Abstract
We present here the summary of main characteristics of ‘Traorella massiliensis’ strain Marseille-P3110T, which was isolated from a left colon liquid sample of a 76-year-old woman.
Keywords: Culturomics, gut microbiota, new species, taxonogenomics, Traorella massiliensis
In May 2016, using the culturomics approach [1] on storied samples from the digestive tract, we isolated the strain Marseille-P3110T from the left colon liquid sample of a 76-year-old woman who underwent colonoscopy for a history of colonic polyp. The patient provided signed informed consent, and the ethics committee of the Institut Fédératif de Recherche IFR48 approved the study under number 2016-010.
The sample was directly inoculated on Columbia agar enriched with 5% of sheep’s blood (COS; bioMérieux, Marcy l’Etoile, France), and strain Marseille-P3110T was obtained after a 3-day incubation at 37°C in an anaerobic atmosphere (anaeroGen Compact; Oxoid, Thermo Scientific, Dardilly, France). Colonies were white, convex and circular with a diameter of 0.8 mm. Cells were Gram-negative bacilli with a mean diameter of 0.3 μm and a length varying from 2.0 to 3.6 μm. Strain Marseille-P3110T was motile and non–endospore forming. This strain exhibited neither catalase nor oxidase activities.
The 16S rRNA gene was sequenced using fD1-rP2 primers as previously described [2] with a 3130-XL sequencer (Applied Biosciences, Saint Aubin, France) because of the impossibility of identifying strain Marseille-P3110T by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Microflex LT; Bruker Daltonics, Bremen, Germany) [3]. Strain Marseille-P3110T exhibited a 93.2% sequence similarity [4] with Dielma fastidiosa type strain JC13T (GenBank accession no. JF824807), the phylogenetically closest species with standing in nomenclature (Fig. 1), which was isolated from the stool of a healthy Senegalese patient for the first time in 2013 [5]. This taxonomic name has been effectively published but not validly published under the rules of the International Code of Nomenclature of Bacteria.
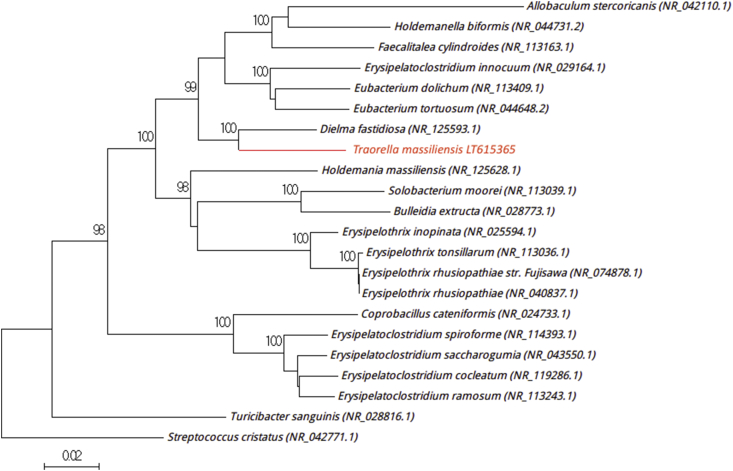
Fig. 1.
Phylogenetic tree showing position of Traorella massiliensis strain Marseille-P3110T relative to other phylogenetically close neighbours. Sequences were aligned using Muscle 3.8.31 with default parameters, and phylogenetic inferences were obtained using neighbour-joining method with 1000 bootstrap replicates within MEGA6 software. Only bootstrap values of >95% are shown. Scale bar represents 2% nucleotide sequence divergence.
Because of a 16S rRNA sequence similarity of <95% between strain Marseille-P3110T and its phylogenetically closest species with standing in nomenclature [6], we propose the creation of a new genus ‘Traorella’ gen. nov. (tra.o.rel′la, N.L. masc. n. Traorella, in honor of a Senegalese scientist, Sory Ibrahima Traore, a pioneer of culturomics). ‘Traorella massiliensis’ gen. nov., sp. nov. (mas.si.li.en′sis, L. masc. adj. massiliensis, ‘Massilia,’ the Roman name of Marseille), is part of the Erysipelotrichaceae family and the Firmicutes phylum. Strain Marseille-P3110 is the type strain of the new species Traorella massiliensis gen. nov., sp. nov.
MALDI-TOF MS spectrum
The MALDI-TOF MS spectrum of ‘Traorella massiliensis’ strain Marseille-P3110T is available online (http://www.mediterranee-infection.com/article.php?laref=256&titre=urms-database).
Nucleotide sequence accession number
The 16S rRNA gene sequence of ‘Traorella massiliensis’ strain Marseille-P3110T was deposited in GenBank under accession number LT615365.
Deposit in a culture collection
‘Traorella massiliensis’ strain Marseille-P3110T was deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR, WDCM 875) under the number P3110 and in the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) under number DSM 103514.
Acknowledgements
This study was funded by the Fondation Méditerranée Infection. The authors thank M. Lardière for English-language review.
Conflict of Interest
None declared.
References
- 1.Lagier J.C., Khelaifia S., Tidjani Alou M., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PubMed] [Google Scholar]
- 2.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64(Pt 2):346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 5.Ramasamy D., Lagier J.C., Nguyen T.T., Raoult D., Fournier P.E. Non-contiguous finished genome sequence and description of Dielma fastidiosa gen. nov., sp. nov., a new member of the Family Erysipelotrichaceae. Stand Genomic Sci. 2013;8:336–351. doi: 10.4056/sigs.3567059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huson D.H., Auch A.F., Qi J., Schuster S.C. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]