Abstract
There is an unmet need for novel metal-based chemotherapeutics with alternative modes of action compared to clinical agents such as cisplatin and metallo-bleomycin. Recent attention in this field has focused on designing intracellular ROS-mediators as powerful cytotoxins of human cancers and identifying potentially unique toxic mechanisms underpinning their utility. Herein, we report the developmental di-manganese(II) therapeutic [Mn2(μ-oda)(phen)4(H2O)2][Mn2(μ-oda)(phen)4(oda)2]·4H2O (Mn-Oda) induces autophagy-promoted apoptosis in human ovarian cancer cells (SKOV3). The complex was initially identified to intercalate DNA by topoisomerase I unwinding and circular dichroism spectroscopy. Intracellular DNA damage, detected by γH2AX and the COMET assay, however, is not linked to direct Mn-Oda free radical generation, but is instead mediated through the promotion of intracellular reactive oxygen species (ROS) leading to autophagic vacuole formation and downstream nuclear degradation. To elucidate the cytotoxic profile of Mn-Oda, a wide range of biomarkers specific to apoptosis and autophagy including caspase release, mitochondrial membrane integrity, fluorogenic probe localisation, and cell cycle analysis were employed. Through these techniques, the activity of Mn-Oda was compared directly to i.) the pro-apoptotic clinical anticancer drug doxorubicin, ii.) the multimodal histone deacetylase inhibitor suberoyanilide hydroxamic acid, and iii.) the autophagy inducer rapamycin. In conjunction with ROS-specific trapping agents and established inhibitors of autophagy, we have identified autophagy-induction linked to mitochondrial superoxide production, with confocal image analysis of SKOV3 cells further supporting autophagosome formation.
Abbreviations: γH2AX, phosphorylated H2AX histone; caspase, cysteine-dependent asparate-specific proteases CAT, catalase; CCCP, carbonyl cyanide m-chlorophenyl hydrazine; Dox, doxorubicin; DSB, double strand breaks; LC-3, microtubule associated protein 1 light weight chain 3; MDC, monodansylcadarevine; Rapa, rapamycin; ROS, reactive oxygen species; SAHA, suberoylanilide hydroxamic acid; SKOV3, human ovarian carcinoma; SOD, superoxide dismutase; SSB, single strand breaks; topo, topoisomerase
Keywords: Cancer, Manganese, Superoxide, Autophagy, Apoptosis
1. Introduction
Metals ions including Mn2+, Mg2+, Cu2+ and Zn2+ are essential in the biochemistry and physiology of living organisms as they are required cofactors for ubiquitous enzymes, transcription factors, transmembrane transporters, growth factors and receptors [1]. Deficiencies in these metal ions result in the onset of neurodegenerative diseases including Alzheimer's, Parkinson's and Menkes syndrome, while Cu2+ overload is associated with Wilson's disease [2]. The incorporation of metal ions into complex scaffolds containing targeting ligands, however, has resulted in the development of anti-tumoural drug candidates [3], [4], [5] capable of mediating apoptotic cell death through a combination of intrinsic and extrinsic pathways [6]. Numerous examples of metal complexes inducing cancer cytotoxicity through apoptosis have been reported in the literature [7]; this fatal mechanism relies on a class of cysteine-dependent asparate-specific proteases (caspases) that activate and execute the apoptotic process leading to characteristic morphological changes and ultimately cell death.
While the majority of cytotoxic metallodrugs act through the induction of apoptosis, the alternative pathway of autophagy has recently emerged as an attractive process to effect cytotoxicity [8]. The phenomenon of autophagy was mechanistically unknown prior to the 1990's until seminal work by Ohsumi revealed the systematic activation and identification of genes essential in the overall pathway [9], [10], [11], [12]. Autophagy is considered as an evolutionary-conserved self-digestion, and quality control mechanism where cell survival and degradation processes compete in order to sustain homeostasis and regulate the longevity of proteins, nucleic acids, whole organelles and pathogenic agents [13]. Under the constraints of increased and/or external stress factors, however, elevated accumulation of autophagic vacuoles and organelle elimination renders the cell incapable of normal function and thus results in cell death [14]. Furthermore, autophagy can be induced and up-regulated in response to intracellular reactive oxygen species (ROS) [15], [16], and acts as a protective antioxidant pathway for oxidative stress associated with neurodegenerative diseases [17]. The hormone therapy agent tamoxifen induces elevated levels of intracellular ROS resulting in positive feedback of Zn2+ accumulation, mediating the initiation of autophagy in breast cancer cell line, MCF-7 [18]. Metal complexes of Ru2+, Pt2+, Mn2+ and Cu2+ are also known to activate the autophagic pathway [8], however, with the exception of selected Pt2+-based complexes, the co-activation of apoptosis and autophagy occurs for almost all other metal compounds and is known to be ROS dependent. The current paradigm of autophagy research (in respect to human cancer) is considered to be multifold and somewhat contradictory: i.) it acts as a suppressor toward tumorgenesis [19]; ii.) it is known to promote tumour survival under starvation or hypoxic conditions of low blood supply and other stress factors attributed to tumour stroma [20], [21]; and iii.) the efficient induction of autophagy can be exploited as a pro-death mechanism, particularly in apoptotic defective cancer cells [22].
This group has recently reported the title compound—a Mn2+ bis−1,10-phenanthroline (phen) di-salt complex, bridged with octanedioate (oda) ([Mn2(μ-oda)(phen)4(H2O)2][Mn2(μ-oda)(phen)4(oda)2]·4H2O (Mn-Oda))—in conjunction with a cationic Cu2+ analogue, [Cu2(μ-oda)(phen)4]2+(Cu-Oda) as potent in vitro anticancer agents with toxicity toward a panel of colorectal cancers (HT29, SW480 and SW620) linked to DNA binding and ROS induction [4]. Comparison of the two agents revealed distinctive modes of action as both complexes were found to bind DNA (Kapp ∼105 M(bp−1)) with fluorescent quenching indicating possible intercalation. However, the Mn-Oda complex was shown to powerfully act as both a superoxide dismutase (SOD) and catalase (CAT) mimetic and elicited exceptional levels of endogenous ROS within cancer cells when examined using the intracellular indicator 2′,7′-dichlorofluorescin diacetate. Given the promising results observed for Mn-Oda, coupled with recent interest in the discovery of new autophagic-activating therapeutic leads, this contribution describes the sequential cytotoxic mechanism of this compound leading to autophagy and intrinsic apoptosis, initiated by superoxide (O2•-) production. To unravel these properties, an extensive range of biophysical, molecular biological, and microscopy experiments were undertaken using a variety of dsDNA polymers along with the ovarian solid epithelial cancer cell line SKOV3. Our motivation for using SKOV3 stems from its intrinsic resistance to cisplatin and our experience in using this cell line to identify the mechanistic profile of the pro-apoptotic anticancer lead [Cu(o-phthalate)(phen)] [6]. Thus, in this work we delineate the cytotoxic properties of Mn-Oda with the Cu2+-phen chemotype and reveal the title complex as an inducer of autophagic cell death via ROS-mediated DNA damage. Further, while the complex was identified as an efficient intercalator of dsDNA, we propose intracellular DNA damage is not induced directly by the Mn-Oda complex, but rather by cytoplasmic exogenous stress factors that contribute toward ROS production—notably autophagosome formation, prior to apoptotic activation.
2. Materials and methods
2.1. Materials and reagents
All chemicals used for complex synthesis were purchased from Sigma Aldrich without further purification. The following assays were purchased from Merck Millipore and procedures were followed as per manufacturer protocols: Guava Nexin® Reagent (4500-0450), Guava EasyCyte™ MitoPotential Kit (4500-0250), Guava Caspase 8 FAM and Caspase 9 SR (4500-0640) and Guava Caspase 3/7. Propidium iodide (PI, BTIU40017) was purchased from VWR. RNase A (12091-021), Alexa Flour 488 goat anti-mouse IgG F(ab)2 fragment (A-11020), Alexa flour 488-phalloidin (A12379), DAPI (D1306) and Mitotracker Deep Red (M22426) were purchased from Biosciences Ireland. Anti-phospho-histone H2AX (05−636) was purchased from Merck Millipore. Salmon testes DNA (D1626) and synthetic double stranded alternating co-polymers, Poly[d(G-C)2] (P9389) and Poly[d(A-T)2] (P0883) used in CD studies were purchased from Sigma Aldrich. pUC19 plasmid DNA (N3041), CutSmart® buffer (B7204), 100X BSA (B9000) and topoisomerase I (E. coli) (M0301) were all purchased from New England Biolabs. LC3 isoform LC3A rabbit monoclonal antibody (Cell Signalling) was kindly donated by Dr. Joanne Keenan while goat anti-rabbit conjugated Alex Fluor-647 (ThermoFisher) was donated by Dr. Clair Gallagher.
2.2. Drug-DNA binding interactions
2.2.1. Circular dichroism spectrometry
Complex-DNA interactions were analysed using Starna quartz cuvettes in 10 mM PBS solution (pH 7.0) in the presence of 25 mM NaCl. Solutions of salmon testes DNA (stDNA, ε260 =12824 M(bp)−1 cm−1), Poly[(d(A-T)2] (ε260 =13100 M(bp)−1 cm−1) and Poly[(d(G-C)2] (ε260 =16800 M(bp)−1 cm−1) were initially heat treated to denature and then allowed to slowly renature prior to quantification using an Agilent Cary 100 dual beam spectrophotometer equipped with a 6 ✕ 6 Peltier multicell system with temperature controller, to give working solutions with a final DNA concentration of 100 µM. Spectra were captured in the range of 200–400 nm and measurements were recorded at a rate of 1 nm per second, where Θ = mdeg. DNA solutions were incubated for 30 min periods at 37 °C with Mn-Oda at varying concentration loadings of 1.0%, 1.5%, 2.0% and 2.5%.
2.2.2. Viscosity
Experiments were conducted using DV-II-Programmable Digital Viscometer equipped with Enhanced Brookfield UL Adapter [23]. Briefly, a concentrated solution of salmon testes dsDNA was prepared by dissolving the fibres in 80 mM of HEPES buffer (pH =7.2). In order to shear dsDNA, a 15 ml solution was passed rapidly through a 19-gauge needle 15 times prior to 90 min sonication. A 15 ml stDNA solution was then prepared at ~1.0 mM in 80 mM HEPES buffer and the complex was added in ratios from 0.10 to 0.20 (where r =[compound/DNA]) and viscosity was recorded as previously reported [23]. Viscosity values, η, (unit: cP) were presented as η/ηo versus [compound]/[DNA] ratio, in which ηo refers to the viscosity of DNA alone and η refers to that of the DNA-complex solution.
2.2.3. Topoisomerase I mediated relaxation
Topoisomerase I relaxation was carried out using a modified method of previously reported protocols [24]. 400 ng of pUC19 plasmid DNA was exposed to varying concentrations of drug (0.1−400 µM) for 30 min at room temperature in a final volume of 20 µl containing 80 mM HEPES buffer (pH 7.2), CutSmart® buffer and 100X BSA. Topoisomerase I (1 unit) was added to the mixture and incubated for 15 min at 37 °C to ensure relaxation of plasmid DNA. The enzymatic reaction was quenched with SDS (0.25%), protein kinase (250 μg/ml) and incubated for 30 min at 50 °C. Samples were loaded onto 1.2% agarose gel with 6X loading buffer. Topoisomers of DNA were separated by electrophoresis in 1X TBE buffer at room temperature for 3 h min at 40 V followed by 2.5 h at 50 V. The agarose gel was post-stained using an ethidium bromide bath (25 µM) for 20 min at room temperature. Finally, the gel was soaked in deionised water for 24 h and imaged using a UV transilluminator.
2.3. In cellulo studies
2.3.1. Cell culture
SKOV3 cells were cultured in RPMI-1640 media, supplemented with 10% FBS and incubated at 37 °C in 5% CO2 and routinely tested for mycoplasma. Cells were seeded and left to adhere and enter cell cycle overnight. Mn-Oda stock solutions were prepared in PBS, carbonyl cyanide m-chlorophenylhydrazone (CCCP), rapamycin (Rapa) and suberoylanilide hydroxamic acid (SAHA) solutions in DMSO and doxorubicin (Dox) in 50:50 DMSO:H2O and further diluted in culture media. Stocks containing DMSO were prepared in the mM range to ensure final incubation concentrations contained <0.1% v/v.
2.3.2. Viability
Cells were seeded at 5×103 cells/well in a 96-well plate overnight and subsequently exposed to drug treatment for 24 h (1 μM Mn-Oda, 1 μM Dox, 75 μM CCCP, 50 μM Rapa and 100 μM SAHA, Fig. S3). For co-treatment experiments, antioxidants and autophagy inhibitors tiron, D-mannitol (Man), histidine (His), sodium pyruvate (Py), 3-methyladenine (3-MA) and NH4Cl, chloroquine (CQ) were treated at 1 mM (with exception of CQ at 10 μM) 2 h prior to drug exposure to facilitate intracellular accumulation. Spent media was removed, cells were detached with trypsin: EDTA (0.25%:0.02% in PBS) and whole samples (100 µl) were added to 100 µl Guava Viacount reagent and incubated for 10 min at room temperature. Viability was acquired on Guava EasyCyte HT with Viacount software.
2.3.3. Nexin® assay
SKOV3 cells were seeded at 3×104 cells/well in 24-well plates overnight and incubated with drug containing media. After the exposure period, spent media was removed, cells were washed with PBS and washings were kept in 1.5 ml microtubes. Subsequently, cells were detached with trypsin and neutralised with fresh media. PBS washings were transferred back into microtubes containing detached cells and centrifuged at 1300 rpm for 5 min and culture media was aspirated. A sufficient volume of media was added to give a concentration range of 2×105 to 1×106 cell/ml. 100 µl of sample was transferred to 96-well round bottom plate and 100 µl of Guava Nexin® Reagent was added and incubated for 20 min at room temperature. Samples were acquired on Guava EasyCyte HT using Nexin software. Innate Dox fluorescence in filter 583/26 nm was accounted for and subtracted within the relevant quadrants.
2.3.4. Mitochondrial membrane potential
Cells were treated as previously described. Cells were resuspended in 600 µl of fresh media to give cell concentration 2×104 to 5×105 cells/ml from which 200 µl was transferred to a 96-well round bottom plate. 50X staining solution (4 µl) containing JC-1 and 7-aminoactinomycin D (7-AAD) was added to each sample, subsequently incubated at 37 °C in darkness (30 min) and acquired on Guava EasyCyte HT using MitoPotential software. Compensation to correct fluorescent overlap between filters was conducted pre-acquisition.
2.3.5. Caspase 8 FAM and 9 SR, Caspase 3/7 FAM
The following fluorescent labelled inhibitors of caspase (FLICA) were used to ascertain the activation of Caspase 3/7, 8 and 9 respectively; FAM-DEVD-FMK, FAM-LETD-FMK and SR-LEHD-FMK. Samples were prepared prior to staining in a similar manner as that described in the Nexin Assay. Cells were transferred to 1.5 ml microtubes and resuspended in 100 µl media. 10 µl of 10X caspase 9 SR working solution and 10 µl of caspase 8 FAM were added and incubated for 1 h at 37 °C in the dark. Cells were resuspended in 200 µl of caspase 7-AAD working solution and transferred to 96-well round bottom plate, which was left to incubate for 10 min at room temperature. Samples were acquired on Guava EasyCyte HT flow cytometer using Guava Caspase software. Compensation to correct fluorescent overlap between filters was conducted pre-acquisition and innate doxorubicin fluorescence in filter 583/26 nm was accounted for and subtracted within the relevant quadrants.
2.3.6. Cell cycle analysis
Cellular DNA content was conducted in a similar manner to previously reported methods [25]. 3×104 cell/well were seeded in 24-well culture plates, drug treated and collected as previously stated. After trypsinization, cells were fixed in 70% ice-cold EtOH and stored at −20 °C (>12 h). Samples were stained with 200 µl PI staining solution (containing 50 μg/ml PI, 100 μg/ml DNase-free RNase A, 0.1% triton X-100 in 1X PBS) (30 min, room temperature). Samples were acquired on Guava EasyCyte HT flow cytometer and normalised to the sum of events in G0/G1, S and G2/M phases.
2.3.7. Immunodetection of γH2AX
Samples were prepared similarly to previously reported methods [26]. 6×104 cells/well were seeded in 12-well plates and treated for 24 h with compounds and fixed with 1.5% formaldehyde (15 min, room temperature) followed by ice-cold 70% ethanol and stored at −20 °C. Samples were resuspended in permeabilisation buffer (0.25% Triton X-100 in PBS) for 30 min on ice and blocked with 2% BSA (30 min, room temperature). Primary antibody (1:500) was incubated for 2 h at room temperature followed by secondary antibody (1:1000) for 1 h at room temperature and co-stained for 10 min with 5 μg/ml propidium iodide. Samples were acquired using ExpressPro software on Guava EasyCyte.
2.3.8. DNA degradation with COMET assay
Cells were seeded at 1.5×105 cells/well in 6-well plates the evening prior to drug addition. After 24 h drug incubation, cells were harvested and 50 µl was resuspended in 500 µl low melting point agarose to give a final density of 1.5×105 cell/ml. 50 µl was spread onto Trevigen COMET slides and allowed to solidify (1 h, 4 °C). Samples were lysed (2.5 M NaCl, 100 mM EDTA, 10 mM Tris. HCl, pH 10) overnight at 4 °C. Slides were allowed to equilibrate in cold electrophoresis buffer (300 mM NaOH, 1.0 mM EDTA, pH 13) for 30 mins at 4 °C. Electrophoresis was run at 300 mA for 30 min with buffer levels adjusted to give a consistent voltage of 25 V. Slides were then washed with water, neutralised in buffer (400 mM Tris. HCl, pH 7.5) (3×5 min), fixed with 70% ethanol (30 min) and dried for desiccated storage (15 min at 37 °C). Prior to scoring, slides where rehydrated for 15 min before staining with propidium iodide solution (10 μg/ml, 10 min room temperature) and imaged through a 10X lens on a Leica DFC 500 epi-fluorescent microscope. Images where then analysed using Open COMET plugin in Image J and plotted using GraphPad Prism software.
2.4. Superoxide detection
SKOV3 cells (6×104) were seeded in 12-well plates and exposed to 1.0 μM of drug for 2, 6, 18 and 24 h. After drug treatment, samples were harvested, washed with PBS, resuspended in 200 µl of 5.0 μM MitoSOX Red or dihyrdoethidium (DHE) and left to incubate for 15 min at 37 °C. Samples where then resuspended in PBS and transferred to 96-well round bottom plates, and acquired on Guava EasyCyte flow cytometer using ExpressPro software.
2.5. Acquisition of confocal images
2.5.1. Cell morphology
Cells (1.4×105) were seeded in 35 mm glass bottom petri dishes and allowed to attach overnight. Samples were then exposed to drugs for 24 h (1 μM Mn-Oda, Rapa 50 μM and SAHA 100 μM). Cells were incubated with media containing MitoTracker Deep Red (150 nM, 30 min, 37 °C) and fixed with 4% paraformaldehyde (PFA) (30 min, room temperature). Samples were permeabilised with 0.25% Triton X-100 (15 min). To avoid non-specific staining, cells were blocked with 1% BSA in PBS solution (30 min, 37 °C) and subsequently stained with Alexa Flour 488-phalloidin (10U/100ul, 30 min), followed by DAPI (4:10,000, 10 min) and mounted in ProLong Gold. Images were acquired on STED-Leica DMi8 confocal microscope equipped with CCD camera and 100X oil-immersion objective. DAPI was excited with 405 nm picoquant laser unit and emission captured between 387−474 nm. Alexa Fluor 488 was excited at 499 nm with emission captured between 490−566 nm, and MitoTracker Deep Red was excited at 653 nm where emission was captured at 658−779 nm. Images were acquired where by combinations of excitation wavelengths and emission filters for specific dyes are applied sequentially.
2.5.2. Autophagic detection with LC3
SKOV-3 cells were seeded as previously described. Samples were fixed with 4% PFA (30 min, room temperature), permeabilised with 0.25% Triton X-100 (30 mins, 4 °C) and blocked with 2% BSA (30 min, room temperature). Cells were incubated with primary antibody (1:500, overnight at 4 °C), secondary antibody goat anti-rabbit Alexa Fluor 647 (1:1000, 1 h at room temperature) and DAPI (4:10,000, 10 min at room temperature) and mounted with ProLong Gold. Images were acquired on STED-Leica DMi8 confocal microscope with 100X lens. Alex Fluor 647 was excited with 653 nm laser and emission captured between 658–783 nm, while DAPI was excited with 405 nm picoquant laser unit and emission captured between 435–560 nm.
2.5.3. Autophagic detection with MDC
Cells were seeded in glass bottom petri dishes as previously described. Following drug exposure, samples were incubated with monodansylcadaverine (MDC) (50 μM, 37 °C, 10 min), washed with PBS and confocal images were immediately acquired on STED-Leica DMi8 confocal microscope equipped with 100X objective. Samples were excited with 405 nm picoquant laser unit and emission captured 470–560 mm. Intensity profiles were analysed using Image J V2.0 on raw images in 8-bit format without further modification. MDC coloured images were enhanced in Adobe Photoshop for printing purposes only.
2.5.4. Statistical analysis
All in cellulo data are presented as mean ± standard deviation where n =3. Unpaired t-tests with Holm-Sidak method were applied to evaluate statistical significance in GraphPad Prism V6 for all data with the exception of superoxide detection studies and viability results in the presence of autophagy inhibitors and antioxidants, where two-way ANOVAs with post-hoc analysis (Dunnett's test) were applied. p≤0.05 were considered to be statistically significant.
3. Results and discussion
3.1. Preparation and structure of Mn-Oda
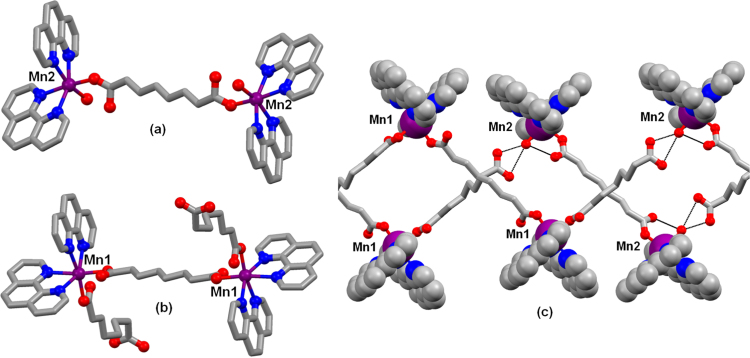
The complex [Mn2(η1η1μ2-oda)(phen)4(H2O)2][Mn2(η1η1μ2-oda)(phen)4(η1-oda)2]·4H2O was prepared according to the method reported by Casey et al. [27] and contains [Mn2(η1η1μ2-oda)(phen)4(H2O)2]2+ cations and [Mn2(η1η1η2-oda)(phen)4(η1-oda)2]2- anions (both of which are centrosymmetric) along with four solvate water molecules. All of the manganese centers are 6-coordinate with two bidentate phen ligands and two oxygen donors in the cis positions. In both complex ions two manganese centers are linked by an oda2- ligand; the sixth site is occupied by water in the cation and by further oda2- groups in the anion (Fig. 1 A and B). Hydrogen bonding between the coordinated water molecule and the uncoordinated carboxylate groups of the anion link the complex ions into zig-zag chains and the chains are linked together by further hydrogen bonding involving the uncoordinated water molecules, generating a 3D network (Fig. 1C).
Fig. 1.
A. The cation [Mn2(η1η1μ2-oda)(phen)4(H2O)2]2+, B. anion [Mn2(η1η1μ2-oda)(phen)4(η1-oda)2]2-, and C. hydrogen bonded chains. Hydrogen atoms omitted for clarity, hydrogen bonds indicated by black dashed lines. Redrawn from coordinates taken from reference 27. Colour key: C (grey), O (red), N (blue) and Mn (purple). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. In vitro Drug-DNA interactions reveal intercalation at the minor groove
4.1. Topoisomerase I mediated relaxation
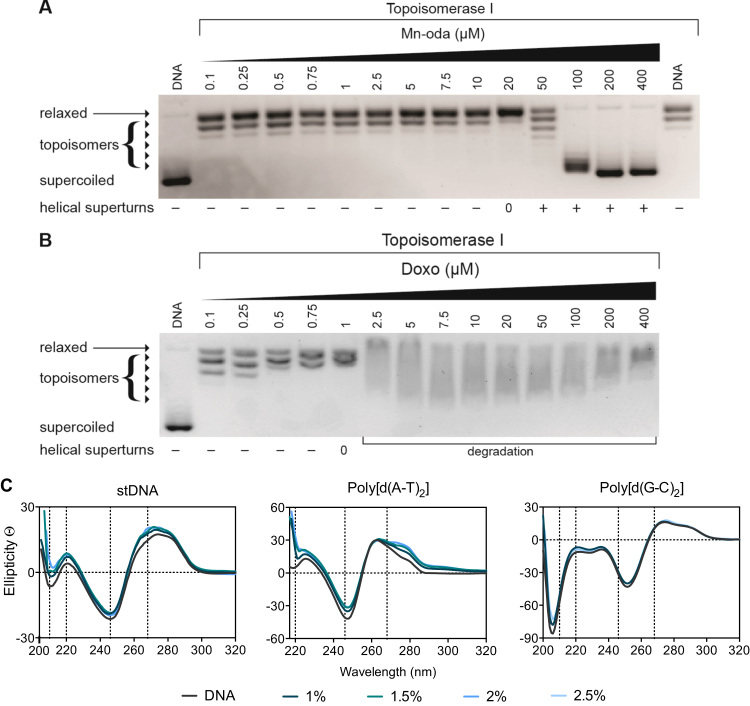
Topoisomerases (Topo) are a specialised class of nuclear enzymes that catalyse the transient cleavage, passage and resealing of either a single strand (topo I) or double strands (topo II) of DNA in order to relax chain intertwinement, release superhelical tension and permit change in topology during replication, transcription and recombination [28], [29]. Topo I, isolated from E. coli, specifically relaxes negatively coiled superhelical plasmid DNA (scDNA) [30], such as the pUC19 substrate used in this study. Topo I mediated relaxation of pUC19 was identified with increasing Mn-Oda (Fig. 2A). The complex was found to completely relax scDNA at 20 μM with positively wound topology of intact scDNA being observed thereafter. This profile is comparable to that of classical intercalating molecules such as ethidium bromide (Fig. S1) which unwinds DNA by 26° yielding similar topoisomeric patterns [31]. Doxorubicin (Dox), a clinically used Topo II poison [32], exhibits enzymatic inhibitory effects on the Topo I with DNA relaxation occurring at 1 μM (Fig. 2B). Samples treated with Dox extending this concentration render DNA degradation and shearing most likely through ROS generation consequent to redox cycling of the quinone moiety [33], [34], [35], and these results in agreement with those described elsewhere [36]. Thus, the Mn-Oda complex is capable of unwinding dsDNA via intercalation but does not induce DNA damage or poison topoisomerase I during this process.
Fig. 2.
A. Release of topological tension of supercoiled plasmid DNA by Mn-Oda and, B. Dox. C. CD profile of Mn-Oda with stDNA and alternating co-polymers poly[d(A-T)2] and poly[d(G-C)2] at drug loadings of 1.0 – 2.5% (respective r values of 0.010 – 0.025).
4.2. Circular dichroism studies
Circular dichroism (CD) spectroscopy is a powerful biophysical technique used to monitor conformational changes, drug-DNA binding interactions and structural dynamics of nucleic acids. The CD profile of classical right handed B-DNA exhibits two positive (220 nm and 268 nm) and two negative (210 nm and 246 nm) elliptical signals, while slight variations in this profile arise when the %A-T content of DNA is varied [37]. Conformations of salmon testes DNA (stDNA) and synthetic alternating copolymers poly[d(A-T)2] and poly[d(G-C)2] were studied with increasing r ([drug]/[DNA]) values, where r =0.010 – 0.025 (experiments containing >0.025 Mn-Oda were found to induce noise in the resulting spectra). Mn-Oda exhibits a concentration-dependent enhancement of the elliptical signals that can be attributed to hydrogen bonding and stacking interactions between nitrogenous bases and the right-handedness of DNA, irrespective of A-T content (Fig. 2C). In the case of stDNA and poly[d(G-C)2], an increase in ellipticity associated with β-N-glycosidic linkages were also noted. Mn-Oda-DNA profiles were compared to that of classical non-covalent intercalating and groove binding molecules (data not shown) with structural perturbations induced by Mn-Oda suggesting an intercalative binding mode (268 nm), particularly at the minor groove, similar to that of EtBr since elliptical signals show an increasing trend at 210, 220 and 268 nm [38]. Additional viscosity analysis further corroborated an intercalating binding motif by Mn-Oda on stDNA (Fig. S2).
5. Genotoxicity studies imply indirect DNA damage
5.1. COMET analysis
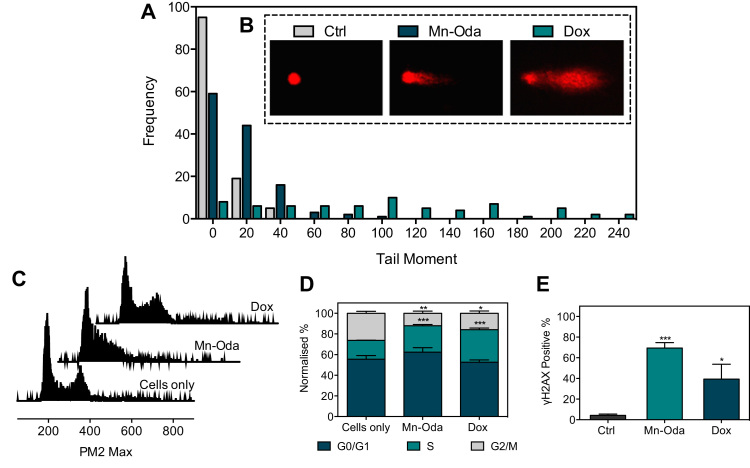
Single cell gel electrophoresis, otherwise known as the COMET assay, was employed to determine intracellular DNA damaging properties of the metal complex. Prior to analysis, viability profiles of Mn-Oda and Dox were identified over 24 h of exposure within the SKOV3 cell line using flow cytometry (Fig. S3). SKOV3 cells were then exposed to 1.0 μM concentrations of both agents, embedded onto agarose coated glass slides and lysed of cellular structure and nucleosome resulting in the nucleoid scaffold that allows DNA migration based on integrity, when subjected to alkaline gel electrophoresis. Single strand breaks (SSB) and double strand breaks (DSBs) were visualised through fluorescence microscopy (Fig. 3B). We selected the clinical anti-tumour antibiotic Dox as a positive control due to its DNA intercalating capacity, ROS generation and topoisomerase poisoning effects. The frequency distribution of the COMET tail moment induced by Mn-Oda (Fig. 3A) showed a departure from the control profile, with a higher number of events occurring between tail moments of 40 and 20 (A.U.). Dox, however, exhibited a dispersed array of tail moments with values reaching 240 a.u. and an elongated tail, extending residual DNA damage and reduced fluorescence intensity of the comet head. These results are in agreement with Manjanatha et al. who reported Dox-induced ROS could generate both direct and indirect DNA damage as indicated by the COMET assay [39].
Fig. 3.
SKOV3 cells were treated with 1.0 μM Mn-Oda and Dox for 24 h and subsequently studied in the following assays. A. Comet assay analysis where the frequency of tail moment (A.U.). B. Examples of typical COMET shapes are represented below respective legends. C. Cell cycle histograms and, D. Cell cycle phase (G0/G1, S and G2/M) distributions. E. Immunodetection of γH2AX positive cells. Not significant p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001.
5.2. Cell cycle analysis
In order to investigate the toxicity mechanism of the di-Mn2+ complex, the effects on SKOV3 cell cycle phase distributions were examined. All cells have an innate growth and replication cycle, the revolution of which yields cell growth (G0/G1), replication of chromosomes (S) and production of daughter cells (G2/M) [40]. As shown in Figs. 3C and 3D, Mn-Oda induces a decrease (14.3%) in the G2/M phase and enrichment within the S (synthesis) phase (7.4%) when compared to the untreated control. A contraction of G2/M phase was also evident in Dox treated samples (10.4%) with escalation of S phase (13.3%) identified. Taken together, data here indicates that at 1.0 μM exposure over 24 h, both tested agents induce cell cycle arrest within SKOV3 in the DNA synthesis phase.
5.3. Immunodetection of γH2AX
A primary response to dsDNA damage is the site selective phosphorylation of histone H2AX that is indiscriminately incorporated during chromatin formation [41], [42]. H2AX differs from other H2A histones through a carboxyl tail containing a 139-serine residue that becomes phosphorylated in the presence of DNA damage and denoted as γH2AX [43]. Phosphorylated H2AX accumulate in the chromatin surrounding the site of damage, thus creating a focus for subsequent recruitment of DNA repair mechanisms. A proportional correlation is observed between the extent of DNA damage and formation of γH2AX foci thus rendering it as a pertinent method for dsDNA damage detection. Following the advent of phosphorylation, the use of recognition antibodies for γH2AX can visualise and quantify this process through fluorogenic conjugation. Immunodetection of γH2AX within SKOV3 cells was quantified using flow cytometry after 24 h of exposure to Mn-Oda and Dox (1.0 μM) (Fig. 3E). Mn-Oda was found to induce remarkably high levels of DSBs (69.4%) in the cellular population. The control agent Dox was also efficient in DSB generation within SKOV3, but results here were notably lower (39.4%).
6. Mn-Oda stimulates mild caspase 9 release but does not trigger early or late-stage apoptosis in SKOV3 cells
6.1. Annexin V
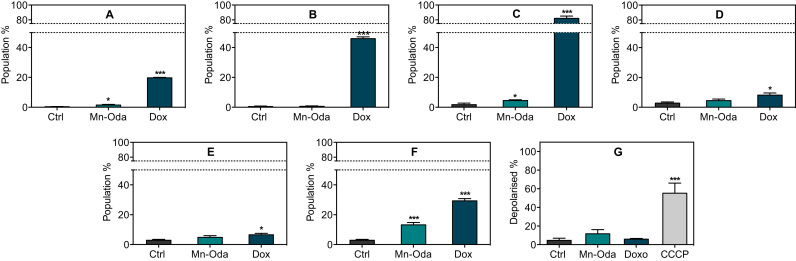
To determine whether Mn-Oda induces apoptotic cell death, a number of critical biomarkers were investigated (Fig. 4). To probe this potential pathway, SKOV3 cells were again exposed to Mn-Oda and Dox (1.0 μM) over 24 h and activation of apoptosis was distinguished from early and late stages through the detection of Annexin V and membrane-impermeable 7-AAD. During apoptosis, membrane-bound protein phosphotadylserine translocates from the inner to outer surface of the cellular membrane and renders the negatively charged phospholipid as an accessible substrate for Annexin V [44], [45]. Interestingly, in these experiments, Mn-Oda induced minimal levels of Annexin V in the early stage of apoptosis (1.5%) with none detectable in the later stages of the 24 h time frame (Fig. 4A and B). This effect contrasts with Dox-treated SKOV3 cells, which induced 19.7% and 45.9% at these respective stages.
Fig. 4.
Apoptotic investigation of Mn-Oda, A. Early and B. late apoptosis measured by the translocation of Annexin V substrate. C. Detection of caspase 3/7 in mid and D. late populations. E. Activation of initiator caspase 8 and F. caspase 9. G. Extent of mitochondrial depolarisation detected through bathochromic shift of JC-1 emission upon formation of J-aggregates. Not significant p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001.
6.2. Caspase activation
In order to confirm Mn-Oda cytotoxicity is provoked through a non-apoptotic cytotoxic pathway, a range of essential components in the form of caspases that contribute to the initiation and execution of this process, were investigated (Fig. 4C-F). Stimulation of the intrinsic apoptotic pathway results in cytosolic release of cytochrome c and thus activation of caspase 9 within the apoptosome [46], [47]. An alternative route of origin also exists through extracellular death factors that stimulate the activation of caspase 8 [48], [49]. Both caspase 8 and 9 result in the consequential activation of executioner caspases such as 3 and 7 that commence an irreversible cascade of proteolytic degradation and membrane collapse, precipitating in programmed cell death [46], [47]. Mn-Oda treated cells did not promote sufficient production of initiator caspase 8 (Fig. 4E) but did activate caspase 9 by 13.2% (Fig. 4F). However, a marginal increase only (4.4%) was observed in early apoptotic detection of executioner caspases 3/7 (Fig. 4C), but not in the latter stages (Fig. 4D). As expected, Dox had a significant effect on the activation of caspase 3/7 (Figure C and D) with activation, and subsequent apoptosis (Fig. 4A and B) originating via the intrinsic pathway (Fig. 4F).
6.3. Mitochondrial depolarisation
Due to the ability of Mn-Oda to generate intracellular ROS [4], changes in mitochondrial transmembrane potential (ΔΨm) were investigated; reduction of the redox potential across inner and outer mitochondrial membranes is also a characteristic of apoptotic induction. Depolarisation measurements were obtained using fluorogenic dye JC-I. The protonophore, and known uncoupler of ΔΨm, carbonyl cyanide m-chlorophenyl hydrazine (CCCP) [29], [30], was employed as a positive control and found to depolarise 55.3% of the sample population while Mn-Oda induced 11.7% depolarisation—a marginal increase in comparison to Dox-treated and non-drug treated cells (6.0% and 4.6% respectively) (Fig. 4G). Rather than Mn-Oda directly influencing deterioration of transmembrane potential, the extent of depolarisation is most likely due to the stimulation of intrinsic apoptosis. Fluorescent quantification of these apoptotic biomarkers, in combination with cytosolic caspases and Annexin V, suggests that Mn-Oda does not directly activate apoptosis as the primary mechanism of cell death to account for the 46% decrease in cellular viability (Fig. S3).
6.4. Mn-Oda stimulates autophagy prior to apoptosis
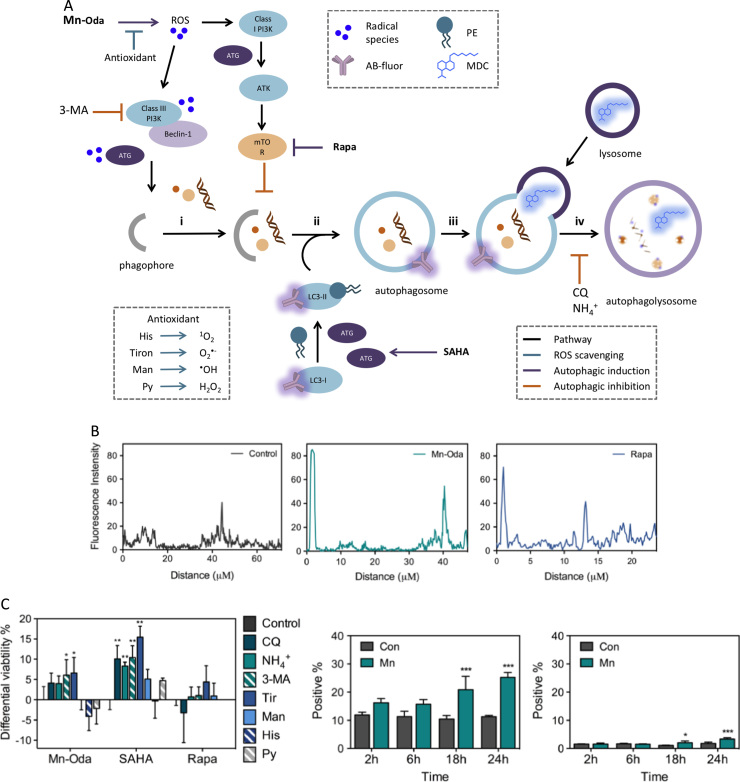
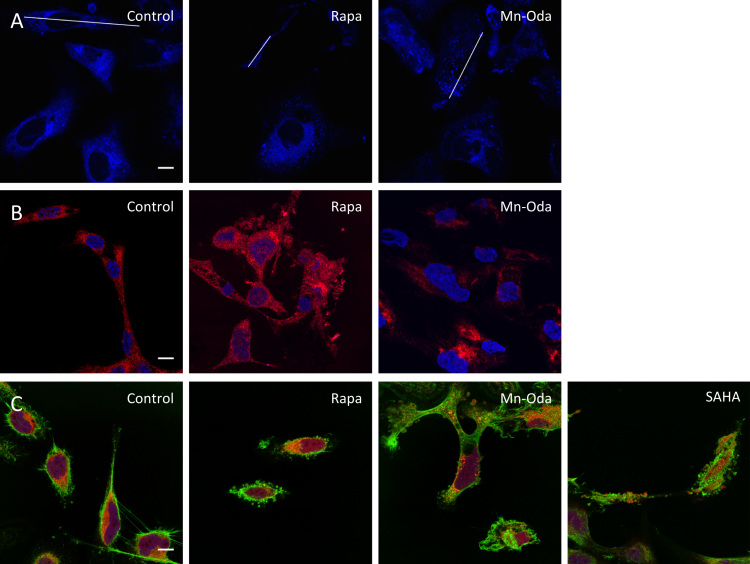
Owing to low levels of caspase 9, the extent of mitochondrial depolarisation and lack of Annexin V and caspase 3/7 detection, there is limited evidence here to suggest that Mn-Oda directly induces cell death via apoptosis. It is likely, therefore, that apoptotic initiation is a downstream effect mediated by an alternative mechanism and this prompted us to investigate the activation of autophagy triggered by Mn-Oda. The autophagy pathway is a lysosomal degradation process, monitoring the homeostasis, longevity, turnover of biomolecules and organelles, while replenishing the nutrient pool particularly under starvation conditions [50]. This complex pathway consists of sequential stages from initiation, nucleation, elongation and finally maturation through the activation and post-transcriptional modification of autophagy related proteins (ATG) (Fig. 6A) [15], [20], [50]. In order to evaluate the mechanistic pathway for Mn-Oda induced toxicity, fluorescent staining with monodansylcadervine (MDC) was primarily monitored. MDC contains a fluorogenic dansyl moiety conjugated to a terminal amine that facilities accumulation and ion trapping in low pH environments such as those found within autophagolysosomes [51]. Autophagy inducers employed in this study were: i.) rapamycin (Rapa), which inhibits mammalian target of rapamycin (mTOR) complex 1 [52], [53], a downstream protein involved in the PI3K-AKT-mTOR regulation pathway, and ii.) suberoyanilide hydroxamic acid (SAHA), an inhibitor of histone deacteylase (HDAC) that induces transcriptional expression of LC3 and mTOR activation [54], independent to apoptosis (Fig. 6A). Non-treated cells demonstrate minimal fluorescence emission of MDC from innately present lysosomes (Fig. 5A). Accumulation of MDC in spherical structures was observed upon Mn-Oda and Rapa treatment with localisation observed in the perinuclear region. Upon complex treatment, quantification of fluorescent intensities (Fig. 6B) revealed enhanced emission profiles due to accumulation of MDC in autophagolysosome when compared to non-treated SKOV3 cells, indicating the induction of Mn-Oda-mediated autophagy. Given that MDC specificity for selective accumulation in autophagic vacuoles has been debated [55], further evidence of Mn-Oda triggered autophagy was identified through aggregation of the autophagic marker LC3. Cytosolic LC3 (LC3-I) is proteolytically cleaved by Atg4, converted to its lipidated form (LC3-II) upon phosphatidylethanolamine (PE) binding [56], and incorporated into the autophagosomal membrane [15]. Non-discriminate immunodetection of LC-3 showed dispersion within the cytosol in the non-treated control (Fig. 5B). Autophagosome and consecutive autophagic vacuole formation is evident in Rapa and Mn-Oda treatments with the identification of LC3-II punta, supporting that the cytotoxic mechanism of Mn-Oda is attributed to autophagy activation.
Fig. 6.
A. Schematic of autophagy pathway [15], [20], [50]. Due to the number of autophagy-related genes (ATGs) and the complexity of their role in the autophagy pathway, the family is represented by ‘ATG’ (purple) for simplicity, i. Initiation begins with the formation of isolation membrane known as a phagophore, engulfing cytoplasmic material, ii. Cytosolic LC3-I is converted to the membrane-associated form LC3-II, through phosphatiylethanolamine (PE) lipidation and incorporated into autophagosomal double-membrane, iii. Docking and fusion of lysosome or late endosomes results in the formation of iv. autophagolysosome. Maturation and catabolic degradation results in recycling and restoration of nutrient stores. Autophagic inducers employed are Rapa and SAHA (purple pathway). Autophagic inhibitors are 3-methyladenine (3-MA), NH4+ and chloroquine (CQ) (orange pathway) while antioxidants utilised are tiron, mannitol (man), histidine (his) and sodium pyruvate (Py) (teal pathway). Induction was probed through immunodetection of LC3 with fluorogenically conjugated secondary antibody and monodansylcadervine (MDC), B. Intensity profiles (indicated in Fig. 5A) were analysed using Image J on raw images in 8-bit format with no further modification. C. Differential viability percentages in the presence of radical scavengers and autophagy inhibitors pre-treated at 1 mM (with the exception of CQ, 10 μM) 2 h prior to drug addition. Mn-Oda, SAHA and Rapa were treated at respective concentrations, 1 μM, 100 μM and 50 μM, over 24 h. D. Percentage of cellular population positive for MitoSOX Red and E. Dihydroehtidium (DHE) fluorescence at 1.0 μM Mn-Oda incubated for 2, 6, 18 and 24 h. Not significant p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
Confocal images of SKOV3 100X treated with Mn-Oda, Rapa or SAHA to examine, A. MDC staining of acidic vacuoles (blue). White lines indicate cross sections for fluorescence quantification profiles (see Fig. 6B). B. Immunofluorescent staining of LC3 (red) and DAPI (blue) control. C. Morphological changes in cellular structure. Nuclei were stained with DAPI (blue), F-actin with Alexa Fluor 488 conjugated to phalloidin (green) and mitochondria with MitoTracker deep red (red). All scale bars (control, bottom right) are 10 µm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
6.5. Cell morphology reveals Mn-Oda-promoted apoptosis
Confocal microscopy experiments were undertaken to further identify the cytotoxic effects of Mn-Oda within SKOV3 cells through the use of location-specific fluorogenic stains to visualise the nucleus, actin cytoskeleton and mitochondria (Fig. 5C). Untreated SKOV3 cells exhibited adherent epithelial morphology with an elongated dome shape. Distinct changes in cellular structure were observed in Rapa treated cells such as adherence contraction, enhanced elongation and cytoskeletal actin encapsulation absent of nuclear or mitochondrial debris. Cells exposed to SAHA exhibit similar contraction and elongation, with clear evidence of apoptotic bodies. Mn-Oda treatment, however, presents alterations in cell morphology and characteristic indicators of apoptosis, particularly with enlargement and fragmentation of the nucleus, organelle contraction, dynamic membrane blebbing and formation of apoptotic bodies [57]. In conjunction with the flow-cytometric detection of caspase 9 and the loss of mitochondrial transmembrane potential, evidence here suggests Mn-Oda induces intrinsic apoptosis as the concluding mechanism of cell death, which is preceded by the activation of autophagy.
6.6. Initiation of autophagy is superoxide-dependent
To investigate the role of Mn-Oda in the activation of autophagy, we further probed cellular viability in the presence of antioxidants and autophagy inhibitors (Fig. 6C). Inhibitors of autophagy are 3-methyladenine (3-MA), which competitively binds to class III phosphatidylinositol 3-kinase (PI3K) [58], [59], and antimalarial agent chloroquine (CQ) which inhibits lysosomal protease degradation through accumulation of the weak base within acidic vacuoles, thus inhibiting autophagolyosome formation (Fig. 6A) [60], [61]. Under physiological conditions ammonium chloride (NH4Cl) becomes protonated, acting as a lysosomotropic agent with similar neutralisation capacity to chloroquine, and causes an increase in local pH [62]. Due to the ROS generation properties of Mn-Oda [4], antioxidants employed in this study consist of a variety of radical-specific trapping agents; tiron for superoxide (O2•−) [63], mannitol for hydroxyl radical (•OH) [64], histidine for singlet oxygen (1O2) [65], [66], and sodium pyruvate for hydrogen peroxide (H2O2) [67]. Upon co-treatment with Mn-Oda, all autophagy inhibitors increased cellular viability with 3-MA and tiron at the highest extent (6.1% and 6.6% respectively). These results in conjunction with fluorescent detection of autophagic vacuoles, suggest that Mn-Oda promotes superoxide-mediated autophagy as hypothesised in Fig. 6A. The combination of SAHA with CQ, NH4Cl and 3-MA significantly increased cell survival by 10.1%, 8.3% and 10.4%, respectively. Interestingly, the most substantial increase was noted for O2•− scavenging agent tiron (15.4%) as SAHA has previously demonstrated significant ROS generation properties [68], [69], which can be attributed to down-regulation of thioredoxin (TRX) a dithiol-reducing redox protein [70], [71], a key response to oxidative stress. The presence of autophagy inhibitors had minimal effect on Rapa viability, which could be due to the limited exposure period of 24 h; rapamycin typically exerts toxic effects within longer time-frames with IC50 concentrations of 25.3 μM in PEO1 ovarian cancer cells after 72 h [72]. Sequestering the basal levels of O2•− enhanced live cell populations by 4.4%, most likely as O2•− is known to inhibit the binding of TORC1 to Rapa: FKBP12 in yeast [73]. Furthermore, although rapamycin can innately generate significant levels of ROS, synergistic co-treatment with a ROS liberating co-factor, such as curcumin analogue (EF24), can lead to excessive production and enhanced cell death [74]. To further probe the subcellular site of Mn-Oda promoted superoxide generation, we employed dihydroethidium (DHE) and the mitochondrial targeting conjugate, MitoSOX Red. Both dyes specifically become oxidised by superoxide [75], resulting in fluorescence upon DNA intercalation (610 nm in chromatin and 580 nm in mitochondria). Over a time-course between 2 and 24 h at 1.0 μM Mn-Oda exposure, intracellular populations demonstrate increasing and selective liberation of O2•− within the mitochondria (Fig. 6D) with 14.0% elevation noted at 24 h. Significantly, no appreciable superoxide was detected within the nucleus (Fig. 6E) with only 1.6% increase at the longest incubation period.
7. Conclusion
Reactive oxygen species (ROS) play an integral role in the regulation and stimulation of autophagy [76], [77]. Previous work by the Gibson group highlighted the fundamental requirement for ROS-mediated autophagic induction under specific starvation conditions leading to the activation of superoxide (O2•−) production, either alone or in combination with hydrogen peroxide (H2O2) [78]. Furthermore, the same study revealed over-expression of the antioxidant enzyme superoxide dismutase could inhibit the activation of this pathway, resulting in catalytic depletion of O2•− to H2O2. The findings of the current study demonstrate clearly that Mn-Oda can intercalate dsDNA at the minor groove, however, unlike doxorubicin or Cu2+-phenanthroline derivatives [4], direct oxidative damage of nucleic acids is abrogated. Instead, there is strong evidence to suggest that DSBs, identified by immunodetection of γH2AX, coupled with nuclear fragmentation observed in the COMET assay, are due to autophagic degradation initiated by complex-mediated intracellular ROS production. Combination of the di-Mn2+ complex along with mediators that hinder radical generation and autophagy, indicate cell death promoted by O2•− production within the mitochondria. Metal-catalysed radical production does not directly influence nucleic acid degradation (evident from the absence of DNA shearing in topoisomerase relaxation), but rather functions as a signalling agent in the activation of autophagy, detected via MDC and immunofluorescence of LC3. Consequentially, intracellular ROS insult by Mn-Oda promotes autophagy, exceeding a critical threshold of adverse conditions, and activates apoptosis via the intrinsic, mitochondrial pathway (caspase 9 and mitochondrial depolarisation) as the lethal effector of cellular death. Further sensitivity may arise from the mutational status of p53 whereby depletion or point mutations can induce autophagy; in the cytosolic setting, baseline levels of p53 can inhibit autophagy under various stress factors such therapeutic, nutrient or endoplasmic reticulum strains [79]. With this in mind, cancers that express mutant or null p53—such as SKOV3 [80]—are more susceptible to autophagy. To our knowledge, Mn-Oda belongs to one of the few transition metal complexes to activate cell death in this sequential manner, where apoptosis is activated only in the final stages of cytotoxicity. In the wider context of metallodrug development and targeting, this complex may serve as a significant milestone in the construction of small molecule therapeutic leads that promotes alternative cytotoxic mechanisms that are not dependent on traditional apoptotic initiation.
Competing interests
Authors declare there are no conflicts of interest.
Acknowledgment
This work was supported by the Irish Research Council (IRC) grants GOIPG/2014/1182 and GOIPG/2013/826. Equipment used in this study was funded under the Programme for Research in Third Level Institutions (PRTLI) Cycle 5. The PRTLI is co-funded through the European Regional Development Fund (ERDF), part of the European Union Structural Funds Programme 2011–2015. A.K. acknowledges funding from the Marie Skłodowska-Curie Innovative Training Network (ITN) ClickGene (H2020-MSCA-ITN-2014−642023). The authors thank Dr. J. Keenan and Dr. C. Gallagher for donating antibodies used in the immunodetection studies and Prof. M. Devereux and Dr. M. McCann for valuable and constructive scientific discussion.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.01.024.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Finney L.A., O'Halloran T.V. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science. 2003;300(5621):931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- 2.Sarkar B. Treatment of Wilson and menkes diseases. Chem. Rev. 1999;99(9):2535–2544. doi: 10.1021/cr980446m. [DOI] [PubMed] [Google Scholar]
- 3.Molphy Z., Prisecaru A., Slator C., Barron N., McCann M., Colleran J., Chandran D., Gathergood N., Kellett A. Copper phenanthrene oxidative chemical nucleases. Inorg. Chem. 2014;53(10):5392–5404. doi: 10.1021/ic500914j. [DOI] [PubMed] [Google Scholar]
- 4.Kellett A., O'Connor M., McCann M., Howe O., Casey A., McCarron P., Kavanagh K., McNamara M., Kennedy S., May D.D., Skell P.S., O'Shea D., Devereux M. Water-soluble Bis(1,10-phenanthroline) octanedioate Cu2+ and Mn2+ complexes with unprecedented nano and picomolar in vitro cytotoxicity: promising leads for chemotherapeutic drug development. Med. Chem. Commun. 2011;2(7):579–584. [Google Scholar]
- 5.Kellett A., O'Connor M., McCann M., McNamara M., Lynch P., Rosair G., McKee V., Creaven B., Walsh M., McClean S., Foltyn A., O'Shea D., Howe O., Devereux M. Bis-phenanthroline copper(ii) phthalate complexes are potent in vitro antitumour agents with “self-activating” metallo-nuclease and DNA binding properties. Dalton Trans. 2011;40(5):1024–1027. doi: 10.1039/c0dt01607a. [DOI] [PubMed] [Google Scholar]
- 6.Slator C., Barron N., Howe O., Kellett A. [Cu(O-phthalate)(phenanthroline)] exhibits unique superoxide-mediated NCI-60 chemotherapeutic action through genomic DNA damage and mitochondrial dysfunction. ACS Chem. Biol. 2016;11(1):159–171. doi: 10.1021/acschembio.5b00513. [DOI] [PubMed] [Google Scholar]
- 7.(a) Farrell N. Metal complexes as drugs and chemotherapeutic agents. Compr. Coord. Chem. II. 2003;9:809–840. [Google Scholar]; (b) Zhang C.X., Lippard S.J. New Metal complexes as potential therapeutics. Curr. Opin. Chem. Biol. 2003;7(4):481–489. doi: 10.1016/s1367-5931(03)00081-4. [DOI] [PubMed] [Google Scholar]; (c) Tan C.-P., Lu Y.-Y., Ji L.-N., Mao Z.-W. Metallomics insights into the programmed cell death induced by metal-based anticancer compounds. Metallomics. 2014;6(5):978–995. doi: 10.1039/c3mt00225j. [DOI] [PubMed] [Google Scholar]; (d) van Rijt S.H., Sadler P.J. Current Applications and future potential for bioinorganic chemistry in the development of anticancer drugs. Drug Discov. Today. 2009;14(23–24):1089–1097. doi: 10.1016/j.drudis.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Santini C., Pellei M., Gandin V., Porchia M., Tisato F., Marzano C. Advances in copper complexes as anticancer agents. Chem. Rev. 2014;114(1):815–862. doi: 10.1021/cr400135x. [DOI] [PubMed] [Google Scholar]
- 8.(a) Shingu T., Chumbalkar V.C., Gwak H.-S., Fujiwara K., Kondo S., Farrell N.P., Bogler O. The Polynuclear platinum BBR3610 induces G2/M arrest and autophagy early and apoptosis late in glioma cells. Neuro-Oncol. 2010;12(12):1269–1277. doi: 10.1093/neuonc/noq095. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Guo W.-J., Zhang Y.-M., Zhang L., Huang B., Tao F.-F., Chen W., Guo Z.-J., Xu Q., Sun Y. Novel monofunctional platinum (II) complex mono-Pt induces apoptosis-independent autophagic cell death in human ovarian carcinoma cells, distinct from cisplatin. Autophagy. 2013;9(7):996–1008. doi: 10.4161/auto.24407. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Trejo-Solís C., Jimenez-Farfan D., Rodríguez-Enríquez S., Fernandez-Valverde F., Cruz-Salgado A., Ruiz-Azuara L., Sotelo J. Copper compound induces autophagy and apoptosis of glioma cells by reactive oxygen species and Jnk activation. BMC Cancer. 2012;12(1):156. doi: 10.1186/1471-2407-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Tan C., Lai S., Wu S., Hu S., Zhou L., Chen Y., Wang M., Zhu Y., Lian W., Peng W., Ji L., Xu A. Nuclear permeable ruthenium(II) Β-carboline complexes induce autophagy to antagonize mitochondrial-mediated apoptosis. J. Med. Chem. 2010;53(21):7613–7624. doi: 10.1021/jm1009296. [DOI] [PubMed] [Google Scholar]; (e) Guo W.-J., Ye S.-S., Cao N., Huang J., Gao J., Chen Q.-Y. ROS-mediated autophagy was involved in cancer cell death induced by novel copper(II) complex. Exp. Toxicol. Pathol. 2010;62(5):577–582. doi: 10.1016/j.etp.2009.08.001. [DOI] [PubMed] [Google Scholar]; (f) Paris I., Perez-Pastene C., Couve E., Caviedes P., Ledoux S., Segura-Aguilar J. Copper dopamine complex induces mitochondrial autophagy preceding caspase-independent apoptotic cell death. J. Biol. Chem. 2009;284(20):13306–13315. doi: 10.1074/jbc.M900323200. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Gorojod R.M., Alaimo A., Porte Alcon S., Pomilio C., Saravia F., Kotler M.L. The autophagic- lysosomal pathway determines the fate of glial cells under manganese- induced oxidative stress conditions. Free Radic. Biol. Med. 2015;87:237–251. doi: 10.1016/j.freeradbiomed.2015.06.034. [DOI] [PubMed] [Google Scholar]; (h) Li X., Zhao K., Guo W., Liu X., Liu J., Gao J., Chen Q., Bai Y. A novel manganese complex LMnAc selectively kills cancer cells by induction of ROS-triggered and mitochondrial-mediated cell death. Sci. China Life Sci. 2014;57(10):998–1010. doi: 10.1007/s11427-014-4682-6. [DOI] [PubMed] [Google Scholar]; (i) Liu J., Guo W., Li J., Li X., Geng J., Chen Q., Gao J. Tumor-targeting novel manganese complex induces ROS-mediated apoptotic and autophagic cancer cell death. Int J. Mol. Med. 2015;35(3):607–616. doi: 10.3892/ijmm.2015.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992;119(2):301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsukada M., Ohsumi Y. Isolation and characterization of autophagy‐defective mutants of saccharomyces cerevisiae. FEBS Lett. 1993;333(1–2):169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 11.Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M.D., Klionsky D.J., Ohsumi M., Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395(6700):395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 12.Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408(6811):488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 13.Klionsky D.J. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shintani T., Klionsky D.J. Autophagy in health and disease: a double-edged sword. Science. 2004;306(5698):990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L., Ishdorj G., Gibson S.B. Reactive oxygen species regulation of autophagy in cancer: implications for cancer treatment. Free Radic. Biol. Med. 2012;53(7):1399–1410. doi: 10.1016/j.freeradbiomed.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 16.Gibson S.B. Investigating the role of reactive oxygen species in regulating autophagy. Meth. Enzymol. 2013;528:217–235. doi: 10.1016/B978-0-12-405881-1.00013-6. [DOI] [PubMed] [Google Scholar]
- 17.Giordano S., Darley-Usmar V., Zhang J. Autophagy as an essential cellular antioxidant pathway in neurodegenerative disease. Redox Biol. 2014;2:82–90. doi: 10.1016/j.redox.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang J.J., Kim H.N., Kim J., Cho D.-H., Kim M.J., Kim Y.-S., Kim Y., Park S.-J., Koh J.-Y. Zinc(II) Ion mediates tamoxifen-induced autophagy and cell death in MCF-7 breast cancer cell line. Biometals. 2010;23(6):997–1013. doi: 10.1007/s10534-010-9346-9. [DOI] [PubMed] [Google Scholar]
- 19.Gozuacik D., Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23(16):2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 20.Poillet-Perez L., Despouy G., Delage-Mourroux R., Boyer-Guittaut M. Interplay between ROS and autophagy in cancer cells, from tumor initiation to cancer therapy. Redox Biol. 2015;4:184–192. doi: 10.1016/j.redox.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazure N.M., Pouysségur J. Hypoxia-induced autophagy: cell death or cell survival? Curr. Opin. Chem. Biol. 2010;22(2):177–180. doi: 10.1016/j.ceb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 22.Kondo Y., Kanzawa T., Sawaya R., Kondo S. The role of autophagy in cancer development and response to therapy. Nat. Rev. Cancer. 2005;5(9):726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 23.McCann M., McGinley J., Ni K., O'Connor M., Kavanagh K., McKee V., Colleran J., Devereux M., Gathergood N., Barron N., Prisecaru A., Kellett A. A new Phenanthroline-Oxazine Ligand: synthesis, coordination chemistry and atypical DNA binding interaction. Chem. Commun. 2013;49(23):2341–2343. doi: 10.1039/c3cc38710k. [DOI] [PubMed] [Google Scholar]
- 24.P. Peixoto C. Bailly M.-H. David-Cordonnier Topoisomerase I-mediated DNA relaxation as a tool to study intercalation of small molecules into supercoiled DNA. In : Drug-DNA Interaction Protocols. K.R. Fox(Ed.), Methods Mol. Biol. 613 2010 235 256. [DOI] [PubMed]
- 25.P. Pozarowski Z. Darzynkiewicz Analysis of cell cycle by flow cytometry In: Checkpoint Controls and Cancer. A.H. Schonthal (Ed.), Methods Mol. Biol. 281 2004 301 311. [DOI] [PubMed]
- 26.Brzozowska K., Pinkawa M., Eble M.J., Müller W.-U., Wojcik A., Kriehuber R., Schmitz S. In vivo versus in vitro individual radiosensitivity analysed in healthy donors and in prostate cancer patients with and without severe side effects after radiotherapy. Int. J. Radiat. Biol. 2012;88(5):405–413. doi: 10.3109/09553002.2012.666002. [DOI] [PubMed] [Google Scholar]
- 27.Casey M.T., McCann M., Devereux M., Curran M., Cardin C., Convery M., Quillet V., Harding C. Synthesis and structure of the MnII,II complex salt [Mn2(η1η1μ2-Oda)(Phen)4(H2O)2][Mn2(η1η1μ2-Oda)(Phen)4(η1-Oda)2]·4h2O (odaH2= Octanedioic Acid): a Catalyst for H2O2 Disproportionation. J. Chem. Soc. Chem. Commun. 1994;22:2643–2645. [Google Scholar]
- 28.Dekker N.H., Rybenkov V.V., Duguet M., Crisona N.J., Cozzarelli N.R., Bensimon D., Croquette V. The mechanism of type IA topoisomerases. Proc. Natl. Acad. Sci. Usa. 2002;99(19):12126–12131. doi: 10.1073/pnas.132378799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khadka D.B., Cho W.-J. Topoisomerase inhibitors as anticancer agents: a patent update. Expert Opin. Ther. Pat. 2013;23(8):1033–1056. doi: 10.1517/13543776.2013.790958. [DOI] [PubMed] [Google Scholar]
- 30.Wang J.C. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 31.Wang J.G. The degree of unwinding of the DNA helix by ethidium: I. Titration of twisted PM2 DNA molecules in alkaline cesium chloride density gradients. J. Mol. Biol. 1974;89(4):783–801. doi: 10.1016/0022-2836(74)90053-9. [DOI] [PubMed] [Google Scholar]
- 32.Capranico G., Kohn K.W., Pommier Y. Local sequence requirements for DNA cleavage by mammalian topoisomerase II in the presence of doxorubicin. Nucleic Acids Res. 1990;18(22):6611–6619. doi: 10.1093/nar/18.22.6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bachur N.R., Gordon S.L., Gee M.V. Anthracycline antibiotic augmentation of microsomal electron transport and free radical formation. Mol. Pharmacol. 1977;13(5):901–910. [PubMed] [Google Scholar]
- 34.Gutteridge J.M., Quinlan G.J. Free radical damage to deoxyribose by anthracycline, aureolic acid and aminoquinone antitumour antibiotics: An essential requirement for iron, semiquinones and hydrogen peroxide. Biochem. Pharmacol. 1985;34(23):4099–4103. doi: 10.1016/0006-2952(85)90200-x. [DOI] [PubMed] [Google Scholar]
- 35.Gewirtz D. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochem. Pharmacol. 1999;57(7):727–741. doi: 10.1016/s0006-2952(98)00307-4. [DOI] [PubMed] [Google Scholar]
- 36.Foglesong P.D., Reckord C., Swink S. Doxorubicin inhibits human DNA topoisomerase I. Cancer Chemother. Pharmacol. 1992;30(2):123–125. doi: 10.1007/BF00686403. [DOI] [PubMed] [Google Scholar]
- 37.Chang Y.-M., Chen C.K.M., Hou M.-H. Conformational changes in DNA upon ligand binding monitored by circular dichroism. Int. J. Mol. Sci. 2012;13(3):3394–3413. doi: 10.3390/ijms13033394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu P.K., Kharatishvili M., Qu Y., Farrell N. A circular dichroism study of ethidium bromide binding to Z-DNA induced by dinuclear platinum complexes. J. Inorg. Biochem. 1996;63(1):9–18. doi: 10.1016/0162-0134(95)00163-8. [DOI] [PubMed] [Google Scholar]
- 39.Manjanatha M.G., Bishop M.E., Pearce M.G. Genotoxicity of doxorubicin in F344 rats by combining the comet assay, flow‐cytometric peripheral blood micronucleus test, and pathway‐focused gene expression profiling. Environ. Mol. Mutagen. 2014;55(1):24–34. doi: 10.1002/em.21822. [DOI] [PubMed] [Google Scholar]
- 40.Bertoli C., Skotheim J.M., de Bruin R.A.M. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013;14(8):518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonner W.M., Redon C.E., Dickey J.S., Nakamura A.J., Sedelnikova O.A., Solier S., Pommier Y. γH2AX and cancer. Nat. Rev. Cancer. 2008;8(12):957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paull T.T., Rogakou E.P., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M. A critical Role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10(15):886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 43.Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 44.Koopman G., Reutelingsperger C.P., Kuijten G.A., Keehnen R.M., Pals S.T., van Oers M.H. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood. 1994;84(5):1415–1420. [PubMed] [Google Scholar]
- 45.Verhoven B., Schlegel R.A., Williamson P. Mechanisms of phosphatidylserine exposure, a phagocyte recognition signal, on apoptotic T lymphocytes. J. Exp. Med. 1995;182(5):1597–1601. doi: 10.1084/jem.182.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hengartner M.O. The biochemistry of apoptosis. Nature. 2000;407(6805):770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 47.Degterev A., Boyce M., Yuan J. A decade of caspases. Oncogene. 2003;22(53):8543–8567. doi: 10.1038/sj.onc.1207107. [DOI] [PubMed] [Google Scholar]
- 48.Kruidering M., Evan G.I. Caspase-8 in apoptosis: the beginning of "the end"? IUBMB Life. 2000;50(2):85–90. doi: 10.1080/713803693. [DOI] [PubMed] [Google Scholar]
- 49.Cohen G.M. Caspases: the executioners of apoptosis. Biochem. J. 1997;326(1):1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaur J., Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 2015;16(8):461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- 51.Biederbick A., Kern H.F., Elsässer H.P. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur. J. Cell Biol. 1995;66(1):3–14. [PubMed] [Google Scholar]
- 52.Benjamin D., Colombi M., Moroni C., Hall M.N. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat. Rev. Drug. Discov. 2011;10(11):868–880. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- 53.Chiarini F., Evangelisti C., McCubrey J.A., Martelli A.M. Current treatment strategies for inhibiting mTOR in cancer. Trends Pharmacol. Sci. 2015;36(2):124–135. doi: 10.1016/j.tips.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 54.Gammoh N., Lam D., Puente C., Ganley I., Marks P.A., Jiang X.J. Role of autophagy in histone deacetylase inhibitor-induced apoptotic and nonapoptotic cell death. Proc. Natl. Acad. Sci. Usa. 2012;109:6561–6565. doi: 10.1073/pnas.1204429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niemann A., Baltes J., Elsässer H.P. Fluorescence properties and staining behavior of Monodansylpentane, a structural homologue of the lysosomotropic agent monodansylcadaverine. J. Histochem. Cytochem. 2001;49(2):177–185. doi: 10.1177/002215540104900205. [DOI] [PubMed] [Google Scholar]
- 56.Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO Journal. 2007;26(7):1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kerr J.F., Wyllie A.H., Currie A.R. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer. 1972;26(4):239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petiot A., Ogier-Denis E., Blommaart E.F., Meijer A.J., Codogno P. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 2000;275(2):992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 59.Blommaart E.F.C., Krause U., Schellens J.P.M., Vreeling-Sindelárová H., Meijer A.J. The Phosphatidylinositol 3‐kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 1997;243(1‐2):240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 60.Poole B., Ohkuma S. Effect of weak bases on the intralysosomal pH in mouse peritoneal macrophages. J. Cell Biol. 1981;90(3):665–669. doi: 10.1083/jcb.90.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Glaumann H., Ahlberg J. Comparison of different autophagic vacuoles with regard to ultrastructure, enzymatic composition, and degradation capacity-formation of crinosomes. Exp. Mol. Pathol. 1987;47(3):346–362. doi: 10.1016/0014-4800(87)90018-9. [DOI] [PubMed] [Google Scholar]
- 62.Kawai A., Uchiyama H., Takano S., Nakamura N., Ohkuma S. Autophagosome-lysosome fusion depends on the pH in acidic compartments in CHO cells. Autophagy. 2007;3(2):154–157. doi: 10.4161/auto.3634. [DOI] [PubMed] [Google Scholar]
- 63.Taiwo F.A. Mechanism of tiron as scavenger of superoxide ions and free electrons. Spectroscopy. 2008;22(6):491–498. [Google Scholar]
- 64.Goldstein S., Czapski G. Mannitol as an OH. Scavenger in aqueous solutions and in biological systems. Int. J. Radiat. Biol. 1984;46(6):725–729. doi: 10.1080/09553008414551961. [DOI] [PubMed] [Google Scholar]
- 65.Matheson I.B.C., Lee J. Chemical reaction rates of amino acids with singlet oxygen. Photochem. Photobiol. 1979;29(5):879–881. [Google Scholar]
- 66.Foote C.S., Clennan E.L. In Active Oxygen in Chemistry; Active Oxygen in Chemistry. Springer Netherlands; Dordrecht: 1996. Properties and reactions of singlet dioxygen; pp. 105–140. [Google Scholar]
- 67.Giandomenico A.R., Cerniglia G.E., Biaglow J.E., Stevens C.W., Koch C.J. The importance of sodium pyruvate in assessing damage produced by hydrogen peroxide. Free Radic. Biol. Med. 1997;23(3):426–434. doi: 10.1016/s0891-5849(97)00113-5. [DOI] [PubMed] [Google Scholar]
- 68.Ruefli A.A., Ausserlechner M.J., Bernhard D., Sutton V.R., Tainton K.M., Kofler R., Smyth M.J., Johnstone R.W. The histone deacetylase inhibitor and chemotherapeutic agent suberoylanilide hydroxamic acid (SAHA) induces a cell-death pathway characterized by cleavage of bid and production of reactive oxygen species. Proc. Natl. Acad. Sci. USA. 2001;98(19):10833–10838. doi: 10.1073/pnas.191208598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petruccelli L.A., Dupéré-Richer D., Pettersson F., Retrouvey H., Skoulikas S., Miller W.H. Vorinostat Induces Reactive Oxygen Species and DNA Damage in Acute Myeloid Leukemia Cells. PLoS ONE. 2011;6(6):e20987. doi: 10.1371/journal.pone.0020987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Butler L.M., Zhou X.B., Xu W.-S., Scher H.I., Rifkind R.A., Marks P.A., Richon V.M. The Histone Deacetylase inhibitor SAHA arrests cancer cell growth, up-regulates thioredoxin-binding protein-2, and down-regulates thioredoxin. Proc. Natl. Acad. Sci. USA. 2002;99(18):11700–11705. doi: 10.1073/pnas.182372299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ungerstedt J.S., Sowa Y., Xu W.-S., Shao Y., Dokmanovic M., Perez G., Ngo L., Holmgren A., Jiang X., Marks P.A. Role of thioredoxin in the response of normal and transformed cells to histone deacetylase inhibitors. Proc. Natl. Acad. Sci. USA. 2005;102(3):673–678. doi: 10.1073/pnas.0408732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Foster H., Coley H.M., Goumenou A., Pados G., Harvey A., Karteris E. Differential expression of mTor signalling components in drug resistance in ovarian cancer. Anticancer Res. 2010;30(9):3529–3534. [PubMed] [Google Scholar]
- 73.Neklesa T.K., Davis R.W. Superoxide anions regulate TORC1 and its ability to bind Fpr1: rapamycin complex. Proc. Natl. Acad. Sci. USA. 2008;105(39):15166–15171. doi: 10.1073/pnas.0807712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen W., Zou P., Zhao Z., Chen X., Fan X., Vinothkumar R., Cui R., Wu F., Zhang Q., Liang G., Ji J. Synergistic antitumor activity of rapamycin and EF24 via increasing ROS for the treatment of gastric cancer. Redox Biol. 2016;10:78–89. doi: 10.1016/j.redox.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Robinson K.M., Janes M.S., Pehar M., Monette J.S., Ross M.F., Hagen T.M., Murphy M.P., Beckman J.S. Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc. Natl. Acad. Sci. Usa. 2006;103(41):15038–15043. doi: 10.1073/pnas.0601945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wen X., Wu J., Wang F., Liu B., Huang C., Wei Y. Deconvoluting the role of reactive oxygen species and autophagy in human diseases. Free Radic. Biol. Med. 2013;65:402–410. doi: 10.1016/j.freeradbiomed.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 77.Chen Y., Gibson S.B. Is mitochondrial generation of reactive oxygen species a trigger for autophagy? Autophagy. 2008;4(2):246–248. doi: 10.4161/auto.5432. [DOI] [PubMed] [Google Scholar]
- 78.Chen Y., Azad M.B., Gibson S.B. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009;16(7):1040–1052. doi: 10.1038/cdd.2009.49. [DOI] [PubMed] [Google Scholar]
- 79.Tasdemir E., Maiuri M.C., Galluzzi L., Vitale I., Djavaheri-Mergny M., D'Amelio M., Criollo A., Morselli E., Zhu C., Harper F., Nannmark U., Samara C., Pinton P., Vicencio J.M., Carnuccio R., Moll U.M., Madeo F., Paterlini-Brechot P., Rizzuto R., Szabadkai G., Pierron G., Blomgren K., Tavernarakis N., Codogno P., Cecconi F., Kroemer G. Vol. 10. 2008. Regulation of autophagy by cytoplasmic P53; pp. 676–687. (Nat. Cell Biol). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yaginuma Y., Westphal H. Abnormal structure and expression of the p53 gene in human ovarian carcinoma cell lines. Cancer Res. 1992;52(15):4196–4199. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material