Abstract
A major goal of synthetic biology is to control cell behavior. RNA-mediated genetic switches (RNA switches) are devices that serve this purpose, as they can control gene expressions in response to input signals. In general, RNA switches consist of two domains: an aptamer domain, which binds to an input molecule, and an actuator domain, which controls the gene expression. An input binding to the aptamer can cause the actuator to alter the RNA structure, thus changing access to translation machinery. The assembly of multiple RNA switches has led to complex gene circuits for cell therapies, including the selective killing of pathological cells and purification of cell populations. The inclusion of RNA binding proteins, such as L7Ae, increases the repertoire and precision of the circuit. In this short review, we discuss synthetic RNA switches for gene regulation and their potential therapeutic applications.
Keywords: synthetic biology, RNA switches, ribozymes, L7Ae, microRNA, RNA-binding proteins
Cell function depends on a network of fundamental biomolecules, including DNA, RNA and proteins. DNA and DNA-binding proteins have made up the basis of artificial devices that synthetic biologists use to control a cell. Although RNA-based systems have comparatively lagged, the realization that RNA carries genetic information like DNA but has a diversity of function more resembling proteins has intensified research on these systems.1) One example is RNA-based genetic switches (RNA switches), which transform an input signal to regulate transcription or translation. For synthetic biologists, post-transcriptional control using synthetic RNA may be advantageous, as it avoids integration into the nucleus and is therefore safer when considering therapeutic applications. In addition, RNA switches are modular, which makes them suitable for complex functions.2,3) For example, by fusing different RNA switches in tandem, synthetic biologists can produce RNA switches that act as band-pass filters.4) Moreover, the RNA motifs in an RNA structure can be replaced with other RNA motifs without changing the surrounding RNA structure, a feature that makes it relatively easy to adjust the function of the RNA switch.
One natural set of RNA switches that exemplifies post-transcriptional control includes the RNAs that cause RNA interference (RNAi).5) Of special interest are microRNAs (miRNAs), which disrupt target mRNAs and/or inhibit translation and are distinctly expressed in healthy and pathological cells.6,7) The use of RNA-binding proteins (RBPs) to alter RNA structures in ways that predictably influence gene expression expands the number of cellular functions that can be regulated while also enhancing the precision of the regulation.8) In this short review, we look at the basic elements of synthetic RNA switches and their application to control cell behavior.
RNA switches that use small molecules as input
Small-molecule responsive RNA switches (i.e., riboswitches) are all-RNA devices that regulate gene expressions at the transcriptional or translational level in response to the binding of small molecules. They possess a universal structure consisting of an aptamer domain, which detects the small molecule, and an actuator domain, which causes the conformational change by responding to ligand binding. Naturally occurring RNA switches have been frequently used to detect and regulate the amount of metabolic products.9,10) Small molecules are attractive inputs, because they are easy to prepare and exogenously introduce into the cell.
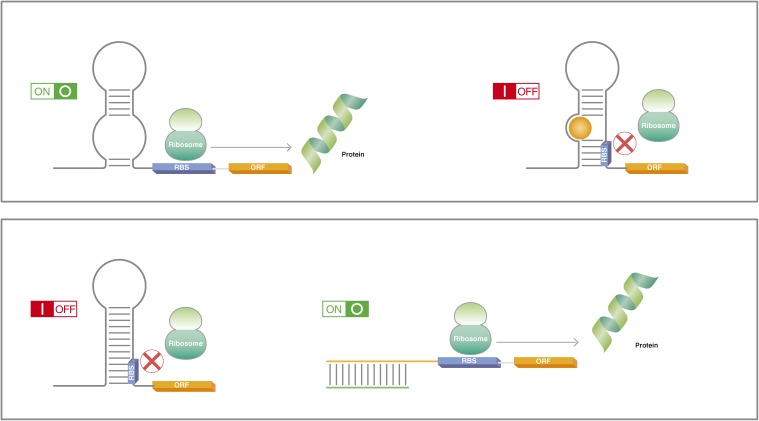
In bacterial cells, translation-regulating RNA switches modulate access to the ribosome binding site (RBS) (Fig. 1).11–13) Although binding to the aptamer triggers the actuator, the two domains are structurally independent, which allows tuning of the RNA switch by modifying the location of each domain in the RNA sequence.14,15) Further, advanced algorithms can predict the RNA structure-function relationship to optimize how the two domains should be combined.16) Recently, synthetic RNA tools have expanded the mode of action in which RNA switches operate. For example, although conventional RNA switches target the RBS to regulate translation, Liu et al. have reported adaptors that convert the regulation to target transcription.17)
Figure 1.
RNA switches are RNAs that alter their structure in response to an input signal. In prokaryotes, the conformational change normally results in the creation or release of hairpin loops or similar structures that obstruct ribosome binding to the RBS. In the upper panel, the ribosome has easy access to the RBS, allowing for the translation of the protein (green ribbon). The binding of a small molecule (orange) to the RNA near the RBS causes a conformation change that obstructs ribosome binding and thus inhibits translation. In the lower panel, the natural conformation of the RNA obstructs ribosome binding. Complementary RNA (green) binds to the hairpin loop structure, exposing the RBS to the ribosome and thus permits translation. RBS, ribosome binding site; ORF, open reading frame.
Essential to their therapeutic potential, RNA switches have also been demonstrated in eukaryotic cells. Werstruck and Green were the first to use aptamers to regulate the expression of a gene in living eukaryotic cells.18) Grate and Wilson went on to show a similar strategy could be used to target endogenous genes by inserting the malachite green aptamer into 5′UTR.19) These studies showed that RNA-ligand binding could inhibit ribosome access to downregulate translation. However, these switches operate as translational inhibitors; RNA switches that upregulate translation in response to an input signal have proven more difficult. An alternative RNA switch that solves this problem is the ribozyme, which is a natural enzyme that cleaves RNA.20,21) Synthetic RNA sequences that carry domains for both aptamers and ribozymes form aptazymes, and their modular and orthogonal nature and ON/OFF ability allows them to be assembled to form various types of Boolean gates in eukaryotic cells.22,23)
Researchers have taken advantage of these features to control diseased or infected cells. By inserting aptazymes into the UTR region of the viral transcription unit, the Nettelbeck lab could shut off viral replication by introducing to the cells theophylline.24) The ability to control viral replication could have significant implications in experimental virotherapies, such as those used to treat cancers.25) In these therapies, virus replication is used to eliminate the cancerous cells, but the replication capacity must be kept in check to avoid undesired side effects. While transcription factors can be used to shut off the replication, a theophylline-responsive aptazyme is likely safer.26) Aptazymes have also been used to regulate key body functions, such as T cell proliferation.27) Here, the ribozyme prevents the synthesis of T cell-activating cytokines, which results in T cell apoptosis. Drug binding at the aptamer inhibits ribozyme activity to allow cytokine expression and T cell expansion. Furthermore, the target cytokine is easily switched between IL-2 and IL-15, which is important for immunotherapies, since these two cytokines have different effects on T cell populations.28) Later, Wei and Smolke used this strategy to control the cell cycle in human cells by designing ribozymes that regulate the expression of key regulators to arrest the cycle at multiple stages.29)
Despite the ability to regulate various cellular phenomena, many aptazymes suffer from poor dynamic ranges, but the range can be improved by taking advantage of the modularity.30) For example, the Hartig lab redesigned ribozymes to achieve a 25-fold dynamic range in mammalian cells.31) Even so, the improved dynamic range does not ameliorate another limitation of synthetic aptazymes: their high basal activity. Beilstein et al. have shown that this problem could be mitigated by disrupting the internal loop, a secondary structure in ribozymes.32) Yet while this approach lowered the basal activity, it also diminished the dynamic range, as the fold difference became 10 times. RNA switches that have both high dynamic range and low basal activity continue to be a challenge in the field, especially in mammalian cells.
RNA switches responsive to small RNA
Although in theory any small molecule can be used as the input for RNA switches, studies have predominantly used theophylline and tetracycline. The diversity and effectiveness of RNA switches, however, would benefit from a larger library of inputs. For example, Klauser and Hartig reported a RNA switch that responds exclusively to RNAs to block translation in bacterial cells.33) Similarly, Lucks et al. used antisense RNAs to signal early termination of transcription by creating a stable hairpin structure in the target mRNA to stall RNA polymerase.34) Shen et al. have reported regazymes, which are RNA switches that use both small molecules and RNAs as their input.35) Here, small molecule binding initiates a specific cellular process that is then sustained by the output RNAs, thus acting as feedback. Using synthetic RNAs as the input signal could be an alternative to gene knockdown experiments.36)
RNA inputs have also been the foundation for a new paradigm of RNA switches. Because the limiting rate of translation initiation depends on the secondary structure of the RBS in prokaryotic cells,37) many RNA switches are designed to alter the RNA structure in this region. However, this strategy imposes several constraints on the design, as the synthetic RNAs must fit into regions upstream of the RBS, limiting the number of switches that can be incorporated.38) Alternatively, Green et al. proposed a system that depends on toehold-mediated linear-linear interactions to raise the degrees of freedom.39) They prepared toehold switches that interact with RNA elements away from the RBS by simple Watson-Click base pairing but still manage to regulate ribosome binding. This switch allowed the authors to regulate 12 genes independently and simultaneously in a single E. coli cell. The diagnostic potential of the toehold switch was demonstrated through its ability to detect different strains of Ebola virus RNA with high sensitivity.40)
In eukaryotic cells, RNAi pathways are mediated by small RNA (sRNAs) that regulate genes post-transcriptionally. There exist several types of sRNAs, including short hairpin RNA (shRNAs), short interfering RNA (siRNAs), and the aforementioned miRNAs. The Benenson lab was the first to demonstrate how sRNAs could act as Boolean gates that respond to endogenous transcription factors and assembled into higher-order gene circuits in mammalian cells.41,42) sRNAs bind to a target mRNA sequence to block translation or degrade the RNA.43) Before reaching their final state, sRNAs undergo several processing steps. For miRNAs, the first occurs in the nucleus and is done by Drosha. The resulting precursor RNA is then exported to the cytoplasm where it is processed by Dicer into the final product. Other sRNA are similarly processed by Dicer in the cytoplasm. Disrupting these two events using RNA switches is one way to regulate RNAi in the cell. The Yokobayashi group disrupted Dicer processing by inserting theophylline aptamers into shRNAs. Theophylline binding to shRNAs caused a conformational change that prevented Dicer from accessing its cleavage site.44,45) Dicer functions outside the nucleus, which means disrupting its activity is effective at regulating cytoplasmic proteins. To target nuclear proteins, aptamers have been incorporated into miRNAs to dose-dependently suppress Drosha processing.46)
There is ample evidence indicating that miRNAs could be effective therapeutic tools. Several pieces of literature have suggested that miRNA circulation is a potential diagnostic for the evaluation of inflammation and cancer,47,48) and tumor cells show lower global miRNA expression levels than normal cells.49) Further, lower expression levels of Drosha and Dicer are consistent with poorer clinical outcomes and advanced tumor stage.50,51) In fact, numerous clinical trials using miRNA-based systems are under exploration.52) Besides cancers, miRNAs show promise as biomarkers for other diseases and cell types.53) They also are influential in cell reprogramming, as they can enhance or suppress the reprogramming mechanism, which makes them valuable resources for cell therapies.54) Indeed, different miRNA expression levels between neurons and astrocytes in a heterogeneous population can be exploited to preferentially perturb the differentiation of progenitor cells.55,56) With regards to clinical use, regulating miRNA expression levels has been found to promote immunologic tolerance, suggesting good safety.57)
Accordingly, researchers have used miRNAs as ligands for synthetic switches to identify and eliminate cancerous cells. Xie et al. prepared a synthetic regulatory circuit they called a cell-type classifier that was designed to activate the synthesis of a pro-apoptosis gene, hBax, in HeLa cells but repress its synthesis in other cells.58) The circuit was designed by preparing two sets of miRNA markers, one set included miRNAs expressed unusually high in HeLa cells and the other included miRNAs expressed unusually low compared with other cells. The study went on to show how this system could selectively force apoptosis in HeLa cells from a heterogeneous population.
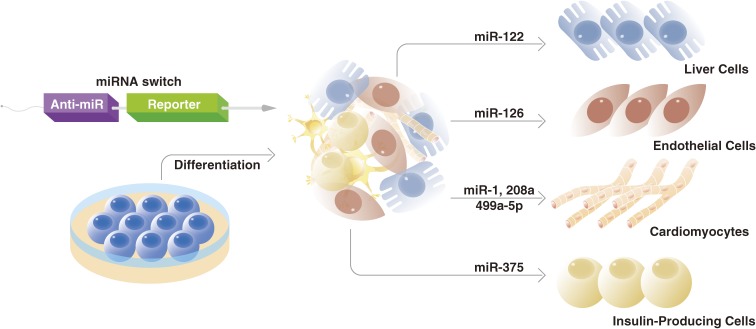
The ability to eliminate cells with high proliferative potential or other undesired characteristics has implications for cell therapies beyond cancer. In many cell therapies, the transplanted cells are prepared from progenitors cells that have proliferative potential, but during the differentiation protocol not all cells are successfully differentiated, resulting in a heterogeneous population that includes tumorigenic undifferentiated cells. Thus, prior to transplantation, the desired cells must be purified. Typically, the purification is done using antibodies that bind to specific surface markers. Yet many cell types, such as cardiomyocytes, lack unique surface markers. In response to this problem, we reported “miRNA switches”, synthetic mRNAs that use miRNAs as the input, as an alternative to antibodies for cell purification (Fig. 2).59) To construct miRNA switches, we inserted a target miRNA-complementary sequence at the 5′UTR region of a fluorescent reporter mRNA and found that co-transfection of the synthetic mRNA with reference fluorescent reporter mRNA enables the distinction of target cells with high sensitivity and efficiency. To demonstrate the capability of miRNA switches for the automatic purification (i.e., without using a cell sorter) of cardiomyocytes derived from human pluripotent stem cells, RNA sequences that included the sequence for Bim, another apoptotic factor, and sequences complementary for cardiomyocyte-specific miRNAs were inserted into the synthetic mRNA and transfected into a heterogeneous cell population. The cardiomyocyte population was purified at levels higher than those using conventional surface markers without sorting and showed good engraftment in a mouse model. The study also showed miRNA switches could select cardiomyocytes at different maturation stages. This ability adds to the advantage miRNA switches could have on cell therapies, since the maturation stage of the cardiomyocytes is associated with the outcome of the cell therapy.60) Furthermore, the original miRNA switch report confirmed that miRNA switches could be used to purify other cell types that also lack reliable surface markers, including endothelial cells, hepatocytes and insulin-producing cells. The success of miRNA switches is premised on the same concept as surface proteins; relatively disproportionate expression levels of a marker in the desired cell type. A later study showed, however, that even if the different cell types express similar levels of a miRNA, because miRNA switches can operate at remarkable resolution, the cells can still be sorted.61) Indeed, that study showed miRNA switches could distinguish cells when active miRNA levels are less than two fold different and further estimated that using just four miRNA switches could resolve hundreds of cell types, in principle.
Figure 2.
miRNA switches are synthetic RNAs that include a RNA sequence complementary to a specific miRNA sequence (anti-miR) and a RNA sequence for the expression of a desired protein (reporter). The reporter will be translated if no miRNA binds to the anti-miR. It has been discovered that many cell types express miRNAs at distinct levels. Therefore, by making the protein a pro-apoptosis factor and the anti-miR complementary to a signature miRNA, it is possible to induce apoptosis in all cell types but the one of interest. miRNA switches have been used to purify a variety of somatic cell types, including cardiomyoctyes, hepatocytes, and endothelial cells among others. They are also capable of identifying different maturation stages (not shown). Both of these abilities have important implications for cell therapies.
RBP-based synthetic RNA devices and circuits
RBPs can bind RNAs to stabilize RNA secondary and tertiary structures. These conformational changes act like RNA switches to regulate gene expressions. Because proteins regulate fundamental cellular functions, synthetic biologists have used RBPs to design protein-responsive RNA switches. The Smolke lab used aptamers that bound to proteins involved in NFκB and Wnt signaling.62) These RNA devices could control cell fate through alternative splicing to regulate the expression of herpes simplex virus-thymidine kinase, which conferred sensitivity to a pro-apoptosis drug. The same study also showed that gene splicing could be controlled with an aptamer for MS2, a bacteriophage coat protein. Bloom et al. used a similar MS2 aptamer to show that RNA switch behavior can be predicted quantitatively.63) They designed a switch in which MS2 binding caused the expression of a fluorescent marker, the level of which could be quantified by the mRNA half life and the number of miRNA target sites. This ability allowed the researchers to measure the concentration of a target nuclear protein non-invasively based on the fluorescence.
While many RBPs have been discovered in eukaryotes, viral and prokaryotic RBPs like MS2 are preferred to control synthetic RNA switches in mammalian cells because they are easier to assemble orthogonally.64) Additionally, the binding of prokaryotic RBPs results in well-defined RNA structures. One example is the k-turn, a dynamic structure that is stabilized by the binding of metal ions and also by the binding of the archaea protein L7Ae.65) We have developed L7Ae-responsive RNA switches where a L7Ae-k-turn interaction on the 5′UTR region of the target mRNA prevents ribosome function and thus prevents translation.66) At the same time, if the k-turn is incorporated into antisense RNA, then L7Ae can be used as input to activate translation by folding the antisense RNA so that the latter cannot engage with its complementary strand. We later showed that the same design could be used to control mammalian cell fate, as the expressions of Bcl-xL and FADD (Fas-Associated protein with Death Domain), both apoptosis genes, were modulated by responding to L7Ae protein levels in mammalian cells.67) Modifying the motifs to which L7Ae binds allows for tuning the translation and even the creation of negative feedback.68,69) A similar approach can be used to regulate RNAi. We showed that Dicer function can be modulated by modifying aptamers that respond to protein ligands.70) To do this, we prepared protein-responsive shRNA devices that were used to detect several proteins, including L7Ae, U1A, and NFκB p50 proteins. The inhibitory effect of the device correlated with the steric hindrance it caused between Dicer and the protein-bound RNA switch, an effect that could be predicted in silico.
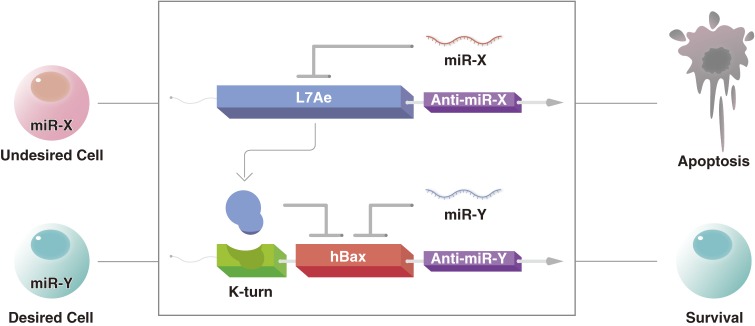
L7Ae has also been used as a basis for biocomputing in mammalian cells. For example, a plug-and-play system was designed where two different inputs were used in parallel to control the output of a reporter gene.71) Here, L7Ae was combined with MS2 and coupled with transcription regulation to generate half-adders and half-subtractors for more sophisticated computing. This circuit was delivered into the cell using plamid DNA. RNA delivery methods were used to introduce a different plug-and-play circuit that was also based on the coupling of L7Ae and MS2.72) This circuit regulated post-transcription events and was delivered using either modified RNA or RNA replicons, making it not only safer compared with DNA-delivery modules, but also indicating that both short- and long-term regulation is possible. L7Ae can further be incorporated into the miRNA-responsive cell classifier circuits described above for better precision of the circuit activity (Fig. 3). While present single-cell biocomputing depends on both transcriptional and translational regulators, it might be possible to replace the transcriptional control with miRNA switches and RNA-based circuits so that advanced biocomputing could be done using only post-translational regulation, an important iteration towards cell therapies in patients.
Figure 3.
RBPs like L7Ae can be used to improve the precision of miRNA switches. Two miRNA switches are transfected into a cell. One contains the RNA sequence for L7Ae synthesis and a sequence complementary to miRNA-X, which is expressed at distinctly high levels in an undesired cell type. The second miRNA switch carries the RNA sequence for hBax synthesis, a sequence complementary to miRNA-Y, which is distinctly expressed by the desired cell type, and the k-turn (green). When the two miRNA switches are transfected into the undesired cell type, miRNA-X prevents the expression of L7Ae. The absence of L7Ae and miRNA-Y makes the probability of hBax expression high. On the other hand, in the desired cell type, the low expression of miRNA-X results in L7Ae expression. This expression, along with the expression of miRNA-Y makes the probability of hBax expression low.
Another protein commonly used in protein-responsive RNA switches is Tetracycline Repressor (TetR). Like L7Ae, TetR binding to RNA blocks translation. The TetR-responsive RNA switch is excellent in terms of control, as it can be regulated by the doxycline inducible system, and the conventional Tet system is easy to incorporate into RNA. Using this system, the Niles lab dose-dependently controlled the translation of a fluorescent protein in yeast.73) Additionally, by modifying the TetR structure, they were able to invert the regulation such that TetR only bound to the RNA and subsequently inhibited translation upon Dox binding. In a later study, they showed this approach can be used to control in human cells the gene expression of Plasmodium falciparum, a parasite that confers malaria.74) Most recently, they demonstrated enhanced dynamic range of this system by fusing TetR with various endogenous proteins that regulate translation.75)
In contrast to the above proteins, which bind to specific RNA motifs, Pumilio and its derivatives, such as Pumilio and FBF (PUF) protein, can bind to arbitrary RNA sequences to regulate translation, primarily by controlling gene splicing. A distinctive feature of PUF domains is their high affinity for short RNA sequences. Like other RBPs, the PUF domain has been used to regulate apoptosis in mammalian cells.76) In these experiments, the PUF domain was fused with functional domains that regulate splicing to form engineered splicing factors (ESF). The PUF domain was designed to have high affinity for a target RNA sequence such that its binding triggered ESF to splice Bcl-X into one of two isoforms, one pro-apoptotic and one anti-apoptotic, thus modulating the sensitivity of the cell to anti-cancer drugs. Fusing the PUF domain with gene regulators was also shown to modulate translation.77) Additionally, PUF can regulate translation by binding to the 5′UTR without a partner protein.78) One difference between the PUF domain and L7Ae is that the former does not require a specific secondary structure to exercise its function.
Like the adaptors described in the above sections, synthetic tools can be designed to change the output behavior of protein-responsive RNA switches. For example, we produced inverters that switch the function of protein-responsive RNA switches from inhibition (OFF) to activation (ON) in mammalian cells.79) The basis of the inverter was nonsense-mediated mRNA decay (NMD). NMD shuts off translation when it detects a premature termination codon. Inserting such codons between the aptamer domain and the first codon of the output protein results in NMD in the absence of an input protein, but no NMD in the presence. Importantly, these new modules can be assembled orthogonally. As more components in the design are added, however, care must be given to the arrangement of the aptamer and actuator domains so that they do not interfere with each other’s function. The Fussenegger lab has described a general strategy for such assemblies.80) They proposed using bioinformatics and RNA libraries to identify tertiary structures that when inserted between the stem loops of the actuator domain enhance the activity of the protein-responsive RNA switch.
Conclusion
RNA has function that goes well beyond its originally defined role as an intermediate for the conversion of gene expression to protein synthesis. By interacting with substrates, RNA can alter its shape to act as a switch inside the cell. The ample progress made in controlling prokaryotic cells with these switches has been followed by consistent gains in eukaryotic cells. These advances suggest promise for new cell therapies. In particular, that RNA switches are sensitive to subtle differences in miRNA levels between cell types suggests they could be used to specifically regulate cells from heterogeneous populations, which has tremendous implications in a number of cell therapies including those for regenerative medicine and cancer. RBPs contribute to the synthetic design by enhancing the dynamic range of the switch and also by enabling higher computational processes for better regulation of cell function.
Acknowledgements
We would like to thank Masaya Todani (CiRA, Kyoto University) for the figures.
Biographies
Profile
Peter Karagiannis graduated from the University of Toronto with a B.S. in theoretical physiology followed by his Ph.D. in biophysics from Case Western Reserve University. He then arrived at Osaka University as a JSPS fellow to research live imaging of single biomolecules. At the end of the fellowship, he continued at Osaka University as an administrator. He joined the Center for iPS Cell Research and Application at Kyoto University two years ago, where he works as the institute’s science writer.
Yoshihiko Fujita studied protein engineering for a temperature-responsive nanostructure in Tokyo Institute of Technology and received Ph.D. degree in bioengineering. After graduation, he worked as a postdoctoral fellow of ICORP RNA synthetic Biology Project and was charge of a protein-responsive riboswitche project with Hirohide Saito. RNA-protein nanostructures that was a second project was studied during assistant professor in Kyoto University. Then, he joined Saito laboratory in Center for iPS Cell Research and Application again to study RNA and nanotechnology for application of iPSC.
Hirohide Saito graduated from The University of Tokyo in 1997. After completing a predoctoral training at SUNY Buffalo in U.S.A., he received his Ph.D. from the Department of Chemistry and Biotechnology, The University of Tokyo in 2002. He has been a professor in Center for iPS Cell Research and Application at Kyoto University since 2014. His research interests include bioengineering and synthetic biology.
References
- 1).Guo P. (2010) The emerging field of RNA nanotechnology. Nat. Nanotechnol. 5, 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Sudarsan N., Hammond M.C., Block K.F., Welz R., Barrick J.E., Roth A., Breaker R.R. (2006) Tandem riboswitch architectures exhibit complex gene control functions. Science 314, 300–304. [DOI] [PubMed] [Google Scholar]
- 3).Sharma V., Nomura Y., Yokobayashi Y. (2008) Engineering complex riboswitch regulation by dual genetic selection. J. Am. Chem. Soc. 130, 16310–16315. [DOI] [PubMed] [Google Scholar]
- 4).Muranaka N., Yokobayashi Y. (2010) A synthetic riboswitch with chemical band-pass response. Chem. Commun. (Camb.) 46, 6825–6827. [DOI] [PubMed] [Google Scholar]
- 5).Hannon G.J. (2002) RNA interference. Nature 418, 244–251. [DOI] [PubMed] [Google Scholar]
- 6).Qi L.S., Arkin A.P. (2014) A versatile framework for microbial engineering using synthetic non-coding RNAs. Nat. Rev. Microbiol. 12, 341–354. [DOI] [PubMed] [Google Scholar]
- 7).Garofalo M., Croce C.M. (2015) Role of microRNAs in maintaining cancer stem cells. Adv. Drug Deliv. Rev. 81, 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Saito H., Inoue T. (2007) RNA and RNP as new molecular parts in synthetic biology. J. Biotechnol. 132, 1–7. [DOI] [PubMed] [Google Scholar]
- 9).Yang J., Seo S.W., Jang S., Shin S.I., Lim C.H., Roh T.Y., Jung G.Y. (2013) Synthetic RNA devices to expedite the evolution of metabolite-producing microbes. Nat. Commun. 4, 1413. [DOI] [PubMed] [Google Scholar]
- 10).Zhou L.B., Zeng A.P. (2015) Exploring lysine riboswitch for metabolic flux control and improvement of L-lysine synthesis in Corynebacterium glutamicum. ACS Synth. Biol. 4, 729–734. [DOI] [PubMed] [Google Scholar]
- 11).Mironov A.S., Gusarov I., Rafikov R., Lopez L.E., Shatalin K., Kreneva R.A., Perumov D.A., Nudler E. (2002) Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111, 747–756. [DOI] [PubMed] [Google Scholar]
- 12).Nahvi A., Sudarsan N., Ebert M.S., Zou X., Brown K.L., Breaker R.R. (2002) Genetic control by a metabolite binding mRNA. Chem. Biol. 9, 1043–1049. [DOI] [PubMed] [Google Scholar]
- 13).Winkler W., Nahvi A., Breaker R.R. (2002) Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419, 952–956. [DOI] [PubMed] [Google Scholar]
- 14).Hanson S., Berthelot K., Fink B., McCarthy J.E., Suess B. (2003) Tetracycline-aptamer-mediated translational regulation in yeast. Mol. Microbiol. 49, 1627–1637. [DOI] [PubMed] [Google Scholar]
- 15).Suess B., Fink B., Berens C., Stentz R., Hillen W. (2004) A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 32, 1610–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Bida J.P., Das R. (2012) Squaring theory with practice in RNA design. Curr. Opin. Struct. Biol. 22, 457–466. [DOI] [PubMed] [Google Scholar]
- 17).Liu C.C., Qi L., Lucks J.B., Segall-Shapiro T.H., Wang D., Mutalik V.K., Arkin A.P. (2012) An adaptor from translational to transcriptional control enables predictable assembly of complex regulation. Nat. Methods 9, 1088–1094. [DOI] [PubMed] [Google Scholar]
- 18).Werstuck G., Green M.R. (1998) Controlling gene expression in living cells through small molecule-RNA interactions. Science 282, 296–298. [DOI] [PubMed] [Google Scholar]
- 19).Grate D., Wilson C. (2001) Inducible regulation of the S. cerevisiae cell cycle mediated by an RNA aptamer-ligand complex. Bioorg. Med. Chem. 9, 2565–2570. [DOI] [PubMed] [Google Scholar]
- 20).Winkler W.C., Nahvi A., Roth A., Collins J.A., Breaker R.R. (2004) Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428, 281–286. [DOI] [PubMed] [Google Scholar]
- 21).Yen L., Svendsen J., Lee J.S., Gray J.T., Magnier M., Baba T., D’Amato R.J., Mulligan R.C. (2004) Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature 431, 471–476. [DOI] [PubMed] [Google Scholar]
- 22).Nomura Y., Zhou L., Miu A., Yokobayashi Y. (2013) Controlling mammalian gene expression by allosteric hepatitis delta virus ribozymes. ACS Synth. Biol. 2, 684–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Win M.N., Smolke C.D. (2008) Higher-order cellular information processing with synthetic RNA devices. Science 322, 456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Ketzer P., Kaufmann J.K., Engelhardt S., Bossow S., von Kalle C., Hartig J.S., Ungerechts G., Nettelbeck D.M. (2014) Artificial riboswitches for gene expression and replication control of DNA and RNA viruses. Proc. Natl. Acad. Sci. U.S.A. 111, E554–E562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Cody J.J., Douglas J.T. (2009) Armed replicating adenoviruses for cancer virotherapy. Cancer Gene Ther. 16, 473–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Ketzer P., Haas S.F., Engelhardt S., Hartig J.S., Nettelbeck D.M. (2012) Synthetic riboswitches for external regulation of genes transferred by replication-deficient and oncolytic adenoviruses. Nucleic Acids Res. 40, e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Chen Y.Y., Jensen M.C., Smolke C.D. (2010) Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc. Natl. Acad. Sci. U.S.A. 107, 8531–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Karagiannis P., Iriguchi S., Kaneko S. (2015) Reprogramming away from the exhausted T cell state. Semin. Immunol. 28, 35–44. [DOI] [PubMed] [Google Scholar]
- 29).Wei K.Y., Smolke C.D. (2015) Engineering dynamic cell cycle control with synthetic small molecule-responsive RNA devices. J. Biol. Eng. 9, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Win M.N., Smolke C.D. (2007) A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl. Acad. Sci. U.S.A. 104, 14283–14288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Klauser B., Atanasov J., Siewert L.K., Hartig J.S. (2015) Ribozyme-based aminoglycoside switches of gene expression engineered by genetic selection in S. cerevisiae. ACS Synth. Biol. 4, 516–525. [DOI] [PubMed] [Google Scholar]
- 32).Beilstein K., Wittmann A., Grez M., Suess B. (2015) Conditional control of mammalian gene expression by tetracycline-dependent hammerhead ribozymes. ACS Synth. Biol. 4, 526–534. [DOI] [PubMed] [Google Scholar]
- 33).Klauser B., Hartig J.S. (2013) An engineered small RNA-mediated genetic switch based on a ribozyme expression platform. Nucleic Acids Res. 41, 5542–5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Lucks J.B., Qi L., Mutalik V.K., Wang D., Arkin A.P. (2011) Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc. Natl. Acad. Sci. U.S.A. 108, 8617–8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35).Shen S., Rodrigo G., Prakash S., Majer E., Landrain T.E., Kirov B., Daros J.A., Jaramillo A. (2015) Dynamic signal processing by ribozyme-mediated RNA circuits to control gene expression. Nucleic Acids Res. 43, 5158–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Peters G., Coussement P., Maertens J., Lammertyn J., De Mey M. (2015) Putting RNA to work: Translating RNA fundamentals into biotechnological engineering practice. Biotechnol. Adv. 33, 1829–1844. [DOI] [PubMed] [Google Scholar]
- 37).de Smit M.H., van Duin J. (1990) Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc. Natl. Acad. Sci. U.S.A. 87, 7668–7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Auslander S., Fussenegger M. (2014) Synthetic biology: Toehold gene switches make big footprints. Nature 516, 333–334. [DOI] [PubMed] [Google Scholar]
- 39).Green A.A., Silver P.A., Collins J.J., Yin P. (2014) Toehold switches: de-novo-designed regulators of gene expression. Cell 159, 925–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Pardee K., Green A.A., Ferrante T., Cameron D.E., DaleyKeyser A., Yin P., Collins J.J. (2014) Paper-based synthetic gene networks. Cell 159, 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Rinaudo K., Bleris L., Maddamsetti R., Subramanian S., Weiss R., Benenson Y. (2007) A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotechnol. 25, 795–801. [DOI] [PubMed] [Google Scholar]
- 42).Leisner M., Bleris L., Lohmueller J., Xie Z., Benenson Y. (2010) Rationally designed logic integration of regulatory signals in mammalian cells. Nat. Nanotechnol. 5, 666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43).He L., Hannon G.J. (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531. [DOI] [PubMed] [Google Scholar]
- 44).An C.I., Trinh V.B., Yokobayashi Y. (2006) Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction. RNA 12, 710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Tuleuova N., An C.I., Ramanculov E., Revzin A., Yokobayashi Y. (2008) Modulating endogenous gene expression of mammalian cells via RNA-small molecule interaction. Biochem. Biophys. Res. Commun. 376, 169–173. [DOI] [PubMed] [Google Scholar]
- 46).Beisel C.L., Chen Y.Y., Culler S.J., Hoff K.G., Smolke C.D. (2011) Design of small molecule-responsive microRNAs based on structural requirements for Drosha processing. Nucleic Acids Res. 39, 2981–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).He X., Li J., Guo W., Liu W., Yu J., Song W., Dong L., Wang F., Yu S., Zheng Y., Chen S., Kong Y., Liu C. (2015) Targeting the microRNA-21/AP1 axis by 5-fluorouracil and pirarubicin in human hepatocellular carcinoma. Oncotarget 6, 2302–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Pua H.H., Ansel K.M. (2015) MicroRNA regulation of allergic inflammation and asthma. Curr. Opin. Immunol. 36, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Lin S., Gregory R.I. (2015) MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 15, 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Lin R.J., Lin Y.C., Chen J., Kuo H.H., Chen Y.Y., Diccianni M.B., London W.B., Chang C.H., Yu A.L. (2010) microRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res. 70, 7841–7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51).Merritt W.M., Lin Y.G., Han L.Y., Kamat A.A., Spannuth W.A., Schmandt R., Urbauer D., Pennacchio L.A., Cheng J.F., Nick A.M., Deavers M.T., Mourad-Zeidan A., Wang H., Mueller P., Lenburg M.E., Gray J.W., Mok S., Birrer M.J., Lopez-Berestein G., Coleman R.L., Bar-Eli M., Sood A.K. (2008) Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med. 359, 2641–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).Bobbin M.L., Rossi J.J. (2016) RNA Interference (RNAi)-Based Therapeutics: Delivering on the Promise? Annu. Rev. Pharmacol. Toxicol. 56, 103–122. [DOI] [PubMed] [Google Scholar]
- 53).Romaine S.P., Tomaszewski M., Condorelli G., Samani N.J. (2015) MicroRNAs in cardiovascular disease: an introduction for clinicians. Heart 101, 921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54).Bernal J.A. (2013) RNA-based tools for nuclear reprogramming and lineage-conversion: towards clinical applications. J. Cardiovasc. Transl. Res. 6, 956–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55).Colin A., Faideau M., Dufour N., Auregan G., Hassig R., Andrieu T., Brouillet E., Hantraye P., Bonvento G., Deglon N. (2009) Engineered lentiviral vector targeting astrocytes in vivo. Glia 57, 667–679. [DOI] [PubMed] [Google Scholar]
- 56).Naka-Kaneda H., Nakamura S., Igarashi M., Aoi H., Kanki H., Tsuyama J., Tsutsumi S., Aburatani H., Shimazaki T., Okano H. (2014) The miR-17/106-p38 axis is a key regulator of the neurogenic-to-gliogenic transition in developing neural stem/progenitor cells. Proc. Natl. Acad. Sci. U.S.A. 111, 1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Annoni A., Brown B.D., Cantore A., Sergi L.S., Naldini L., Roncarolo M.G. (2009) In vivo delivery of a microRNA-regulated transgene induces antigen-specific regulatory T cells and promotes immunologic tolerance. Blood 114, 5152–5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58).Xie Z., Wroblewska L., Prochazka L., Weiss R., Benenson Y. (2011) Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 333, 1307–1311. [DOI] [PubMed] [Google Scholar]
- 59).Miki K., Endo K., Takahashi S., Funakoshi S., Takei I., Katayama S., Toyoda T., Kotaka M., Takaki T., Umeda M., Okubo C., Nishikawa M., Oishi A., Narita M., Miyashita I., Asano K., Hayashi K., Osafune K., Yamanaka S., Saito H., Yoshida Y. (2015) Efficient Detection and Purification of Cell Populations Using Synthetic MicroRNA Switches. Cell Stem Cell 16, 699–711. [DOI] [PubMed] [Google Scholar]
- 60).Funakoshi S., Miki K., Takaki T., Okubo C., Hatani T., Chonabayashi K., Nishikawa M., Takei I., Oishi A., Narita M., Hoshijima M., Kimura T., Yamanaka S., Yoshida Y. (2016) Enhanced engraftment, proliferation, and therapeutic potential in heart using optimized human iPSC-derived cardiomyocytes. Sci. Rep. 6, 19111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61).Endo K., Hayashi K., Saito H. (2016) High-resolution Identification and Separation of Living Cell Types by Multiple microRNA-responsive Synthetic mRNAs. Sci. Rep. 6, 21991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62).Culler S.J., Hoff K.G., Smolke C.D. (2010) Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science 330, 1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63).Bloom R.J., Winkler S.M., Smolke C.D. (2014) A quantitative framework for the forward design of synthetic miRNA circuits. Nat. Methods 11, 1147–1153. [DOI] [PubMed] [Google Scholar]
- 64).Kopniczky M.B., Moore S.J., Freemont P.S. (2015) Multilevel Regulation and Translational Switches in Synthetic Biology. IEEE Trans. Biomed. Circuits Syst. 9, 485–496. [DOI] [PubMed] [Google Scholar]
- 65).Lilley D.M. (2014) The K-turn motif in riboswitches and other RNA species. Biochim. Biophys. Acta 1839, 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66).Saito H., Kobayashi T., Hara T., Fujita Y., Hayashi K., Furushima R., Inoue T. (2010) Synthetic translational regulation by an L7Ae-kink-turn RNP switch. Nat. Chem. Biol. 6, 71–78. [DOI] [PubMed] [Google Scholar]
- 67).Saito H., Fujita Y., Kashida S., Hayashi K., Inoue T. (2011) Synthetic human cell fate regulation by protein-driven RNA switches. Nat. Commun. 2, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68).Endo K., Stapleton J.A., Hayashi K., Saito H., Inoue T. (2013) Quantitative and simultaneous translational control of distinct mammalian mRNAs. Nucleic Acids Res. 41, e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69).Stapleton J.A., Endo K., Fujita Y., Hayashi K., Takinoue M., Saito H., Inoue T. (2012) Feedback control of protein expression in mammalian cells by tunable synthetic translational inhibition. ACS Synth. Biol. 1, 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70).Kashida S., Inoue T., Saito H. (2012) Three-dimensionally designed protein-responsive RNA devices for cell signaling regulation. Nucleic Acids Res. 40, 9369–9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71).Auslander S., Auslander D., Muller M., Wieland M., Fussenegger M. (2012) Programmable single-cell mammalian biocomputers. Nature 487, 123–127. [DOI] [PubMed] [Google Scholar]
- 72).Wroblewska L., Kitada T., Endo K., Siciliano V., Stillo B., Saito H., Weiss R. (2015) Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat. Biotechnol. 33, 839–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73).Goldfless S.J., Belmont B.J., de Paz A.M., Liu J.F., Niles J.C. (2012) Direct and specific chemical control of eukaryotic translation with a synthetic RNA-protein interaction. Nucleic Acids Res. 40, e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74).Goldfless S.J., Wagner J.C., Niles J.C. (2014) Versatile control of Plasmodium falciparum gene expression with an inducible protein-RNA interaction. Nat. Commun. 5, 5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75).Ganesan S.M., Falla A., Goldfless S.J., Nasamu A.S., Niles J.C. (2016) Synthetic RNA-protein modules integrated with native translation mechanisms to control gene expression in malaria parasites. Nat. Commun. 7, 10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76).Wang Y., Cheong C.G., Hall T.M., Wang Z. (2009) Engineering splicing factors with designed specificities. Nat. Methods 6, 825–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77).Cooke A., Prigge A., Opperman L., Wickens M. (2011) Targeted translational regulation using the PUF protein family scaffold. Proc. Natl. Acad. Sci. U.S.A. 108, 15870–15875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78).Cao J., Arha M., Sudrik C., Mukherjee A., Wu X., Kane R.S. (2015) A universal strategy for regulating mRNA translation in prokaryotic and eukaryotic cells. Nucleic Acids Res. 43, 4353–4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79).Endo K., Hayashi K., Inoue T., Saito H. (2013) A versatile cis-acting inverter module for synthetic translational switches. Nat. Commun. 4, 2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80).Auslander S., Stucheli P., Rehm C., Auslander D., Hartig J.S., Fussenegger M. (2014) A general design strategy for protein-responsive riboswitches in mammalian cells. Nat. Methods 11, 1154–1160. [DOI] [PubMed] [Google Scholar]