Summary
Industrial biomining has been extensively used for many years to recover valuable metals such as copper, gold, uranium and others. Furthermore, microorganisms involved in these processes can also be used to bioremediate places contaminated with acid and metals. These uses are possible due to the great metal resistance that these extreme acidophilic microorganisms possess. In this review, the most recent findings related to copper resistance mechanisms of bacteria and archaea related to biohydrometallurgy are described. The recent search for novel metal resistance determinants is not only of scientific interest but also of industrial importance, as reflected by the genomic sequencing of microorganisms present in mining operations and the search of those bacteria with extreme metal resistance to improve the extraction processes used by the biomining companies.
Introduction
Biomining is used to enhance the recovery of metals from ores such as sulfide minerals. Bioleaching solubilizes the metal of interest by biological catalysis as in the case of copper. On the other hand, during biooxidation microorganisms dissolve the mineral matrix occluding the metal to be extracted as it occurs in the case of gold (Kaksonen et al., 2014; Urbieta et al., 2015; Harrison, 2016). These mineral–microbe interactions to recover metals from ores have been widely used in small and large scales (Brierley and Brierley, 2013; Norris et al., 2013; Harrison, 2016). An acidophilic microbial consortium is in charge of metals mobilization. In practice, commercial biomining procedures are mainly used for copper bioleaching and the biooxidation of refractory gold ores (Brierley and Brierley, 2013; Kaksonen et al., 2014; Urbieta et al., 2015). During minerals bioleaching acid mine drainage is generated which can cause serious environmental problems. However, bioremediation or removal of the toxic metals from contaminated soils can be achieved by using abiotic methods or the specific properties of the acidophilic microorganisms interacting with these elements (Johnson and Hallberg, 2005).
Industrial biomining operations are of several kinds depending on the ore type and its geographical location, the metal content and the specific minerals present (metal oxides and mostly secondary metal sulfides such as covellite, chalcocite and others). Metal recovery processes can be carried out by using tank, heap, dump and in situ leaching systems (reviewed in detail by Harrison, 2016). One of the most used setups for the recovery of copper is the irrigation type of processes. These involve the percolation of leaching solutions through the crushed ore that can be contained in a column, heap or dump (Watling et al., 2010; Jerez, 2011; Brierley and Brierley, 2013). The pregnant solution containing copper sulfate generated by the microbial solubilization of the insoluble copper sulfides present in the ore, is subjected to solvent extraction to obtain a highly concentrated copper sulfate solution from which the metal is recovered in an electrowinning plant to generate electrolytic copper of high purity (Watling et al., 2010; Harrison, 2016).
On the other hand, as reactors are expensive to build, they are used with high‐grade ores or with mineral concentrates. The advantages of tank reactors over heaps and dumps, which are ‘open bioreactors’ is that in the tanks conditions can be controlled. The use of hyperthermophilic archaea for mineral sulfide processing has been studied mainly because they appear to have greater tolerances to high copper concentrations and solid particle concentrations that take place in bioreactors compared with other thermoacidophiles (Brierley and Brierley, 2013; Norris et al., 2013; Harrison, 2016). Therefore, the search for key efficient mineral‐oxidizing strains with very high metal resistance may be of great importance to develop improved industrial bioreactors for refractory minerals (Norris et al., 2013; Wheaton et al., 2015; Urbieta et al., 2015).
Thermophilic archaea are able to oxidize mineral sulfides at temperatures higher than 60°C. Among these, Acidianus brierleyi, Sulfolobus metallicus and Metallosphaera have been the most studied (Brierley and Murr, 1973; Lindström and Gunneriusson, 1990; Jones et al., 2011; Norris et al., 2013; Wheaton et al., 2015). When a microbial community is used to start a series of tanks for biomining, metal concentrations in the leachate increase and those species resistant to extremely high metal concentrations (as well as other compounds such as organic carbon) are selected in the final tank (Okibe et al., 2003; Dopson and Holmes, 2014). This suggests that those microbial species with higher metal resistances may be advantageous for the biomining process. In this regard, a spontaneous mutant strain of Metallosphaera sedula (CuR1) with supranormal metal resistance has been reported to leach copper from chalcopyrite (CuFeS2) at an accelerated rate (Maezato et al., 2012) (see the archaeal section below).
Several recent reviews related to biomining methods and the microorganisms involved are available (Brierley and Brierley, 2013; Norris et al., 2013; Vera et al., 2013; Kaksonen et al., 2014; Wheaton et al., 2015; Zhuang et al., 2015; Cárdenas et al., 2016; Harrison, 2016; Hedrich and Schippers, 2016). They cover bacteria and archaea involved in biohydrometallurgy, and describe the new findings obtained by using Omics techniques, such as genomic DNA sequencing of single species or metagenomic sequencing of the microbial communities involved, proteomics and metaproteomics and metabolomics (Martínez et al., 2015). The recently published book ‘Acidophiles: Life in Extremely Acidic Environments’ (Quatrini and Johnson, 2016) is also a reference of great interest as most of its chapters are related to aspects important for bioleaching.
A mini‐review related to several metals resistance in acidophilic microorganisms has been published before (Dopson and Holmes, 2014). In the present review, we concentrate in critically analysing the most recent findings related to copper resistance and its importance in copper sulfides bioleaching.
Microorganisms and bioleaching communities
The most studied leaching bacteria are from the Acidithiobacillia class. Acidithiobacillus ferrooxidans and A. thiooxidans are acidophilic mesophiles and together with the moderate thermophile, A. caldus, are some of the most studied members of the biomining consortium. The role in biomining of many other acidophilic microorganisms present in natural and man‐made acidic environments has recently been reviewed (Hedrich and Schippers, 2016). Although A. ferrooxidans has been described to oxidize metal sulfides even at 0°C (Langdahl and Ingvorsen, 1997), no psychrophilic acidophiles (optimal growth temperature lower than 15°C) have been reported. Some psychrotolerant microorganisms such as Acidithiobacillus ferrivorans have been also isolated that can catalyse metal dissolution from sulfide minerals at 5°C. These bacteria are of importance in biomining operations as several of them are present at high altitudes and in Polar regions and therefore can be used for biotechnological operations in cold environments (Dopson, 2016).
Chalcopyrite is the most abundant copper‐containing mineral (around 70% of the world's copper reserves). However, it is the most difficult mineral to solubilize by mesophilic microorganisms, taking much longer time to recover copper. Therefore, there is actually great interest in developing processes by using thermophilic biomining microorganisms for these recalcitrant minerals.
Genetic manipulation and recent advances using OMICS to analyse biomining microorganisms
It has been postulated that it is important to know the copper resistance mechanisms of bioleaching microorganisms in order to construct strains with high copper tolerance by using genetic modification (Wen et al., 2014). Transformation of some biomining microorganisms has been possible. Gene knockout systems were previously reported in the case of A. ferrooxidans and A. caldus (Liu et al., 2000; Van Zyl et al., 2008). These transformations were performed by using a method based on marker exchange mutagenesis. Later on, an improved method of markerless gene replacement was established for A. ferrooxidans (Wang et al., 2012). However, the reported procedures are difficult to reproduce, very laborious and have very low efficiencies. Recently, a versatile and efficient markerless gene knockout system for A. thiooxidans was reported and used to characterize a copper tolerance related multicopper oxidase (MCO) gene in this microorganism (Wen et al., 2014).
Instead, some authors consider that the engineering constraints of all forms of biomining operations (including stirred tanks) means that it is not possible to prevent the release of microorganisms used in these processes to the environment (Johnson, 2014). An alternative to this situation is the selection of microorganisms from biomining areas that possess a better natural copper resistance together with a higher capacity to bioleach mineral sulfides. A. ferrooxidans strain ATCC 53993 might be an example of this kind of bacteria, as it is able to resist high copper concentrations (10–12 g L−1 range) (Orellana and Jerez, 2011) and capable of oxidizing both reduced inorganic sulfur compounds (RISCs) and ferrous iron. In the case of A. thiooxidans and A. caldus, as they oxidize only RISCs, an additional strain such as Leptospirillum ferrooxidans or a similar ferrous iron oxidizer would be required for suitable bioleaching of metal sulfides. Some biomining companies such as BioSigma, S.A. are currently using this approach and have isolated native species from copper mines such as A. ferrooxidans strain Wenelen (DSM 16786) and others with increased activity for copper leaching from mixed sulfide ores and high copper resistance (higher than 10 g L−1 compared with 5 g L−1 of the type A. ferrooxidans ATCC 23270 strain) for their use in industrial processes (Sugio et al., 2009; Latorre et al., 2016).
Proteomics, metaproteomics, genomics and metagenomics, together with metabolomics, have been widely used in recent years to study the global regulatory responses including metal resistance systems of all kinds of cells individually or in complex microbial communities (Gillan, 2016). By this approach it may be possible to explore the new properties of biomining microorganisms that arise from the interplay of genes, proteins, other macromolecules, small molecules and the environment. These procedures enable to build metabolic models, predict microbial interactions, defining genetic diversity and study microbial evolution. Some of the findings obtained by using these molecular procedures for the study of biomining microorganisms and their metal resistance have been recently reviewed (Martínez et al., 2015; Cárdenas et al., 2016). Genomics has been of great importance to define the biodiversity of biomining processes and has allowed developing both conceptual and metabolic models for some of the acidophilic microorganisms involved. By using metabolomics, some specific metabolites may be used as markers to follow the evolution in time of industrial bioleaching operations (Martínez et al., 2013). On the other hand, proteomics has been useful to study microorganisms–mineral interactions and biofilm formation (Martínez et al., 2015), including copper resistance as described in the next section.
Global changes in gene expression and metabolite levels allow exploring both cannonical and new possible mechanisms of metal resistance and also likely unknown passive or active metal tolerance systems. It is expected that a detailed knowledge of the responses that these environmental microorganisms use to adapt to their harsh niche will help to improve biomining and metal bioremediation in industrial processes (Jerez, 2013; Martínez et al., 2015).
Copper resistance mechanisms
Acid‐leaching solutions are characterized by concentrations of base and transition metals that are toxic to most living organisms, and as might be expected, microorganisms that grow in mineral‐rich environments are, in most cases, remarkably tolerant to a wide range of metal ions and should possess robust metal resistance mechanisms (Orell et al., 2012; Dopson and Holmes, 2014).
Biomining bacteria and archaea resist high levels of copper (100–300 mM range) by using a few ‘canonical systems’ such as active efflux or trapping of the metal ions by metal chaperones. Nonetheless, gene duplications, the presence of genomic islands (GI), the existence of additional strategies such as passive mechanisms for pH and cations homeostasis in acidophiles, and an inorganic polyphosphate (polyP)‐driven metal resistance mechanism have also been proposed (Chi et al., 2007; Orell et al., 2010, 2012; Orellana and Jerez, 2011; Navarro et al., 2013; Dopson and Holmes, 2014; Wen et al., 2014; Wheaton et al., 2015). Next a selection of the most studied biomining microorganisms is analysed regarding their copper resistance capacity.
Copper resistance of biomining bacteria
Acidithiobacillus ferrooxidans
Acidithiobacillus ferrooxidans ATCC 23270 can survive under high copper concentrations. It does so by carrying in its genome more than 10 genes related to copper homeostasis in other bacteria (Rensing and Grass, 2003; Nikaido, 2011). Three of these genes code for putative ATPases related to the transport of copper (copA1 Af, copA2 Af and copB Af), three other genes are related to a system of the RND family, involved in the extraction of copper from the cell by using the proton motive force (PMF; cusA Af, cusB Af, cusC Af) and two additional genes code for periplasmic chaperones for this metal (cusF Af and copC Af) (Navarro et al., 2009). These A. ferrooxidans copper resistance determinants were found to be upregulated when this bacterium was exposed to CuSO4 in the range of 5–25 mM and conferred a greater resistance to copper when expressed in Escherichia coli compared with wild‐type cells, supporting their functionality (Navarro et al., 2009).
Acidithiobacillus ferrooxidans and other acidophiles live at an acid external pH (1–3) and their cytoplasmic pH is up to 5 units higher than the external pH. This generates an elevated pH gradient across the cytoplasmic membrane that contributes to the PMF comprising the membrane potential (ΔΨ) and the transmembrane pH difference (ΔpH) (Baker‐Austin and Dopson, 2007). The RND‐type transporters are antiporters taking advantage of the protons gradient for the efflux of copper with protons entrance to the cytoplasm. Due to its economy from the energetic point of view, the acidophilic microorganisms would preferentially use these systems to remove intracellular copper. A possible cytoplasmic acidification would be expected to take place if the microorganisms would excessively use these efflux pumps in the presence of high metal concentration. However, this acidification could be diminished by the energetic metabolism of the bacterium, as the oxidation of Fe(II) or reduced sulfur compounds by molecular oxygen as the final electron acceptor consumes protons. As the RND systems are introducing protons from the culture medium to the cell during copper detoxification, an increase in the extracellular pH of the growth medium would be expected in the presence of this metal.
The genomic sequence of A. ferrooxidans ATCC 53993 contains all the copper resistance genes present in A. ferrooxidans ATCC 23270 that have been experimentally confirmed as being expressed in the presence of copper, except that strain ATCC 23270 contains a second Cus‐like operon interrupted by a transposase gene which is absent in the genome of strain ATCC 53993 (Luo et al., 2008; Navarro et al., 2009; Orell et al., 2010; Orellana and Jerez, 2011). These ORFs have 100% identity between their corresponding DNA sequences. However, A. ferrooxidans ATCC 53993 contains several additional putative metal resistance ORFs that confer it a higher metal resistance (Orellana and Jerez, 2011). These putative genes are clustered in a 160 kb GI that is absent in the genome of A. ferrooxidans strain ATCC 23270 (Cárdenas et al., 2010; Orell et al., 2010). The GI in A. ferrooxidans ATCC 53993 contains genes encoding heavy metal resistance determinants for copper, mercury detoxification and volatilization, and the transcriptional regulator (merR), as well as a copper translocating P‐type ATPase. On the other hand, strain ATCC 23270 has a 300 kb region that does not contain known metal resistance‐related genes and is absent in the genome of strain ATCC 53993 (Cárdenas et al., 2010; Bustamante et al., 2012). Obviously, the presence of unknown metal resistance genes in this 300 kb GI cannot be excluded at present.
Six ORFs possibly related to Cu resistance are present in the GI of strain ATCC 53993, including a P‐type ATPase annotated as a possible Cu resistance determinant or copA3 Af (Orellana and Jerez, 2011). All Cop‐like proteins found so far in A. ferrooxidans, including CopA3Af contain the metal‐binding motifs that are highly conserved in Cop proteins from other microorganisms (Orell et al., 2010).
Other of the ORFs found in the GI of A. ferrooxidans ATCC 53993 corresponded to members of a putative Cus system (cusA3 Af, cusB3 Af and cusC3 Af). Two additional possible CusF‐like chaperones previously described in A. ferrooxidans ATCC 23270 (Navarro et al., 2009) were found in this GI (Orellana and Jerez, 2011). A comparison of the possible copper resistance genes between these two A. ferrooxidans strains and several other selected biomining microorganisms is shown in Figure 1.
Figure 1.
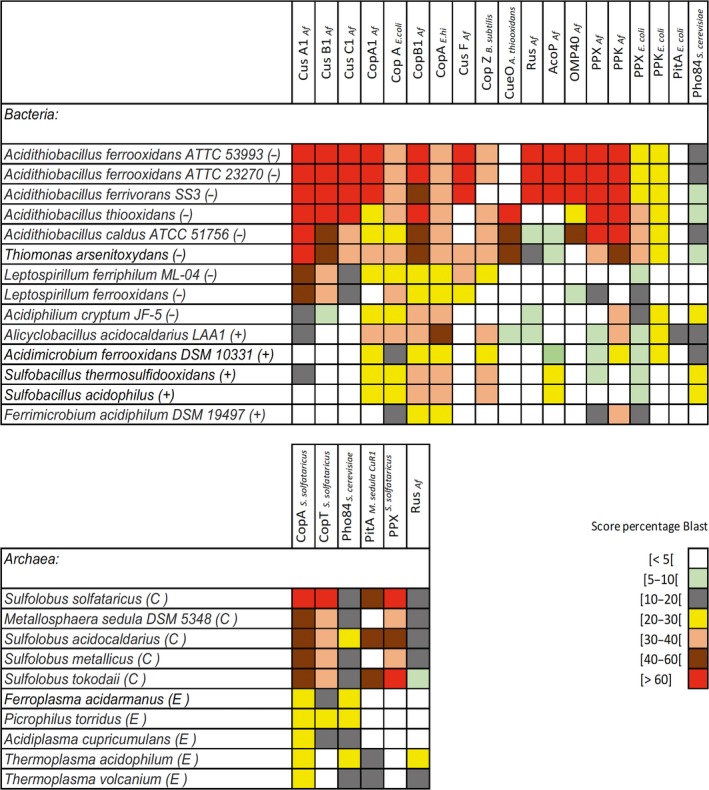
Comparison of the main copper resistance determinants present in a selection of acidophilic bacteria and archaea usually found in biomining environments. All the microorganisms chosen have their genomes sequenced and are publically available. The proteins selected for comparison are those for which at least some experimental evidence as copper resistance determinants exists. Symbols: (−) Gram negative; (+) Gram positive; (C) Crenarchaeota; (E) Euryarchaeota. The Blastp program was used to align protein sequences by means of BLOSUM62 substitution matrix. The colours scale indicate the percentage of the obtained scores related to the maximum score value obtained. The entrance protein sequences are in each column of the table and when more than one protein with a significant score was present in the same microorganism the highest score was taken in each case. Af, Acidithiobacillus ferrooxidans; E. hi, Enteroccoccus hirae.
Acidithiobacillus ferrooxidans ATCC 53993 has a much higher resistance to CuSO4 (>100 mM) than that of strain ATCC 23270 (<40 mM) (Orellana and Jerez, 2011). As it occurs in Cupriavidus metallidurans (Van Houdt et al., 2009), it was expected that the additional multiplicity of possible copper resistance determinants in the GI of A. ferrooxidans ATCC 53993 would confer it a higher tolerance to the metal compared with strain ATCC 23270. Although both strains have a similar growth in the absence of copper, cell numbers of ATCC 23270 were reduced by sevenfold when grown in 100 mM CuSO4, whereas those of ATCC 53993 were diminished by only twofold (Orellana and Jerez, 2011; Jerez, 2013). This additional capacity of strain ATCC 53993 to tolerate copper should confer it an adaptational advantage when growing in a microbial consortium such as the one usually found in their habitat. If these two A. ferrooxidans strains were present in a bioleaching operation such as a heap or more likely in an industrial biooxidation reactor, as copper concentration increases with the dissolution of the mineral, at the end of the operation strain ATCC 53993 should predominate over ATCC 23270. A preliminary test for this idea was carried out by growing a mixture of approximately equivalent numbers of each strain in the presence of copper. The relative proportions of each kind of bacterium were estimated by means of qPCR by using strain‐specific primers. The results obtained clearly indicated that A. ferrooxidans ATCC 53993 was able to outgrow strain ATCC 23270 in the presence of 50 mM CuSO4 (Orellana and Jerez, 2011; Jerez, 2013).
Moreover, an upregulation of the transcriptional expression of most of the additional Cu resistance genes present in the GI of A. ferrooxidans ATCC 53993 was observed when cells were grown in the presence of increasing CuSO4 concentrations. In addition, these genes were functional when expressed in E. coli, strongly supporting the functionality of the copper resistance determinants present in the GI of A. ferrooxidans ATCC 53993 (Orellana and Jerez, 2011). These findings constituted the first experimental evidence for high copper resistance due to the expression of genes present in a GI of an acidophilic chemolithoautotrophic bacterium.
Interestingly, when A. ferrooxidans strains ATCC 53993 vs ATCC 23270 subjected to 40 mM copper were analysed by differential quantitative proteomics, only 49 proteins changed in the former strain vs 120 proteins in the latter one (Martínez‐Bussenius et al., 2016). This greatly diminished response to the metal is in good agreement with the much higher copper resistance of A. ferrooxidans ATCC 53993 already described. These inter‐population interactions may not only lead to changes in the capacities of the bacteria to adapt to their environment but may also help to select the more suitable microorganisms to enhance industrial biomining operations.
Genomic strain variations have also been found in the acidophilic Leptospirillum group II by deep metagenomic genome sequence analysis (Simmons et al., 2008). It is possible that horizontal gene transfer between A. ferrooxidans strains and other microorganisms are key elements to supplement metal resistance and possibly other properties, thus conferring adaptational advantages to all these microorganisms.
One of the most abundant proteins in ferrous iron‐grown A. ferrooxidans is the cupredoxin rusticyanin (Rus) as it occupies 21% of the total volume in the periplasmic space (Li et al., 2015). This periplasmic component forms part of an iron‐oxidizing/oxygen‐reducing supercomplex spanning both inner and outer membranes in A. ferrooxidans (Yarzával et al., 2004; Castelle et al., 2008). Rus and other components of the rus operon have also been found expressed at lower levels in sulfur‐grown cells (Ramírez et al., 2004; Yarzával et al., 2004). However, the role for Rus in sulfur‐grown cells is still not clearly defined (Yarzával et al., 2004). Thus, the overexpression of Rus in sulfur‐grown cells exposed to copper supports the idea that this copper‐binding protein and possibly other components of the rus operon may have an additional role in A. ferrooxidans copper resistance, as previously suggested (Felício et al., 2003), and recently supported by Almárcegui et al. (2014a). In this regard, the analysis of the A. ferrooxidans periplasm by high‐throughput proteomics including high‐resolution linear ion trap‐FT MS allowed the identification of 131 periplasmic proteins (Chi et al., 2007). Among these proteins, several were uncharacterized, including AFE_3151. This protein was later proposed to be part of the iron‐oxidizing/O2‐reducing system in A. ferrooxidans although its role has not been defined so far (Castelle et al., 2008, 2010). This periplasmic protein, now named AcoP, has recently been characterized and it also contains a cupredoxin type copper‐binding site that binds one Cu atom per molecule and likewise forms part of the rus operon and iron respiratory system (Roger et al., 2014). Furthermore, the levels of AcoP also increased in sulfur‐grown cells in the presence of copper (Almárcegui et al., 2014a). Rus and AcoP have conserved copper‐binding sites (HXCX2‐4HX4M and HXCX4‐8HX4M respectively) (Roger et al., 2014) such as those described in CopC, a cupredoxin‐like copper‐binding protein from P. syringae involved in copper homeostasis (Arnesano et al., 2002).
By using high‐throughput quantitative ICPL proteomics, it was found that A. ferrooxidans ATCC 23270 grown in ferrous iron or elemental sulfur and in the presence of 40 mM of copper sulfate changes the expression of several proteins related to copper response, such as the upregulation of a Cus system (Almárcegui et al., 2014a,b), confirming the previously reported transcriptional analysis (Navarro et al., 2009). Furthermore, the upregulation (almost 15‐fold) of a protein coded by AFE_1862, annotated in the genome of A. ferrooxidans ATCC 23270 as a putative heavy metal‐binding protein were also detected by the proteomic approach (Almárcegui et al., 2014b). This protein contains a possible heavy metal‐binding domain (MXCXXC) similar to the one found in CopZ‐like proteins.
The possible role of Rus and AcoP periplasmic cupredoxins and of the putative cytoplasmic CopZ in copper resistance in A. ferrooxidans was recently reported (Navarro et al., 2016). Although CopZ is a known cytoplasmic copper chaperone that binds and delivers copper to CopA in B. subtilis (Radford et al., 2003), the CopZ‐like protein of A. ferrooxidans ATCC 23270 is the first cytoplasmic copper chaperone described in a biomining bacterium with a role in copper resistance. Likewise, Rus and AcoP cupredoxins apparently function not only as components of the ferrous iron oxidation pathway in this acidophilic microorganism but they may also bind excess copper in the periplasm, most likely playing a role in the considerably high copper resistance of A. ferrooxidans (Navarro et al., 2016).
An increased time of growth has been observed for ferrous iron‐grown A. ferrooxidans cells in the presence of copper, suggesting that respiration is affected and the oxidation of ferrous iron occurs more slowly in the presence of the metal (Almárcegui et al., 2014b). It is well documented in the literature that ferrous iron oxidation decreases in several A. ferrooxidans strains when copper concentration increases (Leduc et al., 1997). The results recently reported by Navarro et al., 2016 showed that the two proteins AcoP and Rus are upregulated in A. ferrooxidans cells in the presence of copper, suggesting they might have an additional role in copper tolerance. Rus could be eventually saturated with the bound copper. However, the extremely high concentration of Rus is in the range of copper sulfate that could be present under the usual growth condition for this acidophile. Rus copper‐trapping would be only one of many other components that respond to the presence of the toxic metal in the cells.
It is known that in aerobic systems Cu(II) supplied is reduced to Cu(I) upon contact with the respiratory chain of bacteria (Thauer et al., 1977; Volentini et al., 2011). Almost certainly reduced quinones are the electron donors. As Cu(I) is much more toxic than Cu(II), it could be exported from the periplasm to the outside by Cus‐like RND systems or oxidized back to Cu(II) by CueO/PcoA/CopA‐like periplasmic copper‐containing copper oxidases. A. ferrooxidans has Cus‐like RND systems (Navarro et al., 2009) but it apparently lacks genes coding for periplasmic copper‐containing copper oxidases. A new interesting speculative possibility for this acidophile to prevent Cu(I) extreme toxicity would be to transfer an electron from Cu(I) to the iron oxidation system (Navarro et al., 2016). For this it would be necessary that ApoRus and/or apoAcoP are able to bind Cu(I) at the periplasm. In this connection, it is known that folded aporusticyanin rapidly binds Cu(I) with much higher affinity than Cu(II) (Alcaraz et al., 2007). As proposed by Li et al. (2015), the concentrated Rus in the periplasm would constitute a network where inter‐protein electron transfer interactions across the periplasmic space would effectively function in connection with the respirasome. As previously suggested, it is not unthinkable that this copper‐binding protein may also constitute in part a barrier to trap excess copper in a microorganism normally confronted with high concentrations of this metal (Navarro et al., 2016).
A working model including the possible role of Rus and possibly AcoP in copper resistance is shown in Fig. 2A. Obviously much more experimental work will be required to support these proposals. Not all biomining microorganisms contain Rus (Fig. 1), but in those containing Rus‐like proteins coding genes, such as A. ferrivorans, a similar role for this copper‐binding protein could be possible, especially considering that they possess the same copper‐binding site of type I found in A. ferrooxidans Rus. On the other hand, in sulfur‐oxidizing microorganisms such as A. thiooxidans and A. caldus that lack rus genes (Fig. 1) and are less resistant to copper compared with A. ferrooxidans, different components for copper resistance may be expected.
Figure 2.
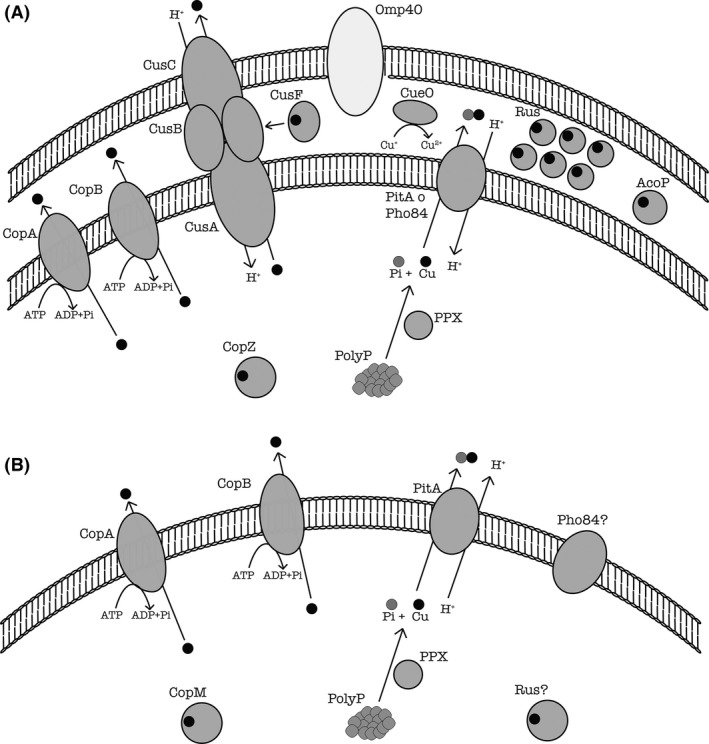
Summary working model of copper resistance determinants in biomining bacteria (A) and archaea (B). Dark grey, proteins that increase their synthesis levels in cells subjected to copper; light grey those down expressed in the presence of copper.
The copper‐responsive protein complement of iron‐grown A. ferrooxidans ATCC 23270 cells was recently evaluated by using ICPL proteomics (Almárcegui et al., 2014b). In addition of the expected upregulation of RND‐type Cus systems, some different RND‐type efflux pumps were overexpressed together with the upregulation of several genes involved in histidine and cysteine synthesis. In response to copper, a downregulation of the major outer membrane porin (Guiliani and Jerez, 2000) and some ionic transporters was observed in A. ferrooxidans ATCC 23270 cells grown in ferrous iron or sulfur (Almárcegui et al., 2014a,b). This result is an example of how a downregulation of outer membrane porins could cause a general decrease in the influx of the metal into the cell.
The native A. ferrooxidans strain Wenelen (DSM 16786) isolated from an industrial copper biomining operation in Chile by Biosigma S.A. was recently reported to have an extreme capacity to resist high concentration of copper (over 10 g L−1) compared with the type strain ATCC 23270 (5 g L−1). Furthermore, the global transcriptional response of this native strain showed the upregulation of several genes related with copper tolerance when the bacterium was grown in the presence of pyrite and CuFeS2 (Latorre et al., 2016).
Current knowledge indicates that in addition to the key elements involved in A. ferrooxidans copper resistance already discussed, an abundant reserve of inorganic polyP used as a polyP‐based copper resistance system and a robust defensive response to oxidative stress are also important (Alvarez and Jerez, 2004; Remonsellez et al., 2006; Orell et al., 2010). Poly P is synthesized in bacteria by a polyphosphate kinase (PPK) and degraded by a polyphosphatase (PPX) (Kornberg et al., 1999). The presence of copper in the cytoplasm would activate PPX, which would degrade polyP to generate free phosphate (see Fig. 2). This phosphate can bind copper to be eliminated to the periplasm or the extracellular medium by means of phosphate transporters such as PitA as proposed before for E. coli (Keasling, 1997; Van Dien et al., 1997) and recently demonstrated for E. coli by Grillo‐Puertas et al. (2014). Figure 2A summarizes a general working model for copper resistance in A. ferrooxidans and other acidophilic bacteria.
Acidithiobacillus ferrivorans
As previously described by González et al. (2014), A. ferrivorans SS3 genome contains four possible cus clusters, three of them cusCBAF type and a fourth one of cusCBA type. All A. ferrivorans SS3 putative Cus systems showed a high score of identity to the A. ferrooxidans system (over 75% Blastp score). Furthermore, upstream of the genomic context of two of the Cus systems of A. ferrivorans SS3, there is a putative copper ATPase (Fig. 3) with 95% Blastp score with CopA from A. ferrooxidans (Fig. 1).
Figure 3.
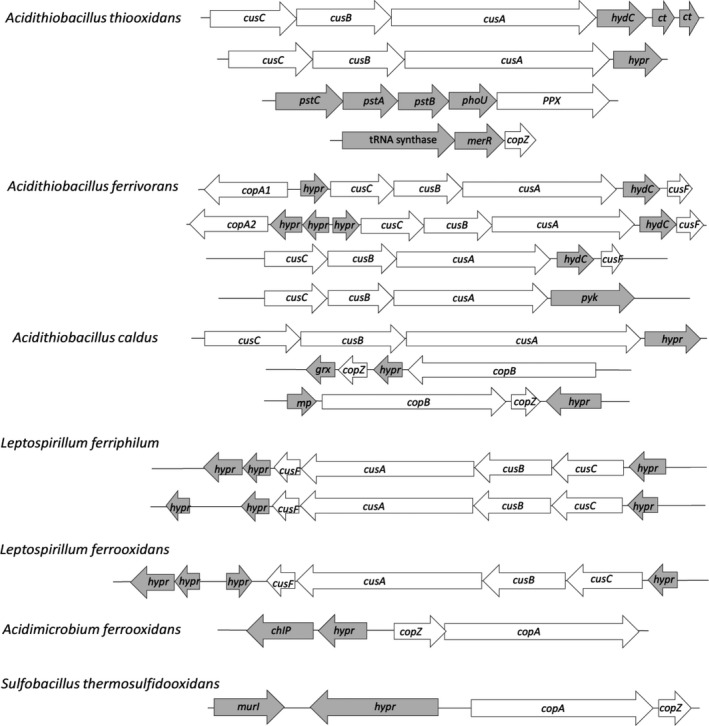
Gene clusters of some copper resistance determinants as predicted by bioinformatics analysis of the indicated selected microorganisms genome sequences. ct, cation transporter; hypr, hypothetical protein; hydC, cytochrome b561; pyk, pyruvate kinase C; merR, transcriptional activators; pstS, phosphate ABC transporter substrate‐binding protein; pstA, phosphate ABC transporter, permease protein; pstB, phosphate ABC transporter ATP‐binding protein; phoU, phosphate transport system regulatory; grx, glutaredoxins; mp, membrane protein; chlP, geranylgeranyl reductase; murl, glutamate racemase.
The A. ferrivorans SS3 genome contains two additional copies of the rus gene (Liljeqvist et al., 2013) and its three Rus proteins not only have a high Blastp score (>85%) to Rus from A. ferrooxidans but all of them also conserve the same copper‐binding site. RusA from A. ferrivorans SS3 is the protein with the highest identity compared with Rus from A. ferroxidans and it forms part of the rus operon from A. ferrivorans together with the gene coding for the AcoP protein (Liljeqvist et al., 2013). In the light of the recent report suggesting that Rus and AcoP are copper resistance determinants in A. ferrooxidans ATCC 23270 (Navarro et al., 2016), it will be of interest to study whether these proteins have a similar role in copper resistance in A. ferrivorans.
González et al. (2014) reported the presence of a ppk2 in a GI of A. ferrivorans SS3. The protein comparisons shown in Fig. 1 support the presence of this PPK2 (with 89% Blastp score) and also indicate that A. ferrivorans SS3 contains a PPX protein (85% Blastp score), suggesting that the polyP‐dependent copper resistance system may also be operative in this psychrotolerant bacterium.
It is also important to consider that not only a higher presence of genes related to metal resistance is the only factor important for extreme metal tolerance but also metal speciation and pH are also important to consider (González et al., 2014). In addition, differences in cell envelope components also may influence metal resistance in bacteria (Harrison et al., 2007; Almárcegui et al., 2014a).
Acidithiobacillus thiooxidans
Studies on copper tolerance mechanisms in A. thiooxidans have also been approached recently (Wen et al., 2014). A putative MCO gene (cueO) was annotated in the genome of A. thiooxidans ATCC 19377 (Valdes et al., 2011). This gene is also present in Acidithiobacillus caldus ATCC 51756, Thiomonas arsenitoxydans and less likely in Alicyclobacillus acidocaldarius LAA1 but is absent in A. ferrooxidans and all the other microorganisms listed in Fig. 1. The MCO has been studied in detail in E. coli (Grass and Rensing, 2001; Roberts et al., 2002; Hoegger et al., 2006). The transcriptional level of A. thiooxidans cueO was upregulated in response to 10 mM copper sulfate and a response to copper of a putative cueO promoter was detected, suggesting it might be stimulated by a putative CueR protein (Wen et al., 2014). By using a modified markerless gene disruption system, these authors generated a cueO mutant of A. thiooxidans. This mutant was more sensitive to external copper and this activity could be restored after complementing the cueO gene, strongly suggesting the involvement of cueO in copper tolerance of this bacterium (Wen et al., 2014).
Figure 1 shows that A. thiooxidans contains several genes coding for proteins with a high identity Blastp score (>70%) with those of the CusCBA systems from A. ferrooxidans. Furthermore, A. thiooxidans may have two Cus systems with similar genetic contexts and the characteristic domains of the previously characterized Cus systems (Fig. 3).
Acidithiobacillus thiooxidans also showed a protein with a high Blastp score (>65%) with CopB from A. ferrooxidans (Fig. 1). This putative copper ATPase shares several domains with CopBAf such as two heavy metal‐binding domains (HMA) and an ATPase E1‐E2 domain, which could be related with the incoming and exporting of copper.
Moreover, Fig. 1 indicates that A. thiooxidans genome has a gene coding for a protein similar to the copper chaperone CopZ from B. subtilis. The A. thiooxidans protein contains the same heavy metal‐binding domain present in CopZB. subtilis with conserved copper‐binding amino acids (MXCXXC). In addition, the bioinformatics analysis of its subcellular location predicted it to be a putative cytoplasmic copper chaperone, suggesting it could be an A. thiooxidans CopZ. Upstream of the gene coding for this protein, there is a putative heavy metals transcriptional regulator that could regulate the expression of this possible A. thiooxidans cytoplasmic metal chaperone (Fig. 3).
As Fig. 1 shows, A. thiooxidans has a high Blastp (>78%) with the genes coding for PPK and PPX from A. ferrooxidans and they share basically the same structural domains. Considering that A. thiooxidans accumulates polyP granules as seen by TEM (Orell et al., 2010), it is possible that a functional polyP‐dependant metal resistance system similar to that previously proposed for A. ferrooxidans (Alvarez and Jerez, 2004) is also present in A. thiooxidans.
The genome sequence of an A. thiooxidans Licanantay strain isolated from a biomining operation suggested the presence of GIs (Travisany et al., 2014) as it has been previously reported for A. ferrooxidans ATCC 53993 strain (Orellana and Jerez, 2011) and for A. ferrivorans SS3 with a predicted GI in its genome (González et al., 2014). Most likely, horizontal gene transfer in mining environments probably generates these genomic elements.
Acidithiobacillus caldus
Regarding copper tolerance of A. caldus, a copper MIC of 24 mM has been reported for strain DSM8584 (Watkin et al., 2008). The response to copper stress of an A. caldus strain isolated in China has been analysed (Xia et al., 2010). The authors found a negative effect on the activity of some of the enzymes involved in sulfur metabolism, such as sulfite oxidase and APS reductase. Nevertheless, no direct evidence was reported to support the idea of overexpression of a Cu‐specific inducible ATPase pump; the authors speculated that would be present under their conditions of study.
Acidithiobacillus caldus ATCC 51756 (Mangold et al., 2013) (see Fig. 1) and strain SM‐1 (You et al., 2011) contain a variety of genes coding for putative copper resistance‐related proteins as they showed a great degree of identity (between 50% and 90%) with those known for A. ferrooxidans (Navarro et al., 2009; Orellana and Jerez, 2011). These two strains of A. caldus share several of these genes in their chromosomal DNA, such as the components of an RND‐type efflux pump (CusABC), two P‐type ATPases (CopB), two cytoplasmic metalochaperones (CopZ) and a copper oxidase (CueO) (see Fig. 1). Interestingly, strain SM‐1 contains a gene coding for a putative periplasmic copper chaperone (CusF), which has been described in E. coli (Loftin et al., 2005) and in A. ferrooxidans ATCC strains 23270 and 53993 (Orell et al., 2010; Orellana and Jerez, 2011).
The search in A. caldus of possible copper resistance determinants similar to those of the CusCBAF system from A. ferrooxidans showed possible homologue proteins although in general they had 40–60% Blastp score (Fig. 1). These proteins possess the characteristic structural domains of these efflux systems and their corresponding genes are contiguous in the genome (Fig. 3). Most likely, they could be related to the transport of heavy metals such as copper, zinc or cadmium.
In addition, A. caldus showed two ATPases with copper‐binding sites (Interpro IPR006122) and with a high Blastp score (>50%) when compared with CopB1 from A. ferrooxidans. Interestingly, a possible cytoplasmic copper chaperone with high similarity to CopZB. subtilis was present in the same DNA region, close the two ATPases already mentioned (Fig. 3). Figure 3 also shows a glutaredoxin in the context of one of the ATPases that could be related to maintain the redox state of the sulfhydryl groups present in cellular proteins damaged by the presence of copper (Almárcegui et al., 2014b).
Figure 1 shows the possible presence of a MCO in A. caldus. This putative protein contains the same domains described for E. coli CueO as noted before (Navarro et al., 2013).
Although a small score (5–10%) indicated the possible presence of Rus and AcoP in A. caldus (Fig. 1), a detailed analysis of the amino acid sequences of these two putative periplasmic proteins indicated that none of them have the domains and copper‐binding sites characteristic of the respirasome‐related proteins from A. ferrooxidans.
As seen in Fig. 1, A. caldus shows the likely presence of PPX and PPK similar to those from A. ferrooxidans (Blastp score >70%), both involved in polyP metabolism and the polyP‐related metal resistance system reported before for A. ferrooxidans (Alvarez and Jerez, 2004; Orell et al., 2010). It has been reported that A. caldus accumulates polyP granules as many other acidophilic microorganisms with high metal resistance (Orell et al., 2012). Furthermore, both A. ferrooxidans and A. caldus have a putative Pho regulon, which includes ppx in its genomic context (Vera et al., 2003; Mangold et al., 2013; Navarro et al., 2013). It is possible then that both acidophilic bacteria would respond in a similar way to the presence of copper (Navarro et al., 2013).
Leptospirillum ferriphilum
Leptospirillum ferriphilum ML‐04 has some identity (20 to 50% depending on the Cus protein) with a CusCBAF system from A. ferrooxidans (Fig. 1) and two copies of this system are present in its genome. As seen in Fig. 3, both Cus systems have a genetic context similar to that described for E. coli, except that CusF is neighbour of CusA. Nevertheless, all of the components of these putative Cus systems contain the characteristic domains of these metal efflux pumps. On the other hand, a copper ATPase with some identity to CopAAf and a gene coding for a protein with 30% identity to CopZB. subtilis were also found. However, this last protein would rather correspond to a mercury reductase and it contains a metal‐binding domain similar to that present in CopZ. Finally a putative PPX coding gene was found in L. ferriphilum but in a different genomic context. In addition, a putative PPK2 was also found in this bacterium but with a low score when compared with the equivalent enzyme from A. ferrooxidans. Without current information of the existence of polyP in L. ferriphilum and the low identity score found for both possible enzymes involved in polyP metabolism, at present it is not possible to speculate on their putative role in metal resistance in this bacterium.
Leptospirillum ferrooxidans
The iron oxidizer L. ferrooxidans shows several putative copper resistance determinants similar to those in A. ferrooxidans (Fig. 1). However, its iron‐oxidizing system lacks rusticyanin (Blake et al., 1993). As seen in Fig. 1, the iron oxidizer L. ferrooxidans contains a possible CusCBAF in which its CusA has a Blastp score over 50% when compared with CusA1 from A. ferrooxidans. In addition, the genetic context of the L. ferrooxidans is in general similar to those described for these Cus systems in other bacteria (Fig. 3). Compared with L. ferriphilum, L. ferrooxidans has only one Cus system. In addition, L. ferrooxidans genome contains a possible metal ATPase with a Blastp score higher than 20% when compared with A. ferrooxidans CopA. Although this is not a very high score value, this protein conserves the metal binding (HMA), E1‐E2_ATPase and ZntA domains typical of CopA.
Regarding the possible existence of a polyP‐dependent metal resistance system in L. ferrooxidans, the bacterium showed two proteins with a low Blastp score (over 10%) compared with the A. ferrooxidans PPX. These two possible PPX enzymes have the Ppx/GppA (IPR003695) domain characteristic of these proteins. On the contrary, no PPK similar proteins were found. This evidence and the lack of information for the presence of polyP do not allow us to suggest the existence of a metal resistance system dependent on polyP in this microorganism.
Acidimicrobium ferrooxidans
The bioinformatics analysis in Fig. 1 indicates that A. ferrooxidans DSM 10331 has a possible copper ATPase with a BlastP score higher than 20% when compared with CopA1 from A. ferrooxidans. Both proteins share several important domains, such as the metal‐binding domain. Interestingly, next to the gene for this putative ATPase the gene coding for a putative small cytoplasmic chaperone similar to CopZ with a copper‐binding site (MXCXXC) characteristic of this protein was found by using the CELLO v.2.5 program (Fig. 3). Being a Gram positive, this bacterium does not contain genes with identity to the Cus system of A. ferrooxidans in Fig. 1.
Acidimicrobium ferrooxidans also has genes coding for putative PPX and PPK both possessing the characteristic domains of these enzymes. Although the presence of inorganic polyP has not been reported in this bacterium, most likely it exists as it occurs in other biomining bacteria to play an important role for oxidative stress response and metal resistance (Orell et al., 2010; Navarro et al., 2013).
Sulfobacillus thermosulfidooxidans
This Gram (+) bacterium is not expected to have a complete Cus system. Figure 1 shows the gene for one protein with a low identity to CusA1 from A. ferrooxidans and no other possible components of a Cus system. However, this possible protein does not contain any of the domains present in CusA1. In contrast, S. thermosulfidooxidans showed four possible ATPases with similarity to CopA1 and CopB1 proteins from A. ferrooxidans. All these putative copper ATPases possess metal binding (HMA), E1‐E2_ATPase and ZntA domains, all of them important for the function of the ATPases and which also form part of CopAAf and CopBAf. As seen in other bacteria (Radford et al., 2003; Navarro et al., 2016), one of these ATPAses is found contiguous to a putative CopZ having an MXCXXC copper‐binding site characteristic of these cytoplasmic chaperones (Fig. 3).
As seen in Fig. 1, S. thermosulfidooxidans has a gene coding for a protein with 20–30% Blastp score compared with AcoP from A. ferrooxidans. There are three copies of this gene coding for a protein that contains a cupredoxin domain to bind copper. However, although these cupredoxin protein genes are neighbours of cytochromes, no rus gene is present in this Gram (+) iron oxidizer (Blake et al., 1993).
Finally, as seen in Fig. 1, clearly Gram (+) bacteria do not contain the proteins related to the Cus systems due to their different envelope structure compared with the Gram (−) bacteria. On the other hand, most Gram (+) bacteria, as expected do not show the presence of periplasmic Rus.
Copper tolerance of biomining Archaea
Concerning archaeal copper resistance mechanisms, a few metal efflux pumps have been identified from sequenced genomes of some members of this domain (Pedone et al., 2004; Wheaton et al., 2015; see Fig. 1). A Cu resistance (cop) loci has been described in Archaea, which includes genes encoding for a new type of archaeal transcriptional regulator (CopT), a putative metal‐binding chaperone (CopM) and a putative Cu‐transporting P‐type ATPase (CopA) (Ettema et al., 2006) (see Fig. 2B).
Single unit, membrane class 1B heavy metal translocating P‐type ATPases are involved in homeostasis and resistance to copper as well as to other metals (Palmgren and Nissen, 2011). Although Sulfolobus solfataricus is not a biomining microorganism, it has been considered a model for archaeal studies. The copTMA operon from S. solfataricus P2 (i.e., copRTA in S. solfataricus 98/2 as reported by Villafane et al., 2009) contains a copper‐responsive regulator (CopT), a copper‐binding protein (CopM) that includes the metal coordinating ligands within the so‐called TRASH domain (‘Trafficking, Resistance and Sensing of Heavy metals’) (Ettema et al., 2003).
CopT usually has an HTH (‘helix–turn–helix’) DNA binding domain in its N‐ terminal end and a TRASH metal‐binding domains in their C‐terminal ends. On the other hand, both CopA and CopM may have metal‐binding domains such as TRASH, YHS (tyrosine, histidine, serine) or HMA (heavy metal‐associated) (Gitschier et al., 1998).
CopA and CopT copper resistance systems are present in most of the selected archaea as shown in Fig. 1. In several cases, there was an overlap between CopA and CopM at the genomic level and therefore they are not annotated in the protein databases. For this reason, only CopA from S. solfataricus was included for comparative purposes in Fig. 1.
CopA is known to impart copper resistance to S. solfataricus (Ettema et al., 2006), being an effective copper pump at low copper concentrations. Other archaeal acidophiles also contain versions of the cop operon (Fig. 1). The search for possible CopT transcriptional regulators in the genomes of the archaea included in Fig. 1 was done by comparison with CopT from S. solfataricus. All archaea where CopT was identified showed high amino acid sequence identity compared with the S. solfataricus regulator and all of these putative transcriptional regulators contained the TRASH and HTH domains characteristic of CopTS. solfataricus.
The genome of Ferroplasma acidarmanus contains a cop operon (copYZB) with similar structure and response to copper as for copTMA (Baker‐Austin et al., 2005). All the putative ATPases identified in the genomes of F. acidarmanus, T. acidophilum, T. volcanium, P. torridus and A. cupricumulans in Fig. 1 contain TRASH domains (data not shown). On the other hand, all the possible CopA found in archaeal genomes in Fig. 1 contained YHS or HMA metal‐binding domains (data not shown).
In silico studies have further identified a CPx‐ATPase, which most likely mediates the efflux of heavy metal cations in the biomining archaeon M. sedula (Auernik et al., 2008). This putative protein has significant identity to a P‐type ATPase from S. solfataricus (CopA) (Ettema et al., 2006). Moreover, M. sedula contains ORFs with significant similarity to both CopM (Msed0491) and CopT (Msed0492) from S. solfataricus (Auernik et al., 2008). Maezato et al. (2012) reported a genetic approach to investigate the specific relationship between metal resistance and lithoautotrophy during biotransformation of CuFeS2 by M. sedula. The functional role of its copTMA operon was demonstrated by cross‐species complementation of a Cu‐sensitive S. solfataricus copR mutant (Maezato et al., 2012).
The previously published genetic context of several archaeal putative CopMA and CopT showed that all of them have the same organization (Ettema et al., 2006; Maezato et al., 2012; Orell et al., 2013). Some of the archaea analysed in Fig. 1 also possess the same genetic organization, indicating that this copper resistance system is highly conserved in most archaea and it could be one of the most relevant ones.
Regarding the polyP‐based copper resistance mechanism, S. metallicus is known to tolerate very high concentrations of copper and accumulates high amounts of polyP granules. Furthermore, the levels of intracellular polyP are greatly decreased when this archaeon is either grown in 200 mM Cu or shifted to 100 mM Cu and an increase in exopolyphosphatase (PPX) activity and Pi efflux due to the presence of Cu suggests a metal tolerance mechanism mediated through polyP in this archaeon (Remonsellez et al., 2006; Orell et al., 2010, 2012).
Acidithiobacillus ferrooxidans (Alvarez and Jerez, 2004) and S. metallicus (Remonsellez et al., 2006) apparently do not contain PitA in their genomes but they have Pho84‐like phosphate transporters similar to Pho‐84 from Saccharomyces cerevisiae which transports metal‐phosphate complexes at acid pH (Fristedt et al., 1999) (see Fig. 1).
Figure 1 clearly shows that all Chrenarchaoeta analysed had a high Blastp score for their putative PPX when compared with the PPX from S. solfataricus. All of these putative PPX enzymes contain the characteristic domains of this PPX (data not shown). Interestingly, all Euryarchaeota in Fig. 1 do not show a protein with identity to PPX from S. solfataricus. This is due to the fact that Euryarchaeota apparently do not have PPX genes identified in their genomes and Crenarchaeota have PPX genes but no genes coding for PPK. As they do synthesize polyP (Orell et al. (2012), a different unknown PPK yet to be identified should be present in these archaeons.
It was recently reported that the wild‐type M. sedula (DSM 5348T) is a pitA mutant, as M. sedula strain CuR1, a spontaneous mutant, had a supranormal metal resistance and was able to leach copper from CuFeS2 at an accelerated rate (McCarthy et al., 2014). This mutant contained a gene orthologue of the bacterial pitA. M. sedula PitA was a low affinity, high velocity secondary transporter implicated in copper resistance and arsenate sensitivity. This archaeal PitA protein would then be a key element for the increased metal resistance of CuR1 and its increased capacity to leach copper (McCarthy et al., 2014). This archaeal Pit transport system would support increased phosphate efflux and metal symport through this Pit‐like system (see Fig. 2B). Previous studies on metal resistance in S. metallicus proposed that another phosphate transporter (Pho84 like) would be present (Remonsellez et al., 2006). Like Pit, Pho84 belongs to the family of phosphate‐proton symporters and the major facilitator superfamily (MFS). As suggested by McCarthy et al. (2014) perhaps PitA and Pho84 comprise dual phosphate transporters that are inherent to these thermoacidophilic archaeons and are critical for their extreme metal resistance.
All archaeons analysed had proteins with around 20% identity to Pho84. Most of these putative proteins are annotated as major facilitator superfamily (MFS) transporters as they contain an MFS domain. In addition, all of them contain a domain annotated as ‘phosphate:H+ symporter’. Considering that both domains are present in Pho84 and that there is evidence for the presence of Pho84‐like and PitA orthologues (Fig. 1) in some archaeons (Remonsellez et al., 2006; McCarthy et al., 2014), the possible function of polyP in metal resistance in these analysed microorganisms cannot be discarded.
Actual evidence suggests that polyP may provide mechanistic alternatives in tuning microbial fitness for the adaptation under stressful environmental situations such as oxidative stress may be of crucial relevance among extremophiles. This is supported by the recent report assigning the universally conserved biopolymer general roles as a protein‐protective chaperone and in regulating general stress response pathways (Gray et al., 2014; Gray and Jakob, 2015).
Figure 1 shows that proteins from some archaeal microorganisms (M. sedula, T. acidophilum, T. volcanium) had a BlastP score of around 20% when compared with the blue‐copper protein Rus from A. ferrooxidans. Other microorganisms such as S. metallicus showed a lower identity (10%). All these archaeal proteins have been annotated as possible rusticyanin, having a type I copper‐binding site and a characteristic cupredoxin domain. In M. sedula, one of these proteins has been suggested to be involved in iron oxidation (Auernik et al., 2008). The proteins from M. sedula, S. acidocaldarius, S. tokodai and S. solfataricus having identity with A. ferrooxidans Rus are sulfocyanins which also have a copper‐binding site. The genetic context for the genes coding these proteins is not identical to that of rus gene from A. ferrooxidans but are close to cytochromes and iron‐sulfur protein genes. As mentioned before, some experimental evidence supports the role of Rus as a possible copper chaperone related with copper resistance in A. ferrooxidans (Navarro et al., 2016). However, whether some of the putative Rus proteins from Fig. 1 other than Rus from A. ferrooxidans are implicated in copper resistance in other bacteria and archaea clearly remains to be demonstrated.
Finally, proteomic studies of S. metallicus subjected to copper, showed 23 downregulated and 30 upregulated proteins, suggesting they are possibly involved in copper resistance (Orell et al., 2013). A similar behaviour was also observed in ‘Fp. acidarmanus’ challenged by either arsenite or copper (Baker‐Austin et al., 2005; Baker‐Austin and Dopson, 2007). Most of these proteins are related to the production and conversion of energy, amino acids biosynthesis and stress responses. Within proteins upregulated in S. metallicus cells exposed to copper, a putative ATP synthase subunit B was detected (Orell et al., 2013). When cells are subjected to some stressing conditions such as the presence of heavy metals, a greater cellular demand for energy has been reported to occur (Baker‐Austin et al., 2005).
Several previous reports have suggested that oxidoreductases contribute to an oxidative protection in both bacteria and archaea in response to heavy metals (e.g. Rodriguez‐Montelongo et al., 2006; Williams et al., 2007). Some proteins involved in oxidative damage repair, such as NADH‐dependent oxidases and thioredoxin reductases, were expressed in cells of ‘Fp. acidarmanus’ exposed to arsenite (Baker‐Austin and Dopson, 2007). The expression of this group of proteins has also been observed when the same microorganism was exposed to copper (Baker‐Austin et al., 2005).
Concluding remarks
Current knowledge indicates that in addition to passive adaptations such as a positive membrane potential and a very high number of basic proteins in the periplasm of Gram (−) bacteria that make difficult cation displacements, key active elements involved in metal resistance in environmental acidophilic microorganisms appear to be a wide repertoire of known regulated copper resistance determinants similar to those present in all microorganisms. The duplications of these copper resistance genes also confer extra metal tolerance to the microorganisms containing them. Global OMICS procedures have allowed determining the presence of novel copper‐chaperones and other possible metal resistance determinants by their overexpression in the presence of the tested metal. The study of novel metal resistance determinants in environmental microorganisms will also help with the functional annotation of these genes in the increasingly available genomic sequences from environmental extremophilic bacteria and archaea.
Horizontal gene transfer by means of mobile genetic elements such as GIs plays an important role in increasing the adaptability and versatility of microorganisms living in environments with high metal concentrations. Most importantly, is to consider that studies of metals resistance in individual microorganisms do not necessarily reflect some interspecific bacterial cooperation that takes place during metal resistance in complex communities. Therefore, future studies on metal resistance determinants in biomining microorganisms should also include detailed analysis of the consortia actually living in industrial bioleaching operations to thoroughly understand the mechanisms that acidophilic bioleaching microorganisms use to adapt to their extreme environments. This knowledge should eventually improve biomining or metal bioremediation processes if the most fitted bacteria are involved in these industrial processes.
Conflict of interest
None declared.
Microbial Biotechnology (2017) 10(2), 279–295
Funding Information
The authors thank projects FONDECYT 1150791 and 1110214 for their research funding.
References
- Alcaraz, L.A. , Gómez, J. , Ramírez, P. , Calvente, J.J. , Andreu, R. , and Donaire, A. (2007) Folding and unfolding in the blue copper protein rusticyanin: role of the oxidation state. Bioinorg Chem Appl 2007: 54232. doi:10.1155/2007/54232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almárcegui, R.J. , Navarro, C.A. , Paradela, A. , Albar, J.P. , von Bernath, D. , and Jerez, C.A. (2014a) a Response to copper of Acidithiobacillus ferrooxidans ATCC 23270 grown in elemental sulfur. Res Microbiol 165: 761–772. [DOI] [PubMed] [Google Scholar]
- Almárcegui, R.J. , Navarro, C.A. , Paradela, A. , Albar, J.P. , von Bernath, D. , and Jerez, C.A. (2014b) b New copper resistance determinants in the extremophile Acidithiobacillus ferrooxidans: a quantitative proteomic analysis. J Proteome Res 13: 946–960. [DOI] [PubMed] [Google Scholar]
- Alvarez, S. , and Jerez, C.A. (2004) Copper ions stimulate polyphosphate degradation and phosphate efflux in Acidithiobacillus ferrooxidans . Appl Environ Microbiol 70: 5177–5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnesano, F. , Banci, L. , Bertini, I. , and Thompsett, A.R. (2002) Solution structure of CopC: a cupredoxin‐like protein involved in copper homeostasis. Structure 10: 1337–1347. [DOI] [PubMed] [Google Scholar]
- Auernik, K.S. , Maezato, Y. , Blum, P.H. , and Kelly, R.M. (2008) The genome sequence of the metal‐mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching‐associated metabolism. Appl Environ Microbiol 74: 682–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker‐Austin, C. , and Dopson, M. (2007) Life in acid: pH homeostasis in acidophiles. Trends Microbiol 15: 165–171. [DOI] [PubMed] [Google Scholar]
- Baker‐Austin, C. , Dopson, M. , Wexler, M. , Sawers, R.G. , and Bond, P.L. (2005) Molecular insight into extreme copper resistance in the extremophilic archaeon Ferroplasma acidarmanus Fer1. Microbiology 151: 2637–2646. [DOI] [PubMed] [Google Scholar]
- Blake, R.C. II , Shute, E.A. , Greenwood, M.M. , Spencer, G.H. , and Ingledew, W.J. (1993) Enzymes of aerobic respiration on iron. FEMS Microbol Rev 11: 9–18. [DOI] [PubMed] [Google Scholar]
- Brierley, C.L. , and Brierley, J.A. (2013) Progress in bioleaching: part B: applications of microbial processes by the minerals industries. Appl Microbiol Biotechnol 97: 7543–7552. [DOI] [PubMed] [Google Scholar]
- Brierley, C.L. , and Murr, L.E. (1973) Leaching: use of a thermophilic and chemoautotrophic microbe. Science 179: 488–490. [DOI] [PubMed] [Google Scholar]
- Bustamante, P. , Covarrubias, P.C. , Levicán, G. , Katz, A. , Tapia, P. , Holmes, D. , et al (2012) ICEAfe1, an actively excising genetic element from the biomining bacterium Acidithiobacillus ferrooxidans . J Mol Microbiol Biotechnol 22: 399–407. [DOI] [PubMed] [Google Scholar]
- Cárdenas, J.P. , Valdés, J. , Quatrini, R. , Duarte, F. , and Holmes, D.S. (2010) Lessons from genomes of extremely acidophilic bacteria and archaea with special emphasis on bioleaching microorganisms. Appl Microbiol Biotechnol 88: 605–620. [DOI] [PubMed] [Google Scholar]
- Cárdenas, J.P. , Quatrini, R. , and Holmes, D.S. (2016) Genomic and metagenomic challenge and opportunities for bioleaching: a mini‐review. Res Microbiol 167: 529–538. [DOI] [PubMed] [Google Scholar]
- Castelle, C. , Ilbert, M. , Infossi, P. , Guiral, M. , Malarte, G. , Ledgham, F. , et al (2008) A new iron‐oxidizing/O2‐reducing supercomplex spanning both inner and outer membranes, isolated from the extreme acidophile Acidithiobacillus ferrooxidans . J Biol Chem 283: 25803–25811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelle, C. , Guiral, M. , Leroy, G. , and Giudici‐Orticoni, M.T. (2010) An unconventional copper protein required for cytochrome c oxidase respiratory function under extreme acidic conditions. J Biol Chem 285: 21519–21525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, A. , Valenzuela, L. , Beard, S. , Mackey, A.J. , Shabanowitz, J. , Hunt, D.F. , and Jerez, C.A. (2007) Periplasmic proteins of the extremophile Acidithiobacillus ferrooxidans . Mol Cell Proteomics 6: 2239–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopson, M. (2016) Physiological and phylogenetic diversity of acidophilic bacteria In Acidophiles. Life in Extremely Acidic Environments. Quatrini R. and Johnson D.B. (eds). Norfolk, UK: Caister Academic Press, pp. 79–91. [Google Scholar]
- Dopson, M. , and Holmes, D.S. (2014) Metal resistance in acidophilic microorganisms and its significance for biotechnologies. Appl Microbiol Biotechnol 98: 8133–8144. [DOI] [PubMed] [Google Scholar]
- Ettema, T.J. , Huynen, M.A. , de Vos, W.M. , and van der Oost, J. (2003) TRASH: a novel metal‐binding domain predicted to be involved in heavy‐metal sensing, trafficking and resistance. Trends Biochem Sci 28: 170–173. [DOI] [PubMed] [Google Scholar]
- Ettema, T.J.G. , Brinkman, A.B. , Lamers, P.P. , Kornet, N.G. , de Vos, W.M. , and van der Oost, J. (2006) Molecular characterization of a conserved archeal copper resistance (cop) gene cluster and its copper‐responsive regulator in Sulfolobus solfataricus P2. Microbiology 152: 1969–1979. [DOI] [PubMed] [Google Scholar]
- Felício, A.P. , Garcia, O. Jr , Bertolini, M.C. , Ottoboni, L.M.M. , and Novo, M.T.M. (2003) The effects of copper ions on the synthesis of periplasmic and membrane proteins in Acidithiobacillus ferrooxidans as analyzed by SDS‐PAGE and 2D‐PAGE. Hydrometallurgy 71: 165–171. [Google Scholar]
- Fristedt, U. , van der Rest, M. , Poolman, B. , Konings, W.N. , and Persson, B.L. (1999) Studies of cytochrome c oxidase‐driven H+‐coupled phosphate transport catalyzed by the Saccharomyces cerevisiae Pho84 permease in coreconstituted vesicles. Biochemistry 38: 16010–16015. [DOI] [PubMed] [Google Scholar]
- Gillan, D.C. (2016) Metal resistance systems in cultivated bacteria: are they found in complex communities? Curr Opin Biotechnol 38: 123–130. [DOI] [PubMed] [Google Scholar]
- Gitschier, J. , Moffat, B. , Reilly, D. , Wood, W.I. , and Fairbrother, W.J. (1998) Solution structure of the fourth metal‐binding domain from the Menkes copper‐transporting ATPase. Nature Struct & Mol Biol 5: 47–54. [DOI] [PubMed] [Google Scholar]
- González, C. , Yanquepe, M. , Cardenas, J.P. , Valdés, J. , Quatrini, R. , Holmes, D.S. , and Dopson, M. (2014) Genetic variability of psychrotolerant Acidithiobacillus ferrivorans revealed by (meta) genomic analysis. Res Microbiol 165: 726–734. [DOI] [PubMed] [Google Scholar]
- Grass, G. , and Rensing, C. (2001) CueO is a multi‐copper oxidase that confers copper tolerance in Escherichia coli . Biochem Biophys Res Commun 286: 902–908. [DOI] [PubMed] [Google Scholar]
- Gray, M.J. , and Jakob, U. (2015) Oxidative protection by polyphosphate – new roles for an old player. Curr Opin Microbiol 24: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, M.J. , Wholey, W.‐Y. , Waqgner, N.O. , Cremers, C.M. , Mueller‐sChickert, A. , Hock, N.T. , et al (2014) Polyphosphate is a primordial chaperone. Mol Cell 53: 689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo‐Puertas, M. , Schurig‐Briccio, L.A. , Rodríguez‐Montelongo, L. , Rintoul, M.R. , and Rapisarda, V.A. (2014) Copper tolerance mediated by polyphosphate degradation and low‐affinity inorganic phosphate transport system in Escherichia coli . BMC Microbiol 14: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiliani, N. , and Jerez, C.A. (2000) Molecular cloning, sequencing, and expression of omp‐40, the gene coding for the major outer membrane protein from the acidophilic bacterium Thiobacillus ferrooxidans . Appl Environ Microbiol 66: 2318–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, S.T.L. (2016) Biotechnologies that utilize acidophiles In Acidophiles. Life in Extremely Acidic Environments. Quatrini R. and Johnson D.B. (eds). Norfolk: UK: Caister Academic Press, pp. 265–283. [Google Scholar]
- Harrison, J.J. , Ceri, H. , and Turner, R.J. (2007) Multimetal resistance and tolerance in microbial biofilms. Nat Rev Microbiol 5: 928–938. [DOI] [PubMed] [Google Scholar]
- Hedrich, S. and Schippers, A. (2016) Distribution of acidophilic microorganisms in natural and man‐made acidic environments In Acidophiles. Life in Extremely Acidic Environments. Quatrini R. and Johnson D.B. (eds). Norfolk, UK: Caister Academic Press, pp. 153–175. [Google Scholar]
- Hoegger, P.J. , Kilaru, S. , James, T.Y. , Thacker, J.R. , and Kües, U. (2006) Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J 273: 2308–2326. [DOI] [PubMed] [Google Scholar]
- Jerez, C.A. (2011) Bioleaching and biomining for the industrial recovery of metals In Comprehensive Biotechnology, 2nd edn Volume 3, Moo‐Young M. (ed.). Boston, MA: Elsevier, pp. 717–729. [Google Scholar]
- Jerez, C.A. (2013) The use of extremophilic microorganisms in industrial recovery of metals In Extremophiles: Sustainable Resources and Biotechnological Implications. Singh, Om V. (ed.). Hoboken, New Jersey: John Wiley & Sons, pp. 319–334. [Google Scholar]
- Johnson, D.B. (2014) Biomining‐biotechnologies for extracting and recovering metals from ores and waste materials. Curr Opin Biotechnol 30: 24–31. [DOI] [PubMed] [Google Scholar]
- Johnson, D.B. , and Hallberg, K.B. (2005) Acid mine drainage remediation options: a review. Sci Total Environ 338: 3–14. [DOI] [PubMed] [Google Scholar]
- Jones, G. , Corin, K.C. , van Hille, R.P. , and Harrison, S.T.L. (2011) The generation of toxic reactive oxygen species (ROS) from mechanically activated sulphide concentrates and its effect on thermophilic bioleaching. Min Eng 24: 1198–1208. [Google Scholar]
- Kaksonen, A.H. , Mudunuru, B.M. , and Hackl, R. (2014) The role of microorganisms in gold processing and recovery—A review. Hydrometallurgy 142: 70–83. [Google Scholar]
- Keasling, J.D. (1997) Regulation of intracellulartoxic metals and other cations by hydrolysis of polyphosphate. Ann NY Acad Sci 829: 242–249. [DOI] [PubMed] [Google Scholar]
- Kornberg, A. , Rao, N.N. , and Ault‐Riché, D. (1999) Inorganic polyphosphate: a molecule of many functions. Ann Rev Biochem 68: 89–125. [DOI] [PubMed] [Google Scholar]
- Langdahl, B.R. , and Ingvorsen, K. (1997) Temperature characteristics of bacterial iron solubilisation and 14C assimilation in naturally exposed sulfide ore material at Citronen Fjord, North Greenland (83°N). FEMS Microbiol Ecol 23: 275–283. [Google Scholar]
- Latorre, M. , Ehrenfeld, N.M. , Cortés, P. , Travisany, D. , Budinich, M. , Aravena, A. , et al (2016) Global transcriptional responses of Acidithiobacillus ferrooxidans Wenelen under different sulfide minerals. Biores Technol 200: 29–34. [DOI] [PubMed] [Google Scholar]
- Leduc, L.G. , Ferroni, G.D. , and Trevors, J.T. (1997) Resistance to heavy metals in different strains of Thiobacillus ferrooxidans . World J Microbiol Biotechnol 13: 453–455. [Google Scholar]
- Li, T.F. , Painter, R.G. , Ban, B. , and Blake, R.C. II (2015) The multicenter aerobic iron respiratory chain of Acidithiobacillus ferrooxidans functions as an ensemble with a single macroscopic rate constant. J Biol Chem 290: 18293–18303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljeqvist, M. , Rzhepishevska, O.I. , and Dopson, M. (2013) Gene identification and substrate regulation provide insights into sulfur accumulation during bioleaching with the psychrotolerant acidophile Acidithiobacillus ferrivorans . Appl Environ Microbiol 79: 951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström, E.B. , and Gunneriusson, L. (1990) Thermophilic bioleaching of arsenopyrite using Sulfolobus and a semi‐continuous laboratory procedure. J Ind Microbiol 5: 375–382. [Google Scholar]
- Liu, Z. , Guiliani, N. , Appia‐Ayme, C. , Borne, F. , Ratouchniak, J. , and Bonnefoy, V. (2000) Construction and characterization of a recA mutant of Thiobacillus ferrooxidans by marker exchange mutagenesis. J Bacteriol 182: 2269–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftin, I.R. , Franke, S. , Roberts, S.A. , Weichsel, A. , Héroux, A. , Montfort, W.R. , et al (2005) A novel copper‐binding fold for the periplasmic copper resistance protein CusF. Biochemistry 44: 10533–10540. [DOI] [PubMed] [Google Scholar]
- Luo, Y. , Liu, Y. , Zhang, C. , Luo, H. , Guan, H. , Liao, H. , et al (2008) Insights into two high homogenous genes involved in copper homeostasis in Acidithiobacillus ferrooxidans . Curr Microbiol 57: 274–280. [DOI] [PubMed] [Google Scholar]
- Maezato, Y. , Johnson, T. , McCarthy, S. , Dana, K. , and Blum, P. (2012) Metal resistance and lithoautotrophy in the extreme thermoacidophilic Metallosphaera sedula . J Bacteriol 194: 6856–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold, S. , Potrykus, J. , Björn, E. , Lövgren, L. , and Dopson, M. (2013) Extreme zinc tolerance in acidophilic microorganisms from the bacterial and archaeal domains. Extremophiles 17: 75–85. [DOI] [PubMed] [Google Scholar]
- Martínez, P. , Gálvez, S. , Ohtsuka, N. , Budinich, M. , Cortés, M.P. , Serpell, C. , Nakahigashi, K. , Hirayama, A. , Toimita, M. , Soga, T. , Martínez, S. , Maass, A. , and Parada, P. (2013) Metabolomic study of Chilean biomining bacteria Acidithiobacillus ferrooxidans strain Wenelen and Acidithiobacillus thiooxidans strain Licanantay. Metabolomics 9: 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez, P. , Vera, M. , and Bobadilla‐Fazzini, R.A. (2015) Omics on bioleaching: current and future impacts. Appl Microbiol Biotechnol 99: 8337–8350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐Bussenius, C. , Navarro, C.A. , Orellana, L. , Paradela, A. , and Jerez, C.A. (2016) Global response of Acidithiobacillus ferrooxidans ATCC 53993 to high concentrations of copper: a quantitative proteomics approach. J Prot 145: 37–45. [DOI] [PubMed] [Google Scholar]
- McCarthy, S. , Chenbing, A. , Wheaton, G. , Tevatia, R. , Eckrich, V. , Kelly, R. , and Blum, P. (2014) Role of an archaeal PitA transporter in the copper and arsenic resistance of Metallosphaera sedula, an extreme thermoacidophile. J Bacteriol 196: 3562–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, C.A. , Orellana, L.H. , Mauriaca, C. , and Jerez, C.A. (2009) Transcriptional and functional studies of Acidithiobacillus ferrooxidans genes related to survival in the presence of copper. Appl Environ Microbiol 75: 6102–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, C.A. , von Bernath, D. , and Jerez, C.A. (2013) Heavy metal resistance strategies of acidophilic bacteria and their acquisition: importance for biomining and bioremediation. Biol Res 46: 363–371. [DOI] [PubMed] [Google Scholar]
- Navarro, C.A. , von Bernath, D. , Martínez‐Bussenius, C. , Castillo, R.A. , and Jerez, C.A. (2016) Cytoplasmic CopZ‐like, periplsasmic Rusticyanin and Acop proteins as possible copper resistance determinants in Acidithiobacillus ferrooxidans ATCC 23270. Appl Environ Microbiol 82: 1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido, H. (2011) Structure and mechanism of RND‐type multidrug efflux pumps. Adv Enzymol Relat Areas Mol Biol 77: 1–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris, P.R. , Burton, N.P. , and Clark, D.A. (2013) Mineral sulfide concentrate leaching in high temperature bioreactors. Min Eng 48: 10–19. [Google Scholar]
- Okibe, N. , Gericke, M. , Hallberg, K.B. , and Johnson, D.B. (2003) Enumeration and characterization of acidophilic microorganisms isolated from a pilot plant stirred‐tank bioleaching operation. Appl Environ Microbiol 69: 19361943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orell, A. , Navarro, C.A. , Arancibia, R. , Mobarec, J.C. , and Jerez, C.A. (2010) Life in blue: copper resistance mechanisms of bacteria and Archaea used in industrial biomining of minerals. Biotechnol Adv 28: 839–848. [DOI] [PubMed] [Google Scholar]
- Orell, A. , Navarro, C.A. , Rivero, M. , Aguilar, J.S. , and Jerez, C.A. (2012) Inorganic polyphosphates in extremophiles and their possible functions. Extremophiles 16: 573–583. [DOI] [PubMed] [Google Scholar]
- Orell, A. , Remonsellez, F. , Arancibia, R. , and Jerez, C.A. (2013) Molecular characterization of copper and cadmium resistance determinants in the biomining thermoacidophilic archaeon Sulfolobus metallicus . Archaea 2013: Article ID 289236. doi:10.1155/2013/289236, 16 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana, L.H. , and Jerez, C.A. (2011) A genomic island provides Acidithiobacillus ferrooxidans ATCC 53993 additional copper resistence: a possible competitive advantage. Appl Microbiol Biotechnol 92: 761–767. [DOI] [PubMed] [Google Scholar]
- Palmgren, M.G. , and Nissen, P. (2011) P‐type ATPases. Annu Rev Biophys 40: 243–266. [DOI] [PubMed] [Google Scholar]
- Pedone, E. , Bartolucci, S. , and Fiorentino, G. (2004) Sensing and adapting to environmental stress: the archaeal tactic. Front Biosci 9: 2909–2926. [DOI] [PubMed] [Google Scholar]
- Quatrini, R. , and Johnson, D.B. (2016) Acidophiles. Life in Extremely Acidic Environments. Norfolk, UK: Caister Academic Press. [Google Scholar]
- Radford, D.S. , Kihlken, M.A. , Borrelly, G.P.M. , Harwood, C.R. , Le Brun, N.E. , and Cavet, J.S. (2003) CopZ from Bacillus subtilis interacts in vivo with a copper exporting Cpox‐type ATPase CopA. FEMS Microbiol Lett 220: 105–112. [DOI] [PubMed] [Google Scholar]
- Ramírez, P. , Guiliani, N. , Valenzuela, L. , Beard, S. , and Jerez, C.A. (2004) Differential protein expression during growth of Acidithiobacillus ferrooxidans on ferrous iron, sulfur compounds or metal sulfides. Appl Environ Microbiol 70: 4491–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remonsellez, F. , Orell, A. , and Jerez, C.A. (2006) Copper tolerance of the thermoacidophilic archaeon Sulfolobus metallicus: possible role of polyphosphate metabolism. Microbiology 152: 59–66. [DOI] [PubMed] [Google Scholar]
- Rensing, C. , and Grass, G. (2003) Escherichia coli mechanism of copper homeostasis in a changing environment. FEMS Microbiol Rev 27: 197–213. [DOI] [PubMed] [Google Scholar]
- Roberts, S.A. , Weichsel, A. , Grass, G. , Thakali, K. , Hazzard, J.T. , Tollin, G. , et al (2002) Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required to copper homeostasis in Escherichia coli . Proc Natl Acad Sci USA 99: 2766–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Montelongo, L. , Volentini, S.I. , Farías, R.N. , Massa, E.M. , and Rapisarda, V.A. (2006) The Cu(II)‐reductase NADH dehydrogenase‐2 of Escherichia coli improves the bacterial growth in extreme copper concentrations and increases the resistance to the damage caused by copper and hydroperoxide. Arch Biochem Biophys 451: 1–7. [DOI] [PubMed] [Google Scholar]
- Roger, M. , Biaso, F. , Castelle, C. , Bauzan, M. , Chaspoul, F. , Lojou, E. , et al (2014) Spectroscopic characterization of a green copper site in a single‐domain cupredoxin. PLoS ONE 9: 1–13. E98941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, S.L. , Dibartolo, G. , Denef, V.J. , Goltsman, D.S.A. , Thelen, M.P. , and Banfield, J.F. (2008) Population genomics analysis of strain variation in Leptospirillum Group II bacteria involved in acid mine drainage formation. PLoS Biol 7: e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio, T. , Miura, A. , Parada Valdecantos, P.A. and Badilla Ohlbaum, R. (2009) Bacteria strain Wenelen DSM 16786, use of said bacteria for leaching of ores or concentrates containing metallic sulfide mineral species and leaching processes based on the use of said bacteria or mixtures that contain said bacteria. United States patent application 7,601,530 B2.
- Thauer, R.K. , Jungermann, K. , and Decker, K. (1977) Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev 41: 100–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travisany, D. , Cortés, M.P. , Latorre, M. , Di Genova, A. , Budinich, M. , Bobadilla‐Fazzini, R.A. , et al (2014) A new genome of Acidithiobacillus thiooxidans provides insights into adaptation to a bioleaching environment. Res Microbiol 165: 743–752. [DOI] [PubMed] [Google Scholar]
- Urbieta, M.S. , Donati, E.R. , Chan, K.‐G. , Shahar, S. , Li Sin, L. , and Goh, K.M. (2015) Thermophiles in the genomic era: biodiversity, science, and applications. Biotechnol Adv 33: 633–647. [DOI] [PubMed] [Google Scholar]
- Valdes, J. , Ossandon, F. , Quatrini, R. , Dopson, M. , and Holmes, D.S. (2011) Draft genome sequence of the extremely acidophilic biomining bacterium Acidithiobacillus thiooxidans ATCC19377 provides insights into the evolution of the Acidithiobacillus genus. J Bacteriol 193: 7003–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dien, S.J. , Keyhani, S. , Yang, C. , and Keasling, J.D. (1997) Manipulation of independent synthesis and degradation of polyphosphate in Escherichia coli for investigation of phosphate secretion from the cell. Appl Environ Microbiol 63: 1689–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt, R. , Monchy, S. , Leys, N. , and Mergeray, M. (2009) New mobile elements in Cupriavidus metallidurans CH34, their possible roles and occurrence in other bacteria. Antoine van Leewenhoek 96: 205226. [DOI] [PubMed] [Google Scholar]
- Van Zyl, L.J. , van Munster, J.M. , and Rawlings, D.E. (2008) Construction of arsB and tetH mutants of the sulfur‐oxidizing bacterium Acidithiobacillus caldus by marker exchange. Appl Environ Microbiol 74: 5686–5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera, M. , Guiliani, N. , and Jerez, C.A. (2003) Proteomic and genomic analysis of the phosphate starvation response of Acidithiobacillus ferrooxidans . Hydrometallurgy 71: 125–132. [Google Scholar]
- Vera, M. , Schippers, A. , and Sand, W. (2013) Progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation‐part A. Appl Microbiol Biotechnol 97: 7529–7541. [DOI] [PubMed] [Google Scholar]
- Villafane, A. , Voskoboynik, Y. , Cuebas, M. , Ruhl, I. , and Bini, E. (2009) Response to excess copper in the hyperthermophile Sulfolobus solfataricus strain 98/2. Biochem Biophys Res Commun 385: 67–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volentini, S.I. , Farias, R.N. , Rodriguez‐Montelongo, L. , and Rapisarda, V.A. (2011) Cu(II) reduction by Escherichia coli cells is dependent on respiratory chain components. Biometals 24: 827–835. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Liu, X. , Liu, S. , Yu, Y. , Lin, J. , Lin, J. , et al (2012) Development of a markerless gene replacement system for Acidithiobacillus ferrooxidans and construction of a pfkB mutant. Appl Environ Microbiol 78: 1826–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkin, E.L.J. , Keeling, S.E. , Perrot, F.A. , Shiers, D.W. , Palmer, M.L. , and Watling, H.R. (2008) Metals tolerance in moderately thermophilic isolates from a spent copper sulfide heap, closely related to Acidithiobacillus caldus, Acidimicrobium ferrooxidans and Sulfobacillus thermosulfidooxidans . J Ind Microbiol Biotechnol 36: 461–465. [DOI] [PubMed] [Google Scholar]
- Watling, H.R. , Watkin, E.L.J. , and Ralph, D.E. (2010) The resilience and versatility of acidophiles that contribute to the bio‐assisted extraction of metals from mineral sulphides. Env Technol 31: 915–933. [DOI] [PubMed] [Google Scholar]
- Wen, Q. , Liu, X. , Wang, H. , and Lin, J. (2014) A versatile and efficient markerless gene disruption system for Acidithiobacillus thiooxidans: application for characterizing a copper tolerance related multicopper oxidase gene. Env Microbiol 16: 3499–3514. [DOI] [PubMed] [Google Scholar]
- Wheaton, G. , Counts, J. , Mukherjee, A. , Kruh, J. , and Kelly, R. (2015) The confluence of heavy metal biooxidation and heavy metal resistance: implications for bioleaching by extreme thermoacidophiles. Minerals 5: 397–451. [Google Scholar]
- Williams, E. , Lowe, T.M. , Savas, J. , and DiRuggiero, J. (2007) Microarray analysis of the hyperthermophilic archaeon Pyrococcus furiosus exposed to gamma irradiation. Extremophiles 11: 19–29. [DOI] [PubMed] [Google Scholar]
- Xia, L. , Yin, C. , Cai, L. , Qiu, G. , Quin, W. , Peng, B. , and Liu, J. (2010) Metabolic changes of Acidithiobacillus caldus under Cu2+ stress. J Basic Microbiol 50: 591–598. [DOI] [PubMed] [Google Scholar]
- Yarzával, A. , Appia‐Ayme, C. , Ratouchniak, J. , and Bonnefoy, V. (2004) Regulation of the expression of the Acidithiobacillus ferrooxidans rus operon encoding two cytochromes c, a cytochrome oxidase and rusticyanin. Microbiol 150: 2113–2123. [DOI] [PubMed] [Google Scholar]
- You, X.Y. , Guio, X. , Zheng, H.J. , Zhang, M.J. , Lie, L.J. , Zhu, Y.Q. , et al (2011) Unraveling the Acidithiobacillus caldus complete genome and its central metabolisms for carbon assimilation. J Genet Genomics 38: 243–252. [DOI] [PubMed] [Google Scholar]
- Zhuang, W.‐Q. , Fitts, J.P. , Ajo‐Franklin, C.M. , Maes, S. , Alvarez‐Cohen, L. , and Hennebel, T. (2015) Recovery of critical metals using biometallurgy. Curr Opin Biotechnol 33: 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]


