Abstract
Introduction
Biomarkers for Alzheimer's disease (AD) pathophysiology have been developed that focus on various levels of brain organization. However, no robust biomarker of large-scale network failure has been developed. Using the recently introduced cascading network failure model of AD, we developed the network failure quotient (NFQ) as a biomarker of this process.
Methods
We developed and optimized the NFQ using our recently published analyses of task-free functional magnetic resonance imaging data in clinically normal (n = 43) and AD dementia participants (n = 28) from the Alzheimer's Disease Neuroimaging Initiative. The optimized NFQ (oNFQ) was then validated in a cohort spanning the AD spectrum from the Mayo Clinic (n = 218).
Results
The oNFQ (d = 1.25, 95% confidence interval [1.25, 1.26]) had the highest effect size for differentiating persons with AD dementia from clinically normal participants. The oNFQ measure performed similarly well on the validation Mayo Clinic sample (d = 1.44, 95% confidence interval [1.43, 1.44]). The oNFQ was also associated with other available key biomarkers in the Mayo cohort.
Discussion
This study demonstrates a measure of functional connectivity, based on a cascading network failure model of AD, and was highly successful in identifying AD dementia. A robust biomarker of the large-scale effects of AD pathophysiology will allow for richer descriptions of the disease process and its modifiers, but is not currently suitable for discriminating clinical diagnostic categories. The large-scale network level may be one of the earliest manifestations of AD, making this an attractive target for continued biomarker development to be used in prevention trials.
Keywords: Alzheimer's disease, Cascading network failure, Biomarker, Default mode network, Connectivity
1. Introduction
Alzheimer's disease (AD) pathophysiology manifests in unique ways throughout the multiscale organization of the central nervous system across the lifespan. A multitude of biomarkers have been developed to measure various levels of this pathophysiology (i.e., molecular, synaptic, cellular, electrophysiological, and global cognitive). We recently described the cascading network failure model of AD pathophysiology in default mode network (DMN) subsystems [1]. Developing a biomarker of this network failure is an important next step to study this model.
In this study, we present a candidate biomarker of the large-scale network-level manifestation of AD pathophysiology. The spatial resolution and the ease of adding a functional magnetic resonance imaging (fMRI) sequence into a routine MRI protocol makes task-free fMRI (TF-fMRI) an attractive modality in which to develop a biomarker of AD pathophysiology at the large-scale systems level. However, measurement error, biological variability, and expertise in processing and interpreting nonlinear patterns of change in connectivity, both increases and decreases, have hampered the development of TF-fMRI as a robust biomarker of network failure in AD.
Functional MRI (fMRI) was initially found to demonstrate decreased connectivity in the DMN associated with AD dementia [2]. Subsequently TF-fMRI studies have also found increases in connectivity in frontal lobe portions of the DMN associated with AD dementia that mirror age-related changes [3] but may eventually decline with disease progression [1], [4]. Increases in functional activation have been observed during task performance in the medial temporal lobe in participants with mild cognitive impairment (MCI) for many years [5], and this has been thought to represent functional compensation. A recent study lends significant support for the compensation hypothesis, given that increases in functional activation were associated with better cognitive performances in amyloid positive cognitively normal individuals [6].
The increases and decreases in DMN connectivity associated with AD dementia, which may take nonlinear trajectories [1], [7], pose a significant challenge in developing a biomarker of large-scale network changes. However, linear increases and decreases may lend themselves to being combined into a more stable ratio of DMN-related connectivity failure across the AD spectrum. In this study, we use the results from our recent TF-fMRI analyses of the Alzheimer's Disease Neuroimaging Initiative (ADNI) as a discovery cohort for the development of such a ratio of increases of connectivity over decreases in connectivity we term the network failure quotient (NFQ). This new optimized biomarker was then validated in a separate cohort from the Mayo Clinic.
2. Methods
Data used in the preparation of this article were obtained from the ADNI database (adni.loni.usc.edu). The ADNI was launched in 2003 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration, private pharmaceutical companies, and nonprofit organizations as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether serial MRI, positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
2.1. ADNI participants
Clinically normal (CN, n = 43) and AD dementia (n = 28) participants that were used in our recent analysis of connectivity within and between DMN subsystems were used as the discovery cohort in these analyses (subject and scan identifiers were previously published as supplementary material in Jones et al. [1]). The basic demographics for this sample are listed in Table 1. The description of the original analyses of these images is as described in Jones et al. [1]. All subjects with AD dementia were amyloid-PET positive. Normal subjects were defined clinically irrespective of amyloid status.
Table 1.
Subject Characteristics
| ADNI CN | ADNI AD | Mayo-matched CN | Mayo AD | Mayo Biomarker CN | Mayo Biomarker CI | |
|---|---|---|---|---|---|---|
| N | 43 | 28 | 42 | 21 | 177 | 41 |
| Age (Q1, Q3) | 73 (69, 78) | 74 (72, 76.5) | 67 (58, 73) | 67 (59, 75) | 67 (58,73) | 68 (63,75) |
| Female % | 25 (58) | 13 (46) | 24 (57) | 12 (57) | 75 (42) | 17 (41) |
| Education (Q1, Q3) | 16 (16, 16) | 15.5 (14, 16) | 14 (12.5, 16) | 16 (16, 16) | 16 (14, 17) | 16 (13, 18) |
| MMSE (Q1, Q3) | 29 (28, 30) | 22 (21, 25) | 29 (29, 29) | 17 (10, 22) | 29 (28, 29) | 22 (17, 26) |
Abbreviations: AD, Alzheimer's disease; ADNI, Alzheimer's Disease Neuroimaging Initiative; CI, clinically impaired; CN, clinically normal; MMSE, Mini-Mental State Examination.
2.2. Mayo participants
All participants in the validation cohort were enrolled in either the Mayo Clinic Study of Aging (MCSA) or the Mayo Clinic Rochester Alzheimer's Disease Research Center. The Alzheimer's Disease Research Center is a longitudinal cohort study that enrolls referral participants at the Mayo Clinic in Rochester, MN. The MCSA is a population-based study of cognitive aging among Olmsted County, MN residents [8]. Enrolled participants are adjudicated to be CN or have MCI by a consensus panel consisting of study coordinators, neuropsychologists, and behavioral neurologists. All participants are followed longitudinally including individuals who progresses to dementia. Methods for defining CN, MCI, and dementia in both these studies conform to standards in the field [9], [10], [11]. All CN participants who had good quality structural MRI, TF-fMRI, tau-PET (AV-1451), and amyloid-PET (PiB) were included in this study. Participants who were clinically impaired (either MCI or dementia) were also included if they had these modalities available at the time of this study and their amyloid-PET standardized uptake value ratio exceeded 1.5 to increase the probability that AD pathophysiology was contributing to their clinical impairment [12]. For investigating the effect size of NFQ in the validation cohort, a subset of participants was used. There were 21 participants with AD dementia, and a subset of CN participants were matched 2:1 on age, gender, and education to the AD dementia participants. See Table 1 for details regarding the cohort and the matched CN and AD subsets.
2.3. Mayo standard protocol approvals, registrations, and patient consents
These studies were approved by the Mayo Clinic and Olmsted Medical Center Institutional Review Boards, and written informed consent was obtained from all participants.
2.4. PET imaging methods
Amyloid-PET imaging was performed with Pittsburgh Compound B [13] and tau-PET with AV-1451 [14], [15]. Computed tomography scan was obtained for attenuation correction. Late uptake amyloid-PET images were acquired from 40 to 60 minutes, fluorodeoxyglucose (FDG) PET from 30 to 40 minutes, and tau-PET from 80 to 100 minutes after injection. PET images were analyzed with our in-house fully automated image processing pipeline [16] where image voxel values are extracted from automatically labeled regions of interest (ROIs) propagated from an MRI template. An amyloid-PET standardized uptake value ratio was calculated as the median uptake in the prefrontal, orbitofrontal, parietal, temporal, anterior and posterior cingulate, and precuneus ROIs normalized to the cerebellar gray median. However, voxels whose probability of being cerebral spinal fluid (CSF) was greater than the probability of being gray matter and greater than the probability of being white matter, based on coregistered segmented MRI, were not included in the calculation. An AD-characteristic FDG-PET meta-ROI was calculated as the average of the median uptake in the angular gyrus, posterior cingulate, and inferior temporal cortical ROIs and normalized to pons and vermis median [17]. FDG-PET data were not partial volume corrected. The average tau-PET signal in an entorhinal cortex (EC) ROI normalized to the cerebellar gray matter was used to compare with connectivity measures. Similar results were obtained when using other ROIs (e.g., inferior temporal ROI).
2.5. Structural MRI
MRI was performed on one of three 3 T systems from the same vendor. The MRI measure was a FreeSurfer (v5.3)-derived AD signature composed of the average of the mean cortical thickness in the following individual ROIs: entorhinal, inferior temporal, middle temporal, and fusiform.
2.6. TF-fMRI preprocessing and network measures
TF-fMRI data were acquired using an eight-channel head coil, gradient echo planar image (EPI), TR = 3000 milliseconds, TE = 30 milliseconds, 90° flip angle, 21 cm field of view, 64 × 64 in-plane matrix, slice thickness 3.3 mm without gap, and 103 volumes were obtained. Participants were instructed to keep their eyes open during scanning. All TF-fMRI data sets with greater than 3 mm of translational movement, 3° of rotational movement, or that failed visual inspection for obvious artifacts were excluded from analysis.
The methods recently developed for analyzing TF-fMRI data in the ADNI were adapted for the TF-fMRI portion of this study [1]. The first three volumes were removed and the time series within each voxel were despiked using AFNI's 3dDespike program (http://afni.nimh.nih.gov). This process was done before realignment, given that realignment and motion correction may be improved by this despiking procedure [18]. Next, we performed slice-timing correction followed by two pass realignment to the mean EPI. The structural images were then coregistered to the mean EPI image.
Unified segmentation and normalization to the MCSA template space was then performed. The MCSA Functional Connectivity Atlas [19] high-dimensional independent components of interest (i.e., ventral DMN [vDMN], posterior DMN [pDMN], anterior ventral DMN [avDMN], and anterior dorsal DMN [adDMN]) were transformed to individual subject space using the inverse warps created during unified segmentation and normalization for each subject. To create an anatomically based “noise ROI” to be used in a component-based noise correction [20] the subject space CSF and white matter segmentations were binarized at a 0.9 probability threshold and eroded by two voxels in each direction to avoid contamination with gray matter voxels. The union of the binarized and eroded images was used to extract the voxelwise time series to be used in a principal component analysis. The first six principal components were combined with the six motion parameters and their first temporal derivatives (18 total regressors) to create a nuisance regressor matrix to be used for further preprocessing.
Finally, AFNI's 3dBandpass program was used to detrend, simultaneously band-pass filter (0.009–0.08 Hz), and perform the nuisance regression using the nuisance regressor matrix. Simultaneous filtering and nuisance regression avoids spectral misspecification of motion artifact further reducing the impact of the motion confound [21]. This program was also used for time series variance normalization, masking, and smoothing with 8 mm full-width half-maximum Gaussian kernel.
After preprocessing, a spatial-temporal regression was performed within a multivariate framework incorporating all four DMN subsystems of interest (see Fig. 1 for component maps) using functions from the group independent component analysis of fMRI toolbox (GICA) of functional MRI Toolbox (GIFT v2.0e) software package [22] as previously described [1]. The four DMN subsystem measures that we had previously shown to be linearly changing (both increases and decreases) in the ADNI cohort across the disease spectrum as part of the cascading network failure (i.e., the pDMN, vDMN, posterior-to-ventral DMN, and posterior-to-anterior dorsal DMN connectivity) were used to create a single summary metric of network failure termed the NFQ.
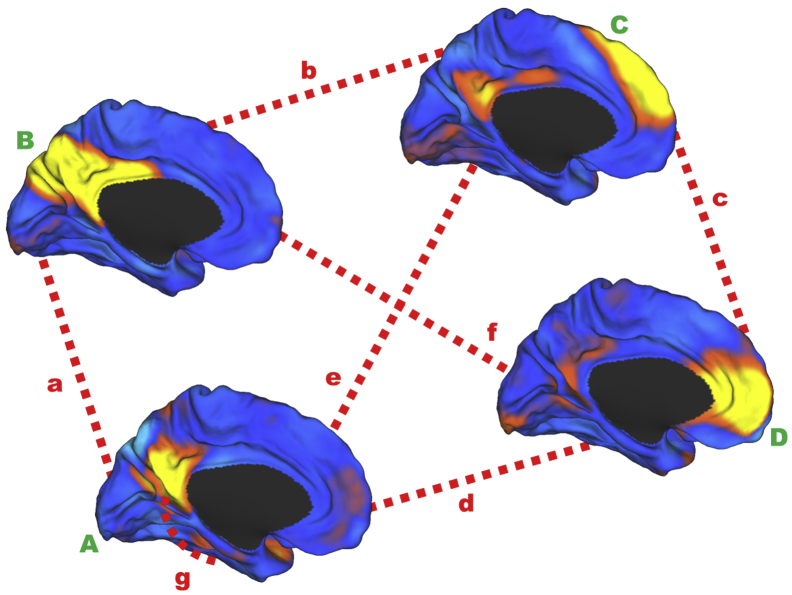
Fig. 1.
Schematic of DMN subsystem elements. The within-DMN subsystems' connectivity for the ventral DMN (A), posterior DMN (B), anterior dorsal DMN (C), and anterior ventral DMN (D) are indicated with green capital letters near the medial surface rendering of their respective intrinsic connectivity networks used to create the DMN connectivity measures. The between-subsystems connectivity is indicated with red dashed lines (a–f). The ventral DMN hippocampal connectivity is indicated by the red dashed line extending from the ventral DMN to the medial temporal lobe (g). The surface renderings of the four independent components from the high-dimensional independent component analysis (ICA) Mayo Clinic Study of Aging Functional Connectivity Atlas [19] were created using the CARET software package http://www.nitrc.org/projects/caret/. The entire component maps were used in the dual-regression analysis. Elements A and B are used in the denominator in the NFQ and elements a and b are used in the numerator. This figure was originally published in the supplementary material of Jones et al. [1]. Abbreviation: DMN, default mode network.
However, each network element is treated equally in the NFQ. Therefore, we also created two different summary measures with weighting of the individual elements. The first used the effect sizes comparing CN with AD dementia subjects in our recent publication [1] as weights for each of the network elements (wNFQ).
We also used an unconstrained nonlinear optimization algorithm (fminsearch function in MATLAB) to optimize the weights that would give the largest effect size in the ADNI data (optimized network failure quotient [oNFQ]). We verified that the algorithm came to a stable solution by assessing the variability of the weights across various bootstrapped samplings of the data.
2.7. Graph theory–based network measures
We compared the NFQ to other network measures based on graph theory. We constructed graphs using the 68 ROIs from the MCSA Functional Connectivity Atlas [19] as nodes and the Fisher transformation of the Pearson correlation coefficient between the preprocessed BOLD time series as the edges. The Brain Connectivity Toolbox was used for graph construction and network measures [23]. The edges were normalized to range from 0 to 1 and weighted graph metrics (global efficiency and nodal strength) were created over the full range of network densities. The NFQ and all four contributing network elements were then correlated with global efficiency across network thresholds. The NFQ was also correlated with each node's centrality (i.e., weighted nodal strength) at a network density of 0.56 given that this was the network density with the peak correlation between NFQ and global efficiency.
2.8. Statistical procedures
A combination of MATLAB-based (Mathworks Inc, Natick, MA) and R-based (http://www.R-project.org) software packages was used to perform all statistical analysis. A bootstrapping procedure was used to derive measures of accuracy on sample statistics. Cohen's d was used as a measure of the effect size in comparing AD dementia with CN participants. Kruskal-Wallis one-way analysis of variance was used for continuous variables, with post hoc Mann-Whitney U tests for pairwise differences. Chi-squared tests were used for categorical variables.
3. Results
3.1. Effect size of the network failure quotient in ADNI data
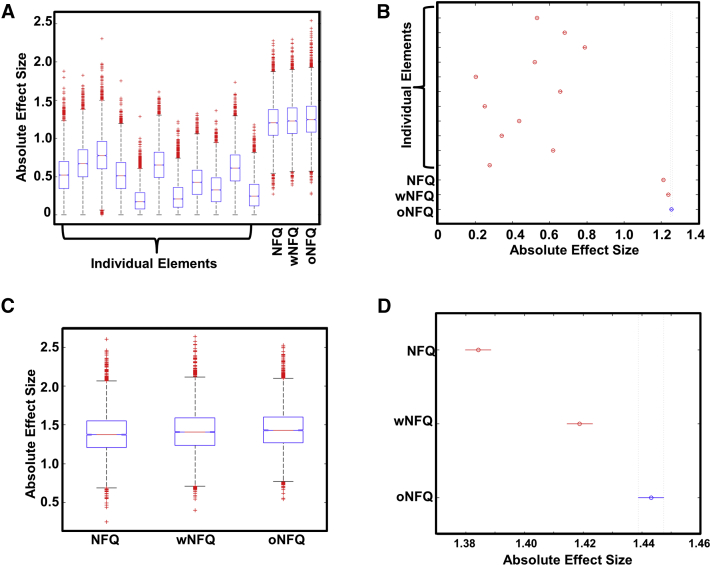
In comparing AD dementia with CN participants in the discovery ADNI data set, there was a main effect of DMN subsystem network element on the observed distribution of effect sizes obtained from 10,000 bootstrapped samples of the data (F (13, 139,986) = 23,984, P = 0) (Fig. 2A). The pDMN connectivity had the greatest absolute effect size of any individual element (d = 0.78, 95% confidence interval [CI] [0.78, 0.79]), but all three NFQ summary metrics of network failure had greater effect sizes than any individual element with oNFQ (d = 1.25, 95% CI [1.25, 1.26]) having the highest effect size of all elements evaluated (Fig. 2B).
Fig. 2.
Superior effect size for the optimized network failure quotient (NFQ) in discovery and validation cohort. (A) Box plots of the absolute effect size comparing clinically normal (n = 43) versus Alzheimer's disease dementia (n = 28) for individual elements of within and between default mode network subsystem connectivity and the proposed combined NFQ metrics described in the main text for 10,000 bootstrapped samples of the Alzheimer's Disease Neuroimaging Initiative discovery cohort. (B) Plot comparing the 95% confidence intervals (CIs) from A, demonstrating no overlap between the optimized NFQ (oNFQ) and any other connectivity measure. (C) Box plots of the absolute effect size comparing clinically normal (n = 42) versus Alzheimer's disease dementia (n = 21) for the proposed combined NFQ metrics described in the main text for 10,000 bootstrapped samples of the Mayo Clinic validation cohort. (D) Plot comparing the 95% CIs from (C), demonstrating no overlap between the oNFQ with the other two measures. In the two box plots, the red line indicates the median, the box indicating the first and third quartile, whiskers marking the minimum and the maximum, notches indicating the 95% CI, and the red cross indicating outliers.
3.2. Effect size of the network failure quotient in Mayo data
Given that the NFQ summary measure was designed to summarize the cascading network failure in DMN subsystems originally observed in the ADNI data set and oNFQ was optimized on this same cohort, we sought to validate these findings in a separate cohort. We selected all the available AD dementia participants (n = 21) in the Mayo multimodal biomarker cohort and matched 1:2 on age, gender, and education to CN participants (Table 1). We then compared the effect sizes of NFQ, wNFQ, and oNFQ across 10,000 bootstrapped samples of this validation cohort. We found that there was a larger effect size for all these summary metrics in this cohort relative to the ADNI cohort (Fig. 2C) with oNFQ (d = 1.44, 95% CI [1.43, 1.44]) modestly outperforming (Fig. 2D) both NFQ (d = 1.38, 95% CI [1.38, 1.39]) and wNFQ (d = 1.42, 95% CI [1.41, 1.42]). For context, we also report the absolute effect size of the other imaging biomarkers in this cohort excluding amyloid-PET, as this was used to define the cohorts: FDG-PET = 3.0, cortical thickness in AD signature regions = 3.3, and tau-PET in the EC = 3.4.
3.3. Network failure quotient versus other biomarkers in Mayo data
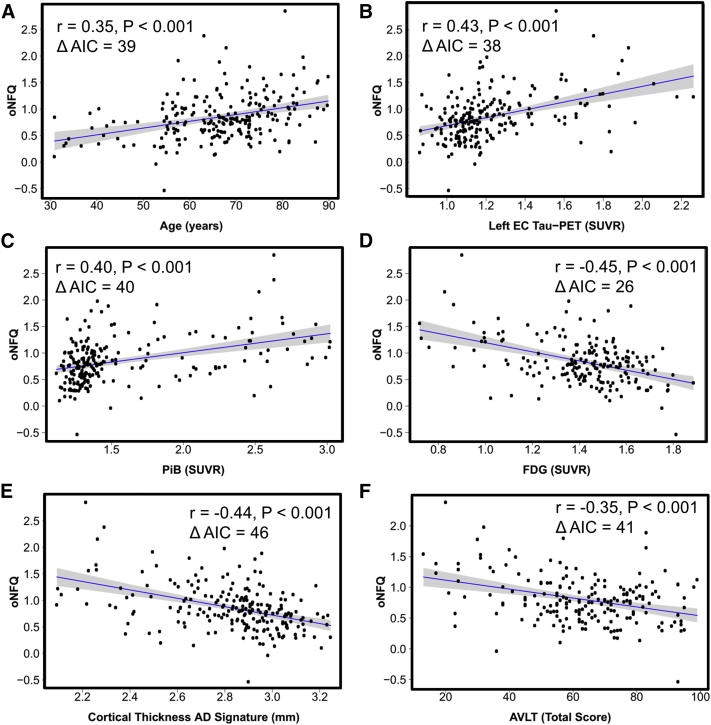
We next used the entire Mayo multimodal biomarker cohort (n = 218) to further evaluate the potential of oNFQ as a biomarker of AD pathophysiology by correlating oNFQ with available key biomarkers of AD pathophysiology (Fig. 3). We found that oNFQ was associated with age (r = 0.35, P < .001), tau-PET in the EC (r = 0.43, P < .001), global PiB (r = 0.40, P < .001), FDG-PET (r = −0.45, P < .001), cortical thickness in AD signature regions (r = −0.44, P < .001), and total score on Rey Auditory Verbal Learning Test (AVLT) (r = −0.35, P < .001). In each instance, models using oNFQ produced better fits than unoptimized NFQ demonstrated by a significant reduction in Akaike's information criterion (reductions ranging from 39 to 46).
Fig. 3.
Validating the biomarker potential of the optimized novel network failure quotient. Our summary measures of large-scale brain network failure (the optimized network failure quotient [oNFQ]) is plotted versus age and five biomarkers of Alzheimer's disease pathophysiology: (A) age, (B) tau-PET SUVR in the entorhinal cortex (EC), (C) amyloid-PET SUVR, (D) FDG-PET SUVR in Alzheimer's disease signature regions, (E) cortical thickness in Alzheimer's disease signature regions, (F) total score on the auditory verbal learning test. The blue line indicates the linear fit with the gray band indicating the 95% confidence interval. Inset in each plot, the correlation coefficient, P value, and the relative reduction in Akaike's information criteria (ΔAIC) between the models using oNFQ versus the ones using NFQ without optimization (see main text for details). Abbreviations: FDG, fluorodeoxyglucose; SUVR, standardized uptake value ratio.
A multivariate model including all these biomarkers as predictors for oNFQ was significant (F (6, 156) = 9.542, P < .001, R2 = 0.27, R2adj = 0.24) with age (β = 0.34, P < .001), entorhinal tau (β = 0.18, P = .08), and FDG (β = −0.19, P = .04) being the most statistically significant contributors to the model.
We repeated these analyses using only CN subjects and found that all the key biomarkers were still correlated with oNFQ. However, in the multivariate model (F (6, 143) = 8.293, P < .001, R2 = 0.26, R2adj = 0.23) age was the only significant predictor (β = 0.39, P < .001).
3.4. Network failure quotient versus global and local graph theory metrics
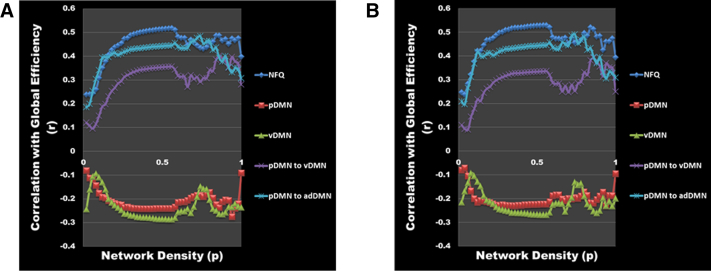
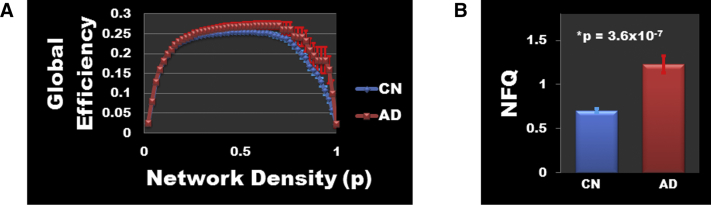
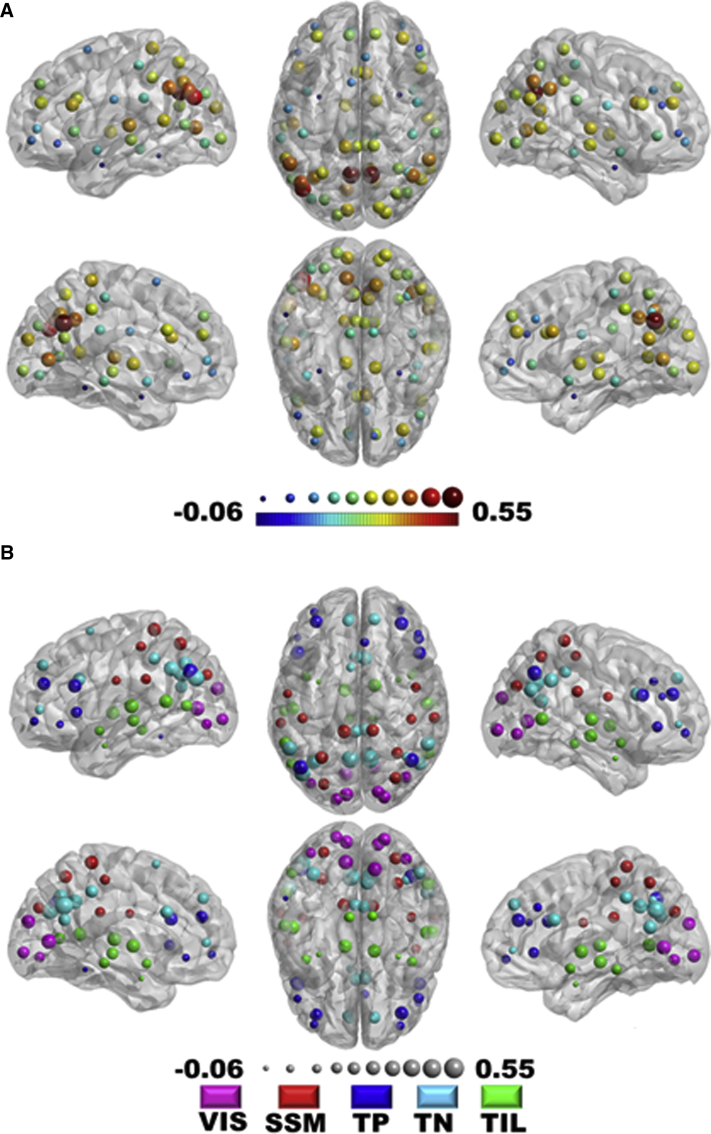
Each of the four elements that comprise the NFQ were correlated with global efficiency, but the combined NFQ showed the strongest correlation with global network efficiency peaking at a network density threshold of 0.54 at Pearson correlation coefficient of 0.52 (P < .001) for the entire cohort (Fig. 4A) and 0.53 (P < .001) for the CN subjects only (Fig. 4B). However, there was no difference in global efficiency between AD dementia participants and matched CN participants (Fig. 5A), but as previously noted, there is a difference in NFQ between these two groups (Fig. 5B). The NFQ is also correlated with the centrality for nodes both inside and outside the DMN (Fig. 6).
Fig. 4.
DMN biomarkers' correlation with global efficiency. The correlation (r) between DMN biomarkers (NFQ [blue], pDMN [red], vDMN [green], pDMN to vDMN [purple], and pDMN to adDMN [cyan]) and global efficiency is plotted across the full range of network densities for (A) all the participants in the biomarker cohort (n = 218) and (B) all the clinically normal participants (n = 177). Abbreviations: adDMN, anterior dorsal DMN; DMN, default mode network; NFQ, network failure quotient; pDMN, posterior DMN; vDMN, ventral DMN.
Fig. 5.
Difference in global efficiency and NFQ between AD dementia and matched CN. (A) Mean global efficiency values are plotted with standard errors of the mean for AD dementia participants (n = 21) and matched CN (n = 42) participants across the full range of network densities. There was no significant difference in global efficiency between AD and CN at any network density. (B) The same comparison is made for NFQ demonstrating the significant increase in NFQ in AD dementia participants relative to the age-, gender-, and education-matched CN participants. The bar plot displays the mean and the standard error of the mean for each group. Note that the NFQ calculation does not require arbitrary network density thresholds as is needed for graph theory–based measures. Abbreviations: AD, Alzheimer's disease; CN, clinically normal; NFQ, network failure quotient.
Fig. 6.
Correlation between NFQ and nodal centrality. The nodal centrality (i.e., weighted strength) was calculated for each node (n = 68) in every subject (n = 218) and correlated with NFQ. (A) The strength of the correlation between each node's centrality and NFQ is encoded in a jet color map and the radius of the node. (B) The same information is encoded in the lower figure in the radius of the nodes, but the color of the nodes indicates the atlas-defined community membership of the node (i.e., visual, VIS; somatic sensory motor, SSM; task-positive, TP; task-negative or default mode network regions, TN; and temporal-insular-limbic, TIL). This demonstrates that the NFQ correlates with nodal centrality in nodes in DMN regions and in non-DMN regions. Abbreviations: DMN, default mode network; NFQ, network failure quotient.
4. Discussion
The NFQ is centered on a model of AD pathophysiology that posits a network failure based on known brain connectivity patterns. We found that the NFQ is a robust biomarker that both differentiates AD dementia from CN people, but is also correlated with a number of other biomarkers of AD pathophysiological severity. The combined ratio has a larger effect size than any individual DMN subsystem (Fig. 2A) and demonstrates a larger effect size in an independent cohort (Fig. 2C). Optimization of the NFQ gives modest, but significant, improvements in the performance demonstrated in the larger effect size comparing CN with AD dementia (Fig. 2B and D) and in the improved model fit in association with other AD biomarkers across the disease spectrum (Fig. 3). Across the disease spectrum in the multivariate model, age was the most significant explanatory variable for increasing NFQ (β = 0.34, P < .001), with smaller contributions from FDG-PET hypometabolism (β = −0.19, P = .04) and trend level contributions from EC tau-PET signal (β = 0.18, P = .08). However, when looking only at the control subjects in a multivariate framework, age was the only significant predictor of the NFQ. This indicates that none of the other key biomarkers of AD pathophysiology are associated with key network physiology in the preclinical disease phase. This highlights the utility of having biomarkers for different levels of AD pathophysiology (e.g., both the molecular scale and the large-scale network scale). Although relevant network changes are related to aging, as is AD risk in general, we have previously shown that these network changes are more advanced in AD dementia [3], and the NFQ is able to distinguished AD dementia from control subjects matched on age as would be expected of a biomarker of AD pathophysiology. Future work will be needed to determine the effect of disease stage on these relationships, with detailed evaluation of the contributions of the increases and decreases in connectivity that constitute the NFQ. In the present study, all four elements of the NFQ appeared to contribute to its ability to distinguish AD dementia from CN people given that none of the elements weights approached zero during the optimization of NFQ. The weightings in the oNFQ were largely consistent with the effect size for the individual elements, in that the pDMN was the most important element. However, the between DMN subsystem connections involving the pDMN had greater weighting than the within vDMN connectivity in the oNFQ relative to the wNFQ. This may reflect the greater generalizability of pDMN-related changes across AD phenotypes. However, more detailed study across disease phenotypes is needed to explore the validity of this hypothesis.
The brain is a complex system with inherent multiscale organization, and AD pathophysiology manifests in unique ways across these levels of brain organization. At the large-scale brain network level, the DMN is affected in complex ways. Given the interdependence of large-scale brain networks, there are AD-related changes in multiple brain networks beyond the DMN subsystems studied here [24], [25]. However, according to the cascading network failure model, the pDMN and the brain-wide compensatory connectivity changes that occur in association with pDMN failure are directly related to β-amyloidosis in hubs of high connectivity. Therefore, a robust biomarker that is able to capture these pDMN changes will be relevant to AD pathophysiology across phenotypic variants that may vary in the extent that they impact other brain networks [26]. In addition, focusing on DMN subsystem elements with attractive biomarker properties, such as having monotonic functional forms across the disease spectrum, will increase the robustness of the measure. Some DMN subsystem elements have nonmonotonic forms and many non-DMN elements are also likely to have nonmonotonic functional forms across the AD spectrum. Disentangling monotonic and nonmonotonic connectivity changes is likely one of the key reasons for the enhanced performance of looking at DMN subsystem elements as biomarkers of AD pathophysiology. In addition, combining the increases and decreases in connectivity into a ratio may help normalize for individual differences in fMRI measures of connectivity and between site variability inherent in the multisite design of studies like ADNI. However, there remains measurement and non-neuronal physiological noise contributing to variability in TF-fMRI that can still be improved by leveraging advances made by the Human Connectome Project in data acquisition and postprocessing [27] that will be possible in ADNI-3. These advances have the potential to significantly improve the biomarker potential of the NFQ for AD pathophysiology.
We believe that the NFQ does capture AD-relevant brain network reorganization, which supports global brain network properties helping to maintain cognition in a compensatory, yet ultimately detrimental, fashion. Graph theoretical measures such as global efficiency, that capture how integrated a network is at the global level, are associated with intelligence [28], and we have observed that the NFQ is associated with global efficiency across the disease spectrum and in CN subjects (Fig. 4). However, global efficiency is maintained in AD dementia subjects (Fig. 5A) while the related NFQ is significantly increased (Fig. 5B). This supports the hypothesis that NFQ captures compensatory brain network rearrangements that help to maintain global network efficiency in setting of advancing AD pathophysiology. This observation also suggests that measures of global efficiency are poor network-level biomarker choices because underlying network rearrangements, captured by the NFQ in a single numeric summary, are taking place to keep global network efficiency at a constant value across the disease course. In addition, we have observed that the NFQ is associated with local network properties beyond DMN regions (Fig. 6), which supports the utility of the NFQ as a biomarker that is capable of capturing AD-related brain network changes that may take place outside the DMN as well. The combined observations in this study strongly support further exploration of the NFQ as a robust biomarker of the network-level pathophysiology of AD.
The current investigation was limited, in that the evaluation of the biomarker potential of the NFQ was focused on a multisite study (i.e., ADNI) with validation in a single site study (i.e., Mayo). Future investigations limiting site-related variability with improved fMRI sequences and postprocessing of data obtained in large sample sizes spanning the AD spectrum will be better suited to investigate the impact of disease stage, AD phenotypes, and for testing complex system models incorporating multiple AD pathophysiological biomarkers across various scales of brain organization.
This study demonstrates the utility of using the cascading network failure pathophysiological model to develop a robust biomarker of the large-scale network effects of AD pathophysiology. Having a robust biomarker of the large-scale network level effects of AD pathophysiology will allow for richer descriptions of the disease process and its modifiers. In addition, the large-scale network level may be one of the earliest manifestations of AD pathophysiology in the preclinical disease phase, as hypothesized by the cascading network failure model. In that context, we observed that no other biomarker tested in this study is associated with preclinical network changes when controlling for age, making the NFQ a very attractive target for continued biomarker development to be used in prevention trials and in studies of healthy brain aging.
Research in context.
-
1.
Systematic review: The authors reviewed the literature using traditional (e.g., PubMed) sources and meeting abstracts and presentations. There has not been a report in the literature of a robust biomarker of connectivity changes in default mode network subsystems associated with cascading network failure observed in Alzheimer's disease (AD).
-
2.
Interpretation: Our findings demonstrate that the network failure quotient is a promising candidate biomarker that measures AD-related changes in large-scale brain networks.
-
3.
Future directions: This article proposes a new biomarker for the large-scale network effects of AD. Having such a biomarker will allow for more complete studies of the AD spectrum, including preclinical disease stages. Including this new measure in multimodal multiscale biomarker studies will allow for the testing of complex system models of AD.
Acknowledgments
The authors would like to thank AVID Radiopharmaceuticals, Inc, for their support in supplying AV-1451 precursor, chemistry production advice and oversight, and FDA regulatory cross-filing permission and documentation needed for this work.
This research was supported by National Institute of Health grants P50 AG016574, R01 NS89757, R01 NS089544, R01 DC10367, U01 AG006786, R21 NS094489, R01 AG11378, R01 AG041851; by the Robert Wood Johnson Foundation; the Elsie and Marvin Dekelboum Family Foundation; the Liston Family Foundation; the Robert H. and Clarice Smith and Abigail van Buren Alzheimer's Disease Research Program; The GHR Foundation; Foundation Dr. Corinne Schuler (Geneva, Switzerland); and the Mayo Foundation.
References
- 1.Jones D.T., Knopman D.S., Gunter J.L., Graff-Radford J., Vemuri P., Boeve B.F. Cascading network failure across the Alzheimer's disease spectrum. Brain. 2016;139:547–562. doi: 10.1093/brain/awv338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greicius M.D., Srivastava G., Reiss A.L., Menon V. Default-mode network activity distinguishes Alzheimer's disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci U S A. 2004;101:4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones D.T., Machulda M.M., Vemuri P., McDade E.M., Zeng G., Senjem M.L. Age-related changes in the default mode network are more advanced in Alzheimer disease. Neurology. 2011;77:1524–1531. doi: 10.1212/WNL.0b013e318233b33d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Damoiseaux J.S., Prater K.E., Miller B.L., Greicius M.D. Functional connectivity tracks clinical deterioration in Alzheimer's disease. Neurobiol Aging. 2012;33:828.e19–828.e30. doi: 10.1016/j.neurobiolaging.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickerson B.C., Salat D.H., Greve D.N., Chua E.F., Rand-Giovannetti E., Rentz D.M. Increased hippocampal activation in mild cognitive impairment compared to normal aging and AD. Neurology. 2005;65:404–411. doi: 10.1212/01.wnl.0000171450.97464.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elman J.A., Oh H., Madison C.M., Baker S.L., Vogel J.W., Marks S.M. Neural compensation in older people with brain amyloid-beta deposition. Nat Neurosci. 2014;17:1316–1318. doi: 10.1038/nn.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sperling R.A., Dickerson B.C., Pihlajamaki M., Vannini P., LaViolette P.S., Vitolo O.V. Functional alterations in memory networks in early Alzheimer's disease. Neuromolecular Med. 2010;12:27–43. doi: 10.1007/s12017-009-8109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts R.O., Geda Y.E., Knopman D.S., Cha R.H., Pankratz V.S., Boeve B.F. The Mayo Clinic Study of Aging: design and sampling, participation, baseline measures and sample characteristics. Neuroepidemiology. 2008;30:58–69. doi: 10.1159/000115751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Kawas C.H. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albert M.S., DeKosky S.T., Dickson D., Dubois B., Feldman H.H., Fox N.C. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen R.C. Mild cognitive impairment as a diagnostic entity. J Intern Med. 2004;256:183–194. doi: 10.1111/j.1365-2796.2004.01388.x. [DOI] [PubMed] [Google Scholar]
- 12.Jack C.R., Jr., Knopman D.S., Weigand S.D., Wiste H.J., Vemuri P., Lowe V. An operational approach to National Institute on Aging-Alzheimer's Association criteria for preclinical Alzheimer disease. Ann Neurol. 2012;71:765–775. doi: 10.1002/ana.22628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klunk W.E., Engler H., Nordberg A., Wang Y., Blomqvist G., Holt D.P. Imaging brain amyloid in Alzheimer's disease with Pittsburgh Compound-B. Ann Neurol. 2004;55:306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- 14.Xia C.F., Arteaga J., Chen G., Gangadharmath U., Gomez L.F., Kasi D. [(18)F]T807, a novel tau positron emission tomography imaging agent for Alzheimer's disease. Alzheimers Dement. 2013;9:666–676. doi: 10.1016/j.jalz.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Schwarz A.J., Yu P., Miller B.B., Shcherbinin S., Dickson J., Navitsky M. Regional profiles of the candidate tau PET ligand 18F-AV-1451 recapitulate key features of Braak histopathological stages. Brain. 2016;139:1539–1550. doi: 10.1093/brain/aww023. [DOI] [PubMed] [Google Scholar]
- 16.Senjem M.L., Gunter J.L., Shiung M.M., Petersen R.C., Jack C.R., Jr. Comparison of different methodological implementations of voxel-based morphometry in neurodegenerative disease. Neuroimage. 2005;26:600–608. doi: 10.1016/j.neuroimage.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landau S.M., Harvey D., Madison C.M., Koeppe R.A., Reiman E.M., Foster N.L. Associations between cognitive, functional, and FDG-PET measures of decline in AD and MCI. Neurobiol Aging. 2011;32:1207–1218. doi: 10.1016/j.neurobiolaging.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jo H.J., Gotts S.J., Reynolds R.C., Bandettini P.A., Martin A., Cox R.W. Effective preprocessing procedures virtually eliminate distance-dependent motion artifacts in resting state FMRI. J Appl Math. 2013;2013 doi: 10.1155/2013/935154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones D.T., Vemuri P., Murphy M.C., Gunter J.L., Senjem M.L., Machulda M.M. Non-stationarity in the “resting brain's” modular architecture. PLoS One. 2012;7:e39731. doi: 10.1371/journal.pone.0039731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behzadi Y., Restom K., Liau J., Liu T.T. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage. 2007;37:90–101. doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hallquist M.N., Hwang K., Luna B. The nuisance of nuisance regression: spectral misspecification in a common approach to resting-state fMRI preprocessing reintroduces noise and obscures functional connectivity. Neuroimage. 2013;82:208–225. doi: 10.1016/j.neuroimage.2013.05.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calhoun V.D., Adali T., Pearlson G.D., Pekar J.J. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001;14:140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubinov M., Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Agosta F., Pievani M., Geroldi C., Copetti M., Frisoni G.B., Filippi M. Resting state fMRI in Alzheimer's disease: beyond the default mode network. Neurobiol Aging. 2012;33:1564–1578. doi: 10.1016/j.neurobiolaging.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 25.Brier M.R., Thomas J.B., Snyder A.Z., Benzinger T.L., Zhang D., Raichle M.E. Loss of intranetwork and internetwork resting state functional connections with Alzheimer's disease progression. J Neurosci. 2012;32:8890–8899. doi: 10.1523/JNEUROSCI.5698-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitwell J.L., Jones D.T., Duffy J.R., Strand E.A., Machulda M.M., Przybelski S.A. Working memory and language network dysfunction in logopenic aphasia: a task-free fMRI comparison to Alzheimer's dementia. Neurobiol Aging. 2015;36:1245–1252. doi: 10.1016/j.neurobiolaging.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glasser M.F., Sotiropoulos S.N., Wilson J.A., Coalson T.S., Fischl B., Andersson J.L. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van den Heuvel M.P., Stam C.J., Kahn R.S., Hulshoff Pol H.E. Efficiency of functional brain networks and intellectual performance. J Neurosci. 2009;29:7619–7624. doi: 10.1523/JNEUROSCI.1443-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]