component in construction of in vitro cancer models.11,12,94 Bioprinting has the unique capability to introduce well-organized blood vessel-like structures into engineered 3D tissues. One of the most successful strategies lies in the use of a technique referred to as sacrificial bioprinting.85,86,95–97 In a typical procedure, a sacrificial template is first deposited using the bioprinter in arbitrary patterns resembling the vascular network, after which a hydrogel matrix is casted around the template, and finally the template is selectively removed leaving perfusable channels embedded within the hydrogel. These hollow channels can be further endothelialized to mimic the vascular functionality. Several classes of materials have been adopted as the template to achieve sacrificial bioprinting of vasculature. In an earlier example, Chen and co-workers used 3D printed carbohydrate glass fibers coated with a thin layer of PLGA as the template, which were then embedded into a variety of hydrogels including agarose, alginate, PEG, fibrin, or Matrigel encapsulating fibroblasts; the carbohydrate glass fibers could dissolve by perfusion with the cell culture medium to generate the lumens and allow subsequent seeding of endothelial cells on the surface of the channels (Figure 4A).95 Alternatively, hydrogels may also be used as the sacrificial templates. These examples include: (i) Pluronic solution that exhibits a sol–gel transition at reduced temperature, allowing bioprinting at room temperature but dissolution of the Pluronic at a temperature close to 0 °C (Figure 4B);31,86 (ii) gelatin solution that presents a sol–gel transition at elevated temperature, enabling dissolution of the template when bioprinted constructs are transferred to an incubator at 37 °C (Figure 4C);97 and (iii) agarose solution that forms stiff hydrogel microfibers that can be directly extracted from the hydrogel matrix under mild vacuum (Figure 4D).96 In all these cases the obtained interconnected microchannels may be endothelialized to recapitulate the biological functions of blood vessels.
Figure 4. Examples of bioprinted vascular structures.
(A) Sacrificial bioprinting by templating against an open, interconnected, self-supporting carbohydrate glass lattice, where endothelialization of channel walls and formation of intervessel junctions could be achieved. The carbohydrate template was dissolved in culture medium. Adapted with permission from ref 95. Copyright 2012 Nature Publishing Group. (B) Sacrificial bioprinting by templating against pluronic. A heterogeneous engineered tissue construct was produced, in which blue, red, and green filaments corresponded to bioprinted 10T1/2 fibroblast (blue), human neonatal dermal fibroblast cells (green), and HUVECs (red). The pluronic template was liquefied at <4 °C and removed by vacuum suction. Adapted with permission from ref 85. Copyright 2014 Wiley–VCH. (C) Sacrificial bioprinting by templating against gelatin, where endothelial sprouting into the surrounding matrix was shown to occur. The gelatin template was liquefied at <37 °C and removed spontaneously. Adapted with permission from ref 97. Copyright 2014 Springer. (D) Sacrificial bioprinting by templating against agarose, where the channels could be subsequently endothelialized. The solidified agarose template was removed by pulling or vacuum suction. Adapted with permission from ref 96. Copyright 2014 Royal Society of Chemistry. (E) Schematic showing a microscale capillary tip sweeping out a complex pattern as the bioink was injected into the granular shear-thinning gel medium. Freeform vascular patterns could be generated using this embedded bioprinting approach. Top image dapted with permission from ref 98. Copyright 2015 American Association for the Advancement of Science. Bottom image adapted with permission from ref 82. Copyright 2015 Wiley–VCH.
More recently, a novel bioprinting method relying on depositing the sacrificial bioink into a supporting hydrogel matrix has enabled fabrication of highly complex vascular structures within a 3D volume (Figure 4E).82,98,99 In this case, a medium consisting of a hydrogel is used as the support to prevent the collapse of the deposited bioink during the bioprinting procedure. The medium is usually shear-thinning, allowing movement of the nozzle and fast extrusion of the bioink into the matrix while self-healing to ensure integrity of the embedding hydrogel. By taking advantage of this technique, vascular structures of arbitrary 3D shapes and architecture can be produced, which are not easily achieved using conventional 3D bioprinting approaches.
Recent advancement in bioprinting has further enabled alternative approaches for direct deposition of hollow tubular structures mimicking the blood vessels. A coaxial extrusion device has been proposed to deliver the bioink in the sheath flow and the cross-linking solution in the core, where in situ cross-linking of the bioink and stabilization of its shape immediately occur upon extrusion to obtain tubular structures possessing hollow lumens.100,101 The fabricated hollow tubes are continuous and perfusable. Endothelial cells may also be encapsulated within the bioink to fill the wall thickness of the bioprinted hollow tubes, which overtime will spread and proliferate to achieve biological functions of the blood vessels.
Cancer Bioprinting
Current 3D cancer models present multiple limitations such as batch-to-batch variability, limited control over cell patterning, and low throughput. Therefore, 3D bioprinting as a versatile platform that enables deposition of volumetric structures and architecture at high precision and fidelity, provides a novel solution to address these issues. However, only few applications of bioprinting in construction of biomimetic cancer models have been demonstrated so far since it has entered this field of research only very recently.
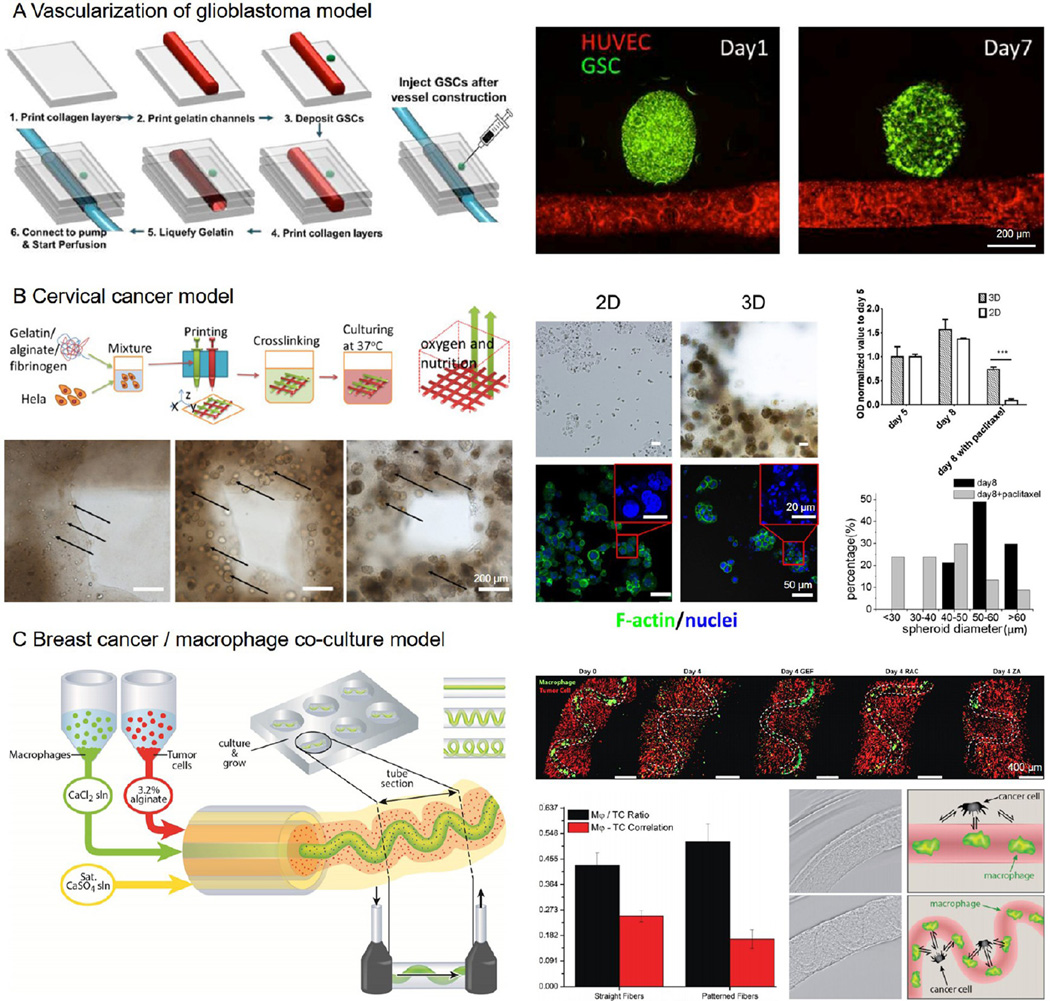
As aforementioned vascularization plays a critical role in growth of most cancer types by supplying sufficient nutrients and oxygen.11,12,94 To this end, Yoo et al. created a glioblastoma vascularization model combining the sacrificial bioprinting strategy with multicellular spheroids.102 A microchannel seeded with human umbilical vein endothelial cells (HUVECs) was bioprinted within a collagen block, after which a spheroid consisting of glioma stem cells obtained from a patient was injected beside the vascular channel to mimic the in vivo scenario (Figure 5A). It was noted that the glioma spheroid gradually remodeled over a course of 7 days showing signs of sprouting and potential vascularization. This model is general, and can be conveniently extended to a variety of other cancer types to study the tumor-vascular niche.
Figure 5. Cancer bioprinting.
(A) Construction of a glioblastoma vascular niche by injecting a spheroid of glioma stem cells (green) besides a sacrificially bioprinted perfusable vascular channel (red). Adapted with permission from ref 102. Copyright 2015 Institute of Electrical and Electronics Engineers. (B) Fabrication of a cervical cancer model by direct 3D bioprinting of Hela cell-laden bioink. The cells proliferated and clustered (black arrows) within the bioprinted microfibrous structure. The bioprinted 3D cervical cancer model showed drug-induced toxicity at higher resistance than the 2D cultures. Adapted with permission from ref 103. Copyright 2014 IOPscience. (C) Bioprinting of a breast cancer/ macrophage coculture model, where the relative geometry of the two cell populations was shown to affect the tumor-immune cell interactions. Adapted with permission from ref 106. Copyright 2015 Wiley–VCH.
Apart from focusing on the vascularization, studies have further investigated the tumor microenvironment, which is also playing a crucial role in tumor development. For example, Sun et al. encapsulated HeLa cells in a composite bioink of gelatin/ alginate/fibrinogen to directly bioprint a 3D cervical cancer model for drug testing (Figure 5B).103,104 It was demonstrated that the bioprinted 3D model of cervical cancer was more realistic than the conventional 2D cultures in terms of cancer cell proliferation, viability, and response to chemotherapy. The deposited network of cancer cell-embedding microfibrous matrix may be designed to assume any shape and architecture to meet the requirement of different cancer cell types. Using a droplet bioprinter, Demirci et al. created a coculture model using OVCAR-5 ovarian cancer cells and normal fibroblasts in Matrigel. The bioprinted pattern permitted an efficient coculture as well as a perfused system.105 This 3D droplet-based cancer model enabled a better control of the spatial repartition of the cells as well as the cell density, hence enabling a more reproducible cell patterning and controlled tumor microenvironment.
Immune cells are proven to be another critical component in the tumor microenvironment, which actively participate in the remodeling of local cancer tissues.10 Kilian et al. developed a coextrusion bioprinting model to analyze the interactions between MDA-MB 231 breast cancer cells and macrophages (Figure 5C),106 which are known to create a paracrine loop inducing an increased motility for both cell types thereby potentially leading to tumor cell extravasation into the bloodstream.10 The two cell types were loaded into the individual cartridges of the bioprinter and coextruded in a spatially controlled manner, where the breast cancer cells were embedded in an alginate bioink and delivered through the sheath flow while the macrophages were suspended in a CaCl2 solution and dispensed via the core to cross-link the outer alginate layer. It was discovered that, the macrophages initially residing in the core microchannel could gradually migrate out to interact with the surrounding breast cancer cells, and such interaction (indicated by the correlation of the two cell types) was dependent on the geometry of the bioprinted macrophage channels within the tumor matrix. This model provides a convenient method to study the interactions of tumor cells with the surrounding stromal cells in the vicinity. Bioprinting tumor cells within a carefully designed niche also enables further studies on cancer metastasis and migration.107
CONCLUSIONS AND PERSPECTIVES
Advances in the 3D bioprinting technologies has enabled in the past decade the fabrication of in vitro 3D tissue models with improve biomimetic properties and functionality, by providing the ability to precisely engineer tissue composition, architecture, and vascularization at good fidelity and potentially high throughput. However, it was not until recently that the community started to focus on 3D cancer models, which are strongly desired for fundamental biological studies as well as for more accurate anticancer drug screening to eventually achieve personalized cancer treatment.
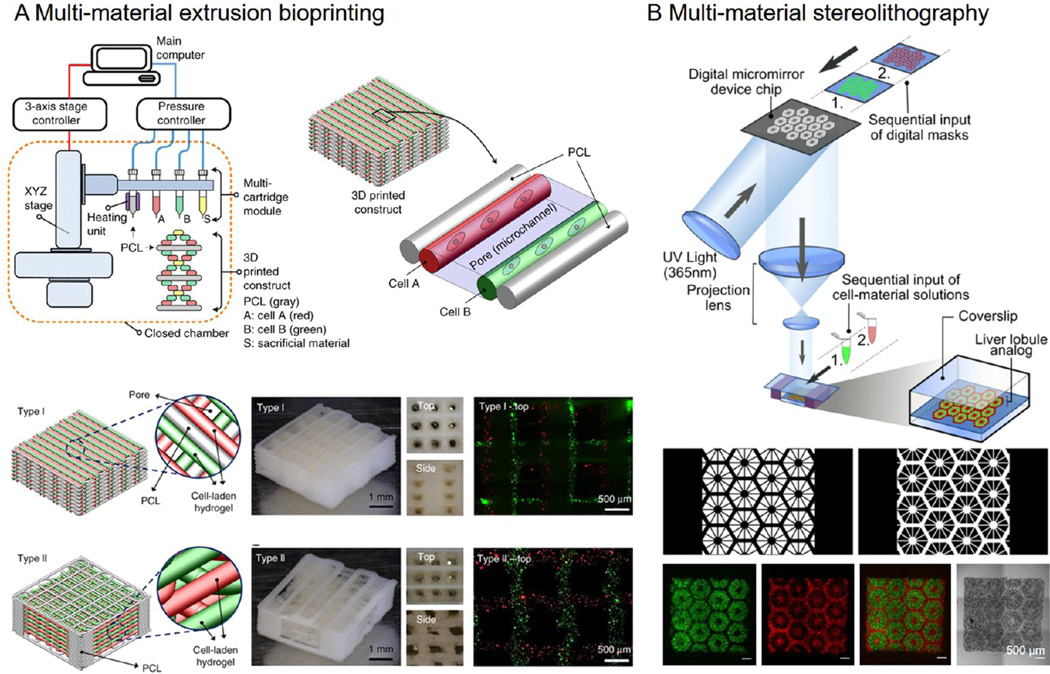
Although current bioprinting strategies have evolved with great sophistication, one major future direction that we envision is the combination of various but complementary approaches capable of recapitulating the extremely complex and heterogeneous cancer microenvironment. In addition to the need for vascularization, we further propose the requirement for compositional accuracy of the cancer microenvironment. To this end, it is believed that the recent development of multimaterial bioprinting systems will become very useful tools in advancing this new field of cancer bioprinting.85,108,109 For example, recently a hybrid bioprinting platform that integrates a heating unit for extrusion of the polymeric scaffold together with three additional cartridges capable of depositing different bioinks of interest was reported (Figure 6A).110 With this multimaterial extrusion-based bioprinting, several types of functional tissues were produced. In comparison, a multi-material stereolithography-based bioprinting platform has also been recently developed, where sequentially presented digital masks were used to pattern biological structures containing more than one bioinks (Figure 6B).61 Using a microfluidic printhead heterogeneous patterns and gradient structures may also be directly produced.76,111,112 Using these novel multi-material bioprinting systems, it is possible to simultaneously deposit desired cell types critical for recapitulation of the cancer microenvironment at the stage of the disease, such as stromal cells, immune cells, and cancer-associated fibroblasts and microvascular cells, in addition to the cancer epithelial cells. Moreover, cell type-specific ECMs, or artificial bioinks mimicking the ECMs, can be used for encapsulating corresponding types of the cancer-related cells to match the requirements of these different cells to achieve their respective optimal functions.
Figure 6. Multimaterial bioprinting.
(A) Extrusion-based multimaterial bioprinting system capable of depositing multiple bioinks along with polymeric scaffold for fabrication of large-scale vascularized functional tissue constructs. Adapted with permission from ref 110. Copyright 2016 Nature Publishing Group. (B) Multimaterial DMD stereolithography bioprinting system capable of high-resolution microscale patterning of the complex tissue microarchitecture to achieve functionality. Adapted with permission from ref 61. Copyright 2016 the National Academy of Sciences of the United States.
Maintaining sufficiently high viability and native phenotypes of cancer and cancer-associated cells posts another major challenge that needs to be overcome to facilitate the application of 3D bioprinting in cancer tissue engineering. To achieve personalized cancer modeling, it is of great importance to be able to preserve the cell bioactivity and phenotypic expressions in a way they behave in their native niches. To this end the exposure to the shear stress during the bioprinting process (especially in the case of extrusion-based bioprinting) may lead to reduced viability of the cells. Sometimes even though the viability is not significantly affected, the phenotypes, or functional behaviors of the deposited cells may still alter over the subsequent period of culture as a result of the stress that the cells experience as they are bioprinted.113–116 Therefore, the proper selection of the bioprinting parameters, such as the viscosity of the bioink, the extrusion rate of the bioink, and the moving speed of the nozzle become particularly important in an effort to achieve unaltered cell viability and phenotypes in bioprinted cancer models.
We believe that, with further development of the tumor-specific bioinks with optimized bioprinting conditions, as well as the convergence of these various bioprinting techniques, advances in the field of cancer bioprinting will significantly improve the biological and physiological relevance of in vitro cancer models. In combination with patient-derived somatic, stromal, and cancerous cells these advances will facilitate progress toward truly personalized cancer treatment.
Acknowledgments
The authors acknowledge funding from the National Institutes of Health (AR057837, DE021468, D005865, AR068258, AR066193, EB022403, EB021148), and the Office of Naval Research Presidential Early Career Award for Scientists and Engineers (PECASE). Y.S.Z. acknowledges the National Cancer Institute Pathway to Independence Award (1K99CA201603-01A1).
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Stewart BW, Wild CP, editors. World Cancer Report 2014. International Agency for Research on Cancer. Lyon, France: World Health Organization; 2014. [Google Scholar]
- 2.Harris AL. Hypoxia—a key regulatory factor in tumour growth. Nature Reviews Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 3.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat. Rev. Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 4.Brown JM, Wilson W. R Exploiting tumour hypoxia in cancer treatment. Nat. Rev. Cancer. 2004;4:437–447. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Calmels TP, Mattot V, Wernert N, Vandenbunder B, Stéhelin D. Invasive tumors induce c-ets1 transcription factor expression in adjacent stroma. Biol. Cell. 1995;84:53–61. doi: 10.1016/0248-4900(96)81318-9. [DOI] [PubMed] [Google Scholar]
- 7.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 9.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat. Rev. Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turley SJ, Cremasco V, Astarita JL. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat. Rev. Immunol. 2015;15:669. doi: 10.1038/nri3902. [DOI] [PubMed] [Google Scholar]
- 11.Cao Y. Tumor angiogenesis and therapy. Biomed. Pharmacother. 2005;59:S340–S343. doi: 10.1016/s0753-3322(05)80070-8. [DOI] [PubMed] [Google Scholar]
- 12.Fukumura D, Jain RK. Tumor microenvironment abnormalities: Causes, consequences, and strategies to normalize. J. Cell. Biochem. 2007;101:937–949. doi: 10.1002/jcb.21187. [DOI] [PubMed] [Google Scholar]
- 13.Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Res. 2012;72:4875–4882. doi: 10.1158/0008-5472.CAN-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 15.Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walter T, Shattuck DW, Baldock R, Bastin ME, Carpenter AE, Duce S, Ellenberg J, Fraser A, Hamilton N, Pieper S, et al. Visualization of image data from cells to organisms. Nat. Methods. 2010;7:S26–S41. doi: 10.1038/nmeth.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Longo DL. Tumor heterogeneity and personalized medicine. N. Engl. J. Med. 2012;366:956–957. doi: 10.1056/NEJMe1200656. [DOI] [PubMed] [Google Scholar]
- 18.Thibaudeau L, Taubenberger AV, Holzapfel BM, Quent VM, Fuehrmann T, Hesami P, Brown TD, Dalton PD, Power CA, Hollier BG, Hutmacher DW. A tissue-engineered humanized xenograft model of human breast cancer metastasis to bone. Dis. Models & Mech. 2014;7:299–309. doi: 10.1242/dmm.014076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi SYC, Lin D, Gout PW, Collins CC, Xu Y, Wang Y. Lessons from patient-derived xenografts for better in vitro modeling of human cancer. Adv. Drug Delivery Rev. 2014;79–80:222–237. doi: 10.1016/j.addr.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Aparicio S, Hidalgo M, Kung AL. Examining the utility of patient-derived xenograft mouse models. Nat. Rev. Cancer. 2015;15:311–316. doi: 10.1038/nrc3944. [DOI] [PubMed] [Google Scholar]
- 21.Eagle H, Foley GE. Cytotoxicity in human cell cultures as a primary screen for the detection of anti-tumor agents. Cancer Res. 1958;18:1017–1025. [PubMed] [Google Scholar]
- 22.Hirschberg E. Tissue culture in cancer chemotherapy screening. Cancer Res. 1958;18:869–878. [PubMed] [Google Scholar]
- 23.Alemany-Ribes M, Semino CE. Bioengineering 3D environments for cancer models. Adv. Drug Delivery Rev. 2014;79–80:40–49. doi: 10.1016/j.addr.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Unger C, Kramer N, Walzl A, Scherzer M, Hengstschläger M, Dolznig H. Modeling human carcinomas: Physiologically relevant 3D models to improve anti-cancer drug development. Adv. Drug Delivery Rev. 2014;79–80:50–67. doi: 10.1016/j.addr.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 25.Pickl M, Ries C. Comparison of 3D and 2D tumor models reveals enhanced HER2 activation in 3D associated with an increased response to trastuzumab. Oncogene. 2009;28:461–468. doi: 10.1038/onc.2008.394. [DOI] [PubMed] [Google Scholar]
- 26.Weigelt B, Lo AT, Park CC, Gray JW, Bissell MJ. HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment. Breast Cancer Res. Treat. 2010;122:35–43. doi: 10.1007/s10549-009-0502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Tan Y, Zhang H, Zhang Y, Xu P, Chen J, Poh Y-C, Tang K, Wang N, Huang B. Soft fibrin gels promote selection and growth of tumorigenic cells. Nat. Mater. 2012;11:734–741. doi: 10.1038/nmat3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hickman JA, Graeser R, de Hoogt R, Vidic S, Brito C, Gutekunst M, van der Kuip H. Three-dimensional models of cancer for pharmacology and cancer cell biology: Capturing tumor complexity in vitro/ex vivo. Biotechnol. J. 2014;9:1115–1128. doi: 10.1002/biot.201300492. [DOI] [PubMed] [Google Scholar]
- 29.Yeon S-E, Lee S-H, Nam SW, Oh I-H, Lee J, Kuh H-J. Application of concave microwells to pancreatic tumor spheroids enabling anticancer drug evaluation in a clinically relevant drug resistance model. PLoS One. 2013;8:e73345. doi: 10.1371/journal.pone.0073345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelm JM, Timmins NE, Brown CJ, Fussenegger M, Nielsen LK. Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol. Bioeng. 2003;83:173–180. doi: 10.1002/bit.10655. [DOI] [PubMed] [Google Scholar]
- 31.Carver K, Ming X, Juliano RL. Multicellular tumor spheroids as a model for assessing delivery of oligonucleotides in three dimensions. Mol. Ther.-Nucleic Acids. 2014;3:e153. doi: 10.1038/mtna.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song K, Liu T, Li X, Cui Z, Sun X, Ma X. Three-dimensional expansion: in suspension culture of SD rat’s osteoblasts in a rotating wall vessel bioreactor. J. Biomed. Mater. Res., Part A. 2007;20:91. [PubMed] [Google Scholar]
- 33.Lee JM, Mhawech-Fauceglia P, Lee N, Parsanian LC, Lin YG, Gayther SA, Lawrenson K. A three-dimensional microenvironment alters protein expression and chemosensitivity of epithelial ovarian cancer cells in vitro. Lab. Invest. 2013;93:528–542. doi: 10.1038/labinvest.2013.41. [DOI] [PubMed] [Google Scholar]
- 34.Friedrich J, Seidel C, Ebner R, Kunz-Schughart LA. Spheroid-based drug screen: considerations and practical approach. Nat. Protoc. 2009;4:309–324. doi: 10.1038/nprot.2008.226. [DOI] [PubMed] [Google Scholar]
- 35.Vinci M, Gowan S, Boxall F, Patterson L, Zimmermann M, Lomas C, Mendiola M, Hardisson D, Eccles SA. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012;10:29. doi: 10.1186/1741-7007-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim B, Han G, Toley BJ, Kim C-k, Rotello VM, Forbes NS. Tuning payload delivery in tumour cylindroids using gold nanoparticles. Nat. Nanotechnol. 2010;5:465–472. doi: 10.1038/nnano.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat. Rev. Cancer. 2014;14:430–439. doi: 10.1038/nrc3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, Polverini PJ, Mooney DJ. Engineering tumors with 3D scaffolds. Nat. Methods. 2007;4:855–60. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 39.Song H-HG, Park KM, Gerecht S. Hydrogels to model 3D in vitro microenvironment of tumor vascularization. Adv. Drug Delivery Rev. 2014;79–80:19–29. doi: 10.1016/j.addr.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loessner D, Meinert C, Kaemmerer E, Martine LC, Yue K, Levett PA, Klein TJ, Melchels FPW, Khademhosseini A, Hutmacher DW. Functionalization, preparation and use of cell-laden gelatin methacryloyl-based hydrogels as modular tissue culture platforms. Nat. Protoc. 2016;11:727–746. doi: 10.1038/nprot.2016.037. [DOI] [PubMed] [Google Scholar]
- 41.Chwalek K, Tsurkan MV, Freudenberg U, Werner C. Glycosaminoglycan-based hydrogels to modulate heterocellular communication in in vitro angiogenesis models. Sci. Rep. 2014;4:4. doi: 10.1038/srep04414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nietzer S, Baur F, Sieber S, Hansmann J, Schwarz T, Stoffer C, Häfner H, Gasser M, Waaga-Gasser AM, Walles H, Dandekar G. Mimicking Metastases Including Tumor Stroma: A New Technique to Generate a Three-Dimensional Colorectal Cancer Model Based on a Biological Decellularized Intestinal Scaffold. Tissue Eng., Part C. 2016 doi: 10.1089/ten.tec.2015.0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szot CS, Buchanan CF, Freeman JW, Rylander MN. 3D in vitro bioengineered tumors based on collagen I hydrogels. Biomaterials. 2011;32:7905–7912. doi: 10.1016/j.biomaterials.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeon JS, Bersini S, Gilardi M, Dubini G, Charest JL, Moretti M, Kamm RD. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc. Natl. Acad. Sci. U. S. A. 2015;112:214–219. doi: 10.1073/pnas.1417115112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 2007;1:84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kleinman HK, Martin GR. Matrigel: Basement Membrane Matrix with Biological Activity. Semin. Cancer Biol. 2005;2005:378–386. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Zhang YS, Xia Y. Multiple facets for extracellular matrix mimicking in regenerative medicine. Nanomedicine. 2015;10:689–692. doi: 10.2217/nnm.15.10. [DOI] [PubMed] [Google Scholar]
- 48.Ma PX. Biomimetic materials for tissue engineering. Adv. Drug Delivery Rev. 2008;60:184–198. doi: 10.1016/j.addr.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knowlton S, Onal S, Yu CH, Zhao JJ, Tasoglu S. Bioprinting for cancer research. Trends Biotechnol. 2015;33:504–513. doi: 10.1016/j.tibtech.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 50.Malda J, Visser J, Melchels FP, Jüngst T, Hennink WE, Dhert WJ, Groll J, Hutmacher DW. 25th anniversary article: engineering hydrogels for biofabrication. Adv. Mater. 2013;25:5011–5028. doi: 10.1002/adma.201302042. [DOI] [PubMed] [Google Scholar]
- 51.Murphy SV, Atala A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014;32:773–785. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 52.Zhang YS, Yue K, Aleman J, Mollazadeh-Moghaddam K, Bakht SM, Dell’Erba V, Assawes P, Shin SR, Dokmeci MR, Oklu R, Khademhosseini A. 3D Bioprinting for Tissue and Organ Fabrication. Ann. Biomed. Eng. 2016 doi: 10.1007/s10439-016-1612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakamura M, Kobayashi A, Takagi F, Watanabe A, Hiruma Y, Ohuchi K, Iwasaki Y, Horie M, Morita I, Takatani S. Biocompatible inkjet printing technique for designed seeding of individual living cells. Tissue Eng. 2005;11:1658–1666. doi: 10.1089/ten.2005.11.1658. [DOI] [PubMed] [Google Scholar]
- 54.Boland T, Xu T, Damon B, Cui X. Application of inkjet printing to tissue engineering. Biotechnol. J. 2006;1:910–917. doi: 10.1002/biot.200600081. [DOI] [PubMed] [Google Scholar]
- 55.Khalil S, Sun W. Biopolymer deposition for freeform fabrication of hydrogel tissue constructs. Mater. Sci. Eng., C. 2007;27:469–478. [Google Scholar]
- 56.Chang CC, Boland ED, Williams SK, Hoying JB. Direct-write bioprinting three-dimensional biohybrid systems for future regenerative therapies. J. Biomed. Mater. Res., Part B. 2011;98:160–170. doi: 10.1002/jbm.b.31831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guillotin B, Souquet A, Catros S, Duocastella M, Pippenger B, Bellance S, Bareille R, Rémy M, Bordenave L, Amédée J, Guillemot F. Laser assisted bioprinting of engineered tissue with high cell density and microscale organization. Biomaterials. 2010;31:7250–7256. doi: 10.1016/j.biomaterials.2010.05.055. [DOI] [PubMed] [Google Scholar]
- 58.Gaebel R, Ma N, Liu J, Guan J, Koch L, Klopsch C, Gruene M, Toelk A, Wang W, Mark P, et al. Patterning human stem cells and endothelial cells with laser printing for cardiac regeneration. Biomaterials. 2011;32:9218–9230. doi: 10.1016/j.biomaterials.2011.08.071. [DOI] [PubMed] [Google Scholar]
- 59.Gauvin R, Chen Y-C, Lee JW, Soman P, Zorlutuna P, Nichol JW, Bae H, Chen S, Khademhosseini A. Microfabrication of complex porous tissue engineering scaffolds using 3D projection stereolithography. Biomaterials. 2012;33:3824–3834. doi: 10.1016/j.biomaterials.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soman P, Chung PH, Zhang AP, Chen S. Digital microfabrication of user-defined 3D microstructures in cell-laden hydrogels. Biotechnol. Bioeng. 2013;110:3038–3047. doi: 10.1002/bit.24957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma X, Qu X, Zhu W, Li Y-S, Yuan S, Zhang H, Liu J, Wang P, Lai CSE, Zanella F, Feng G-S, Sheikh F, Chien S, Chen S. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc. Natl. Acad. Sci. U. S. A. 2016;113:2206–2211. doi: 10.1073/pnas.1524510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cui X, Breitenkamp K, Finn M, Lotz M, D’Lima DD. Direct human cartilage repair using three-dimensional bioprinting technology. Tissue Eng., Part A. 2012;18:1304–1312. doi: 10.1089/ten.tea.2011.0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cui X, Gao G, Qiu Y. Accelerated myotube formation using bioprinting technology for biosensor applications. Biotechnol. Lett. 2013;35:315–321. doi: 10.1007/s10529-012-1087-0. [DOI] [PubMed] [Google Scholar]
- 64.Gou M, Qu X, Zhu W, Xiang M, Yang J, Zhang K, Wei Y, Chen S. Bio-inspired detoxification using 3D–printed hydrogel nanocomposites. Nat. Commun. 2014;5:3774. doi: 10.1038/ncomms4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee MP, Cooper GJ, Hinkley T, Gibson GM, Padgett MJ, Cronin L. Development of a 3D printer using scanning projection stereolithography. Sci. Rep. 2015;5:9875. doi: 10.1038/srep09875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Majumder B, Baraneedharan U, Thiyagarajan S, Radhakrishnan P, Narasimhan H, Dhandapani M, Brijwani N, Pinto DD, Prasath A, Shanthappa BU, Thayakumar A, Surendran R, Babu GK, Shenoy AM, Kuriakose MA, Bergthold G, Horowitz P, Loda M, Beroukhim R, Agarwal S, Sengupta S, Sundaram M, Majumder PK. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat. Commun. 2015;6:6169. doi: 10.1038/ncomms7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zaman MH, Trapani LM, Sieminski AL, MacKellar D, Gong H, Kamm RD, Wells A, Lauffenburger DA, Matsudaira P. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pathak A, Kumar S. Independent regulation of tumor cell migration by matrix stiffness and confinement. Proc. Natl. Acad. Sci. U. S. A. 2012;109:10334–10339. doi: 10.1073/pnas.1118073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang S, Ingber DE. Cell tension, matrix mechanics, and cancer development. Cancer Cell. 2005;8:175–176. doi: 10.1016/j.ccr.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 71.Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat. Rev. Cancer. 2009;9:108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Skardal A, Atala A. Biomaterials for Integration with 3-D Bioprinting. Ann. Biomed. Eng. 2015;43:730–746. doi: 10.1007/s10439-014-1207-1. [DOI] [PubMed] [Google Scholar]
- 73.Chimene D, Lennox KK, Kaunas RR, Gaharwar AK. Advanced Bioinks for 3D Printing: A Materials Science Perspective. Ann. Biomed. Eng. 2016;44:2090–2102. doi: 10.1007/s10439-016-1638-y. [DOI] [PubMed] [Google Scholar]
- 74.Jungst T, Smolan W, Schacht K, Scheibel T, Groll Jr. Strategies and Molecular Design Criteria for 3D Printable Hydrogels. Chem. Rev. 2016;116:1496–1539. doi: 10.1021/acs.chemrev.5b00303. [DOI] [PubMed] [Google Scholar]
- 75.Murphy SV, Skardal A, Atala A. Evaluation of hydrogels for bio-printing applications. J. Biomed. Mater. Res., Part A. 2013;101A:272–284. doi: 10.1002/jbm.a.34326. [DOI] [PubMed] [Google Scholar]
- 76.Colosi C, Shin SR, Manoharan V, Massa S, Costantini M, Barbetta A, Dokmeci MR, Dentini M, Khademhosseini A. Microfluidic bioprinting of heterogeneous 3D tissue constructs using low viscosity bioink. Adv. Mater. 2016;28:677–684. doi: 10.1002/adma.201503310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Augst AD, Kong HJ, Mooney DJ. Alginate hydrogels as biomaterials. Macromol. Biosci. 2006;6:623–633. doi: 10.1002/mabi.200600069. [DOI] [PubMed] [Google Scholar]
- 78.Christensen K, Xu C, Chai W, Zhang Z, Fu J, Huang Y. Freeform inkjet printing of cellular structures with bifurcations. Biotechnol. Bioeng. 2015;112:1047–55. doi: 10.1002/bit.25501. [DOI] [PubMed] [Google Scholar]
- 79.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Cell-laden microengineered gelatin meth-acrylate hydrogels. Biomaterials. 2010;31:5536–44. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yue K, Trujillo-de Santiago G, Alvarez MM, Tamayol A, Annabi N, Khademhosseini A. Synthesis, properties, and biomedical applications of gelatin methacryloyl (GelMA) hydrogels. Biomaterials. 2015;73:254–71. doi: 10.1016/j.biomaterials.2015.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Malda J, Visser J, Melchels FP, Jungst T, Hennink WE, Dhert WJ, Groll J, Hutmacher DW. 25th anniversary article: Engineering hydrogels for biofabrication. Adv. Mater. 2013;25:5011–28. doi: 10.1002/adma.201302042. [DOI] [PubMed] [Google Scholar]
- 82.Highley CB, Rodell CB, Burdick JA. Direct 3D Printing of Shear-Thinning Hydrogels into Self-Healing Hydrogels. Adv. Mater. 2015;27:5075–5079. doi: 10.1002/adma.201501234. [DOI] [PubMed] [Google Scholar]
- 83.Hoffman AS. Hydrogels for biomedical applications. Adv. Drug Delivery Rev. 2012;64:18–23. doi: 10.1016/s0169-409x(01)00239-3. [DOI] [PubMed] [Google Scholar]
- 84.Seliktar D. Designing cell-compatible hydrogels for biomedical applications. Science. 2012;336:1124–1128. doi: 10.1126/science.1214804. [DOI] [PubMed] [Google Scholar]
- 85.Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA. 3D Bioprinting of Vascularized, Heterogeneous Cell-Laden Tissue Constructs. Adv. Mater. 2014;26:3124–3130. doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 86.Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA. Three-dimensional bioprinting of thick vascularized tissues. Proc. Natl. Acad. Sci. U. S. A. 2016;113:3179–3184. doi: 10.1073/pnas.1521342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang YS, Aleman J, Arneri A, Bersini S, Piraino F, Shin SR, Dokmeci MR, Khademhosseini A. From Cardiac Tissue Engineering to Heart-on-a-Chip: Beating Challenges. Biomed. Mater. 2015;10:034006. doi: 10.1088/1748-6041/10/3/034006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Peppas NA, Hilt JZ, Khademhosseini A, Langer R. Hydrogels in Biology and Medicine: From Molecular Principles to Bionanotechnology. Adv. Mater. 2006;18:1345–1360. [Google Scholar]
- 89.Khademhosseini A, Langer R. Microengineered hydrogels for tissue engineering. Biomaterials. 2007;28:5087–5092. doi: 10.1016/j.biomaterials.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 90.Pati F, Jang J, Ha DH, Won Kim S, Rhie JW, Shim JH, Kim DH, Cho DW. Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink. Nat. Commun. 2014;5:3935. doi: 10.1038/ncomms4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Murphy SV, Atala A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014;32:773–85. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 92.Skardal A, Devarasetty M, Kang H-W, Mead I, Bishop C, Shupe T, Lee SJ, Jackson J, Yoo J, Soker S, Atala A. A hydrogel bioink toolkit for mimicking native tissue biochemical and mechanical properties in bioprinted tissue constructs. Acta Biomater. 2015;25:24–34. doi: 10.1016/j.actbio.2015.07.030. [DOI] [PubMed] [Google Scholar]
- 93.Skardal A, Devarasetty M, Kang H-W, Seol Y-J, Forsythe SD, Bishop C, Shupe T, Soker S, Atala A. Bioprinting Cellularized Constructs Using a Tissue-specific Hydrogel Bioink. J. Visualized Exp. 2016:e53606–e53606. doi: 10.3791/53606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 95.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen D-HT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, et al. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat. Mater. 2012;11:768–774. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bertassoni LE, Cecconi M, Manoharan V, Nikkhah M, Hjortnaes J, Cristino AL, Barabaschi G, Demarchi D, Dokmeci MR, Yang Y, Khademhosseini A. Hydrogel bioprinted microchannel networks for vascularization of tissue engineering constructs. Lab Chip. 2014;14:2202–2211. doi: 10.1039/c4lc00030g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee VK, Kim DY, Ngo H, Lee Y, Seo L, Yoo S-S, Vincent PA, Dai G. Creating perfused functional vascular channels using 3D bio-printing technology. Biomaterials. 2014;35:8092–8102. doi: 10.1016/j.biomaterials.2014.05.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bhattacharjee T, Zehnder SM, Rowe KG, Jain S, Nixon RM, Sawyer WG, Angelini TE. Writing in the granular gel medium. Science Advances. 2015;1:e1500655. doi: 10.1126/sciadv.1500655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hinton TJ, Jallerat Q, Palchesko RN, Park JH, Grodzicki MS, Shue H-J, Ramadan MH, Hudson AR, Feinberg AW. Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Science Advances. 2015;1:e1500758. doi: 10.1126/sciadv.1500758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gao Q, He Y, Fu J-z, Liu A, Ma L. Coaxial nozzle-assisted 3D bioprinting with built-in microchannels for nutrients delivery. Biomaterials. 2015;61:203–215. doi: 10.1016/j.biomaterials.2015.05.031. [DOI] [PubMed] [Google Scholar]
- 101.Zhang Y, Yu Y, Chen H, Ozbolat IT. Characterization of printable cellular micro-fluidic channels for tissue engineering. Biofabrication. 2013;5:025004. doi: 10.1088/1758-5082/5/2/025004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee VK, Dai G, Zou H, Yoo S-S. 41st Annual Northeast, Biomedical Engineering Conference. Piscataway, NJ: IEEE; 2015. Generation of 3-D Glioblastoma-Vascular Niche Using 3-D Bioprinting; pp. 1–2. [Google Scholar]
- 103.Zhao Y, Yao R, Ouyang L, Ding H, Zhang T, Zhang K, Cheng S, Sun W. Three-dimensional printing of Hela cells for cervical tumor model in vitro. Biofabrication. 2014;6:035001. doi: 10.1088/1758-5082/6/3/035001. [DOI] [PubMed] [Google Scholar]
- 104.Ling K, Huang G, Liu J, Zhang X, Ma Y, Lu T, Xu F. Bioprinting-Based High-Throughput Fabrication of Three-Dimensional MCF-7 Human Breast Cancer Cellular Spheroids. Engineering. 2015;1:269–274. [Google Scholar]
- 105.Xu F, Celli J, Rizvi I, Moon S, Hasan T, Demirci U. A three-dimensional in vitro ovarian cancer coculture model using a high-throughput cell patterning platform. Biotechnol. J. 2011;6:204–212. doi: 10.1002/biot.201000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grolman JM, Zhang D, Smith AM, Moore JS, Kilian KA. Rapid 3D extrusion of synthetic tumor microenvironments. Adv. Mater. 2015;27:5512–5517. doi: 10.1002/adma.201501729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huang TQ, Qu X, Liu J, Chen S. 3D printing of biomimetic microstructures for cancer cell migration. Biomed. Microdevices. 2014;16:127–132. doi: 10.1007/s10544-013-9812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Khalil S, Nam J, Sun W. Multi-nozzle deposition for construction of 3D biopolymer tissue scaffoldsnull. Rapid Prototyping Journal. 2005;11:9–17. [Google Scholar]
- 109.Chang R, Nam J, Sun W. Effects of Dispensing Pressure and Nozzle Diameter on Cell Survival from Solid Freeform Fabrication-Based Direct Cell Writing. Tissue Eng., Part A. 2008;14:41–48. doi: 10.1089/ten.a.2007.0004. [DOI] [PubMed] [Google Scholar]
- 110.Kang H-W, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A. A 3D bioprinting system to produce human-scale tissue constructs with structural integrity. Nat. Biotechnol. 2016;34:312. doi: 10.1038/nbt.3413. [DOI] [PubMed] [Google Scholar]
- 111.Hardin JO, Ober TJ, Valentine AD, Lewis JA. Microfluidic Printheads for Multimaterial 3D Printing of Viscoelastic Inks. Adv. Mater. 2015;27:3279–3284. doi: 10.1002/adma.201500222. [DOI] [PubMed] [Google Scholar]
- 112.Ober TJ, Foresti D, Lewis JA. Active mixing of complex fluids at the microscale. Proc. Natl. Acad. Sci. U. S. A. 2015;112:12293–12298. doi: 10.1073/pnas.1509224112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Blaeser A, Duarte Campos DF, Puster U, Richtering W, Stevens MM, Fischer H. Controlling Shear Stress in 3D Bioprinting is a Key Factor to Balance Printing Resolution and Stem Cell Integrity. Adv. Healthcare Mater. 2016;5:326–333. doi: 10.1002/adhm.201500677. [DOI] [PubMed] [Google Scholar]
- 114.Blaeser A, Campos D, Fischer H. 3D–Bioprinting Induced Shear Stress Strongly Impacts Human MSC Survival and Proliferation Potential. Tissue Eng., Part A. 2015;21:S322–S323. [Google Scholar]
- 115.Ning L, Guillemot A, Zhao J, Kipouros G, Chen D. Influence of flow behavior of alginate-cell suspensions on cell viability and proliferation. Tissue Eng., Part C. 2016 doi: 10.1089/ten.TEC.2016.0011. [DOI] [PubMed] [Google Scholar]
- 116.Li M, Tian X, Kozinski JA, Chen X, Hwang DK. Modeling mechanical cell damage in the bioprinting process employing a conical needle. J. Mech. Med. Biol. 2015;15:1550073. [Google Scholar]