Abstract
We report here the main characteristics of “Negativibacillus massiliensis” strain Marseille-P3213T, isolated from a human left-colon wash sample.
Keywords: Culturomics, human gut, microbiota, “Negativibacillus massiliensis”, taxonogenomics
In 2016, as a part of a culturomics study [1] focused on the modifications of the human gut microbiome along the whole gastrointestinal tract, we isolated from the left colon of a 76-year-old patient a bacterial strain that escaped our systematic matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) screening on a Microflex spectrometer (Bruker Daltonics, Bremen, Germany) [2]. The patient, who underwent simultaneous upper and lower endoscopy for medical reasons, provided signed informed consent, and the study was validated by the ethics committee of the Institut Fédératif de Recherche IFR48 under number 09-022.
Strain Marseille-P3213T growth was obtained on 5% sheep's blood–Columbia agar medium (bioMérieux, Marcy l’Etoile, France) under anaerobic atmosphere (anaeroGEN, Oxoid, Dardilly, France) after a 14-day enrichment of the fresh left-colon sample in an anaerobic blood culture bottle (Becton Dickinson, Pont de Claix, France) added with 5 mL sheep’s blood (bioMérieux) and 5 mL 0.2 μm filtered (Thermo Fisher Scientific, Villebon-sur-Yvette, France) rumen at 37°C. After a 5-day anaerobic incubation on 5% sheep’s blood–enriched Columbia agar (bioMérieux), colonies were approximatively circular, raised with undulated edges, whitish and not haemolytic. The mean diameter was 0.5 to 3 mm.
Bacterial cells were Gram-negative, nonmotile rods 0.5 to 0.8 μm wide by 3.0 to 4.5 μm long. Strain Marseille-P3213T tested catalase and oxidase negative. Sporulation was obtained in 20 minutes at 80°C, while no growth was observed under aerobic or microaerophilic (campyGen, Oxoid) conditions. Growth was obtained on blood-enriched agar under an anaerobic atmosphere at 37°C and 45°C (no growth at 20°C, 28°C and 55°C).
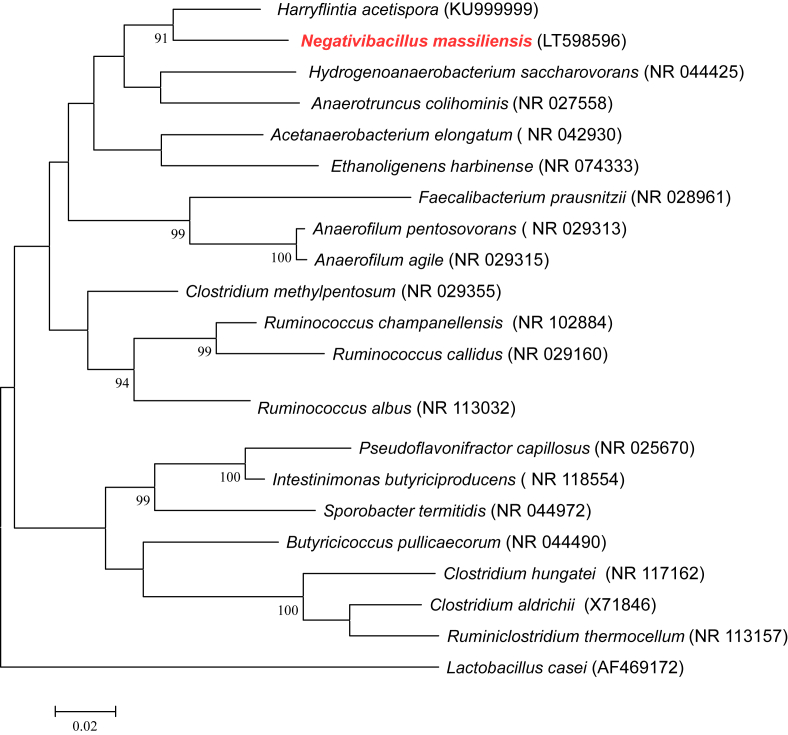
The 16S rRNA gene was sequenced using fD1-rP2 primers as previously described [3], using a 3130-XL sequencer (Applied Biosciences, Saint Aubin, France). Strain Marseille-P3213T exhibited a 93.52% sequence identity with Harriflyntia acetispora strain V20-281aT (GenBank accession no. KU999999), the phylogenetically closest species with standing in nomenclature (Fig. 1), which putatively classifies strain Marseille-P3213T as a member of a new genus within the Clostridiales cluster IV in the phylum Firmicutes.
Fig. 1.
Phylogenetic tree showing position of “Negativibacillus massiliensis” strain Marseille-P3213T relative to other phylogenetically close neighbours. Sequences were aligned using CLUSTALW and phylogenetic inferences obtained using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 500 times to generate majority consensus tree. Only bootstrap scores of at least 90 were retained.
Clostridiales cluster IV was created in 1994 and groups microorganisms that exhibit Clostridium and non-Clostridium-like characteristics [4], including Gram-negative microorganisms [5]. Harriflyntia acetispora, the phylogenetically closest species to strain Marseille-P3213T, is a Gram-negative, endospore-forming rod isolated from chicken gut. Unlike strain Marseille-P3213T, it is catalase positive [6].
On the basis of the phenotypic (catalase activity) and 16S rRNA gene sequence divergence of strain Marseille-P3213T with the phylogenetically closest species with standing in nomenclature [7], we propose here the creation of the new genus “Negativibacillus” (Ne.ga.ti.vi.ba.cil′lus, L. adj. negativus, ‘negative’; L. masc. n. bacillus, ‘a small staff’; N.L. masc. n. Negativibacillus, bacillus with a Gram-negative cell wall structure). Strain Marseille-P3213T (= CSURP3213 = DSM103594) is the type strain of “Negativibacillus massiliensis” gen. nov., sp. nov., (mas.si.li.en’sis, L. masc. adj. massiliensis from Massilia, the Roman name of Marseille).
MALDI-TOF MS spectrum
The MALDI-TOF MS spectrum of “Negativibacillus massiliensis” strain Marseille-P3213T is available online (http://www.mediterranee-infection.com/article.php?laref=256&titre=urms-database).
Nucleotide sequence accession number
The 16S rRNA gene sequence was deposited in GenBank under accession number LT598596.
Deposit in a culture collection
Strain Marseille-P3213T was deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR, WDCM 875) under number P3213 and in the Deutsche Sammlung von Mikroorganismen und Zellkulturen under number DSM 103594.
Acknowledgement
This study was funded by the Fondation Méditerranée Infection.
Conflict of Interest
None declared.
References
- 1.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins M.D., Lawson P.A., Willems A., Cordoba J.J., Fernandez-Garayzabal J., Garcia P. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 5.Carlier J.P., Bedora-Faure M., K'ouas G., Alauzet C., Mory F. Proposal to unify Clostridium orbiscindens Winter et al 1991 and Eubacterium plautii (Séguin 1928) Hofstad and Aasjord 1982, with description of Flavonifractor plautii gen. nov., comb. nov., and reassignment of Bacteroides capillosus to Pseudoflavonifractor capillosus gen. nov., comb. nov. Int J Syst Evol Microbiol. 2010;60:585–590. doi: 10.1099/ijs.0.016725-0. [DOI] [PubMed] [Google Scholar]
- 6.Petzoldt D., Breves G., Rautenschlein S., Taras D. Harryflintia acetispora gen. nov., sp. nov., isolated from chicken caecum. Int J Syst Evol Microbiol. 2016;66:4099–4104. doi: 10.1099/ijsem.0.001317. [DOI] [PubMed] [Google Scholar]
- 7.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]