Abstract
An unbalanced microbiome may lead to disease by creating aberrant immune responses. A recent association of cellular rejection with the development of interstitial fibrosis and tubular atrophy (IFTA) suggests the role of immune-mediated tissue injury. We hypothesized that developing IFTA correlates with altered urinary tract microbiomes (UMBs).
UMBs at two serial time points, 1 and 6-8 months post-transplant, were assessed by 16S microbial ribosomal gene sequencing in 25 patients developing biopsy-proven IFTA compared to 23 transplant patients with normal biopsies and excellent function (TX) and 20 healthy non-transplant controls (HC).
Streptococcus, the dominant genera in HC males, was lower in IFTA and TX males at 1 month compared to HCs. At 6-8 months, Streptococcus was further decreased in IFTA males, but normalized in TX. IFTA males and females had increases in number of genera per sample at 6-8 months. UMB composition varied substantially between individuals in all groups.
Despite the wide variation in UMBs between individuals, IFTA associated with a loss in dominant resident urinary microbes in males, and a parallel increase in non-resident, pathogenic bacteria in males and females. UMB changes may contribute to IFTA development by alteration of the host immune response.
INTRODUCTION
The human microbiome, defined as the aggregate of microorganisms that inhabit the human body, likely plays a role in training and maintenance of the host immune response. The microbiome is important in human health (1). Recent evidence suggests that altered or unbalanced microbial communities are associated with a dysregulated immune responses (2, 3) that may contribute to the development of diseases such as IBD (4, 5), asthma (6-8) and transplant rejection (9). Sensitive DNA-based sequencing techniques have revealed the existence of bacteria in body areas originally believed to be sterile (10-12), including the existence of a urinary tract microbiome (UMB) (13, 14). Kidney transplantation alters the UMB by surgical stress, perioperative immunosuppression and prophylaxis and/or therapy with antibiotics.
Interstitial fibrosis and tubular atrophy (IFTA) is a replacement of normal structures by interstitial fibrosis secondary to tissue injury. IFTA not associated with another known etiology (i.e. recurrent disease, BK nephropathy) occurs in ~25% of 1-year surveillance transplant biopsies (15-17) as well as correlating with decreased graft survival (18). Inflammation with histological evidence of IFTA has been correlated with increased graft loss (15, 19-26). However, our latest data indicates that any case of histological IFTA represents a risk for immune-mediated graft loss (18). Thus, we believe that IFTA is the result of chronic rejection and there is an etiologic role confirmed by molecular analysis of biopsies and immune pathway mapping of differentially expressed genes for both T cell and antibody-mediated immune mechanisms.
Independent of immunosuppression, infections of the urinary tract or post-transplant alterations of the UMB may well lead to additional immune dysregulation and consequent immune-mediated graft injury. In fact, two recent papers from our laboratory group demonstrate that antibodies produced by B cell clusters commonly found in the biopsies of IFTA subjects are not recognizing donor HLA antigens but rather are specific for E. coli LPS, a powerful immune activator through the canonical TLR4 receptor (27, 28). Another of our studies identified the role of a new mycoplasma protein, obtained from a potential human urinary pathogen, as a powerful, polyclonal B cell activator (28). In the present study, we aimed to determine if post-transplant changes in the UMB were correlated with biopsy-proven IFTA samples without a known etiology (i.e. chronic rejection).
DNA sequencing of bacterial 16S rRNA was used to assess the UMBs in 25 patients at 1 and 6-8 months post-transplant that demonstrated development of IFTA by surveillance biopsies and 23 patients enrolled in parallel but with normal biopsies and excellent function (TX) at both time points. UMBs were also obtained on 10 healthy male and 10 healthy female non-transplant volunteers. We found that the UMBs of healthy males and healthy females were different and each contained a relatively small number of dominant microbial species. In contrast, UMBs in the transplant patients were more complex and evolved over time in a way that correlated with the development of IFTA. We theorize that post-transplant alterations of the UMB may contribute to IFTA development by dysregulated stimulation of the host immune system secondary to disruption of normal microbiomes or replacement of bacteria in genitourinary niches with pathological bacteria.
METHODS
Study Population
Clean catch urine samples were obtained from 25 patients at two time points after transplantation: approximately 1 month and 6-8 months. These patients demonstrated development of IFTA on 6-8 month surveillance biopsies compared to their biopsies done at 1 month. Moderate to severe IFTA was defined according to the Banff 2007 schema as tubulo-interstitial changes in greater than 25% of the cortical area examined. Cases where these changes could be linked to known mechanisms of kidney injury like recurrent or concomitant disease were excluded. Urine samples at the same time points were obtained from 23 patients with normal surveillance biopsies and stable kidney function (TX). These samples were collected at Emory University with Institutional Review Board approval. Additional urine samples were obtained from 10 healthy male and 10 healthy female non-transplant volunteers to determine normal microbial populations. The only exclusions to study entry were individuals with biopsies that did not conform to the study’s inclusion/exclusion criteria for IFTA with unknown cause (Supplement 1 Expanded Methods), such as the presence of BK nephritis or recurrent disease. Biopsies with borderline changes, higher grades of T-cell/antibody mediated rejection, or focal/minimal C4d were all excluded.
Statistical Analysis
ANOVA or Pearson’s chi-squared tests were used to detect differences in continuous and categorical variables between phenotypes. All p-values were adjusted with Bonferroni correction for multiple hypothesis testing when appropriate.
16S Microbial Ribosomal DNA Sequencing
A minimum of 50 ml of urine was spun for 30 minutes at 1800G before adding 1mL of RNAlater (Qiagen Inc., Valencia, CA) to the pelleted sample and storing at −80° C. TRIzol ® Reagent (ThermoFisher, Waltham, MA) was used to isolate DNA. Sequencing libraries were prepped using the Ion 16S™ Metagenomics Kit (ThermoFisher). The 16S hypervariable region was amplified using two primer sets: 1) V2, V4 and V8, 2) V3, V6, V7, and V9. The resulting cDNAs were barcoded with Ion Torrent Tags 1-16 to allow multiplex sequencing. Optimal DNA integrity was ensured by an analysis using the Agilent® 2200 TapeStation System and the Agilent® High Sensitivity D1000 ScreenTape System (Agilent Technologies, Santa Clara, CA, USA). Sequencing was performed using the Ion PGM™ System (ThermoFisher). The template preparation was performed and loaded onto Ion 318™ v2 Chips using the Ion PGM™ OT2 400 kit and the Ion OneTouch™ 2 System. The chips were then sequenced by the Ion PGM™ using the Ion PGM™ Sequencing 400 kit.
Mapping Gene Sequences to Urine 16S rRNA Databases
Ion ReporterTM software was then used to map and annotate 16S gene sequences. This software uses a two-step Basic Local Alignment Search Tool (BLAST) that maps to two separate reference libraries. Reads are first aligned to a vendor-specific microbiome library named MicroSEQ. For reads not aligned to the MicroSEQ library, a second alignment is then made to the publically available Greengenes database.
Normalization of Count Data
For each urine sample, the number of reads that mapped to each bacteria phyla, classes, orders, families, and genera were totaled. These bacteria read counts were divided by the total number of reads mapped in each sample (i.e. number of bacteria-specific sequence/total number of sequences mapped for a sample) (29) (i.e. normalization). These fractions were converted to percentages, providing a bacteria composition for each sample.
RESULTS
UMBs were assessed at 1 month and then again approximately 6-8 months post-transplant. UMBs were also obtained on 10 healthy male and 10 healthy female non-transplant volunteers to establish the composition of normal UMBs. The IFTA group had a higher percentage of African Americans than TX (68% vs. 57%, p=0.04). The study overall contained more males than females, but no differences in the percent of females in comparison of IFTA vs. TX groups (Table 1). There were also no differences in age, immunosuppressive regimen, EBV and CMV positivity, and incidence of type 1 or type 2 diabetes. The immunosuppressive regimen was the standard of care for Emory’s transplant program: basiliximab induction followed by tacrolimus, MMF and prednisone maintenance.
Table 1.
Demographics of study population†
| IFTA | TX | Comparison (p-value)* |
|
|---|---|---|---|
| Total number | 25 | 23 | |
| Age (mean ± SE) | 56 ± 3 | 51 ± 3 | 0.18 |
| Sex (Female/Male) | 12/13 | 6/17 | 0.11 |
| Race (African American/Caucasian/Other) |
17/8/0 | 8/14/1 | 0.04 |
| Type 1 Diabetes† (yes/no) | 2/19 | 4/18 | 0.60 |
| Type 2 Diabetes† (yes/no) | 4/17 | 3/19 | 0.89 |
| 2nd organ transplant (liver/pancreas/kidney) |
1/0/2 | 1/5/3 | 0.83 |
| CMV Serology† (Positive/Negative) | 20/5 | 19/3 | 0.56 |
| EBV Serology† (Positive/Negative) | 24/0 | 23/0 | NA |
| Tacrolimus treatment† (yes/no) | 23/0 | 23/0 | 0.17 |
| Mycophenolate treatment† (yes/no) | 20/0 | 23/0 | NA |
Analysis method: ANOVA for quantitative data (Probability > F statistic), Pearson’s chi-squared test for dichotomous data.
Missing data: Type 1 diabetes (n=3, 6%), Type 2 diabetes (n=2, 4%), CMV serology (n=1, 2%), EBV serology (n=1, 2%), Tacrolimus treatment (n=2, 4%), Mycophenolate treatment (n=5, 10%)
Abbreviations: IFTA: Interstitial fibrosis and tubular atrophy, mm: mismatch, NA: not applicable, SE: standard error, TX: Treatment group with excellent functioning,
Microbiome differences in male vs. female healthy non-transplant controls
There were no statistically significant differences in bacterial compositions at the level of phyla for comparisons of UMBs in male vs. female healthy controls. Firmicutes made up 65% of male vs. 73% of female UMBs. The following phyla combined with Firmicutes to comprise near 97% of all phyla in healthy men and women: Actinobacteria (15% males vs. 19% females), Bacteroidetes (10% males vs. 3% females) and Proteobacteria (8% males vs. 3% females).
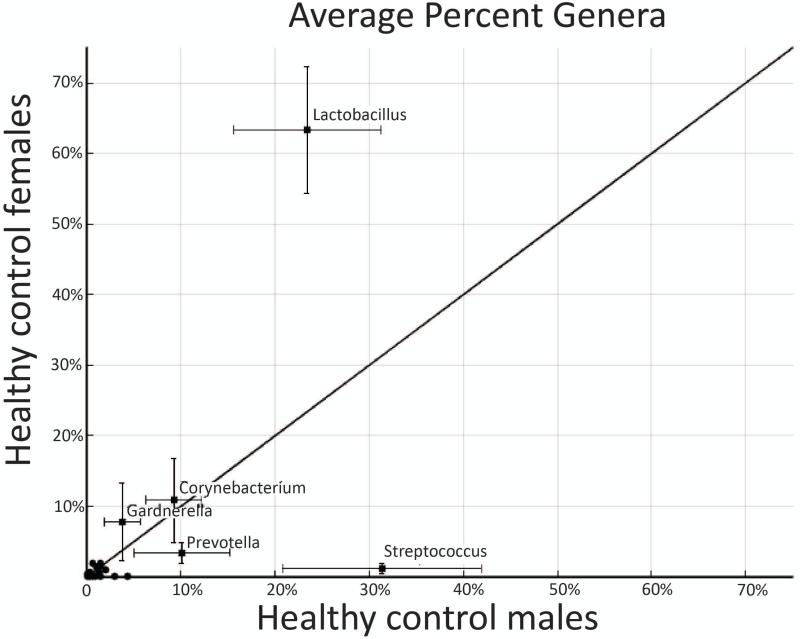
In contrast, there were statistically significant differences between males and females at the bacteria genus level. In healthy male controls, 10 genera comprised over 90% of the bacterial population. The top 5 bacteria in order of abundance were Streptococcus (31%), Lactobacillus (23%), Prevotella (10%), Corynebacterium (9%), and Pseudomonas (4%). In the healthy female controls, 10 genera comprised over 94% of the bacteria population. The top five bacteria in females according to prevalence were Lactobacillus (63%), Corynebacterium (11%), Gardnerella (8%), Prevotella (3%) and Bacillus (2%). Figure 1 plots the composition of genera in healthy males vs. females. There was a statistically higher percentage of lactobacillus in females compared to males (p=0.01). Alternatively, there was statistically higher amount of Streptococcus in males (p=0.006).
Figure 1. Healthy control male vs. female urinary microbiomes.
The xy graph plots the average percent genera in healthy control (HC) males vs. HC females. The error bars at each point represent the standard error. The diagonal line represents the line of no difference between the two sexes. There was a statistically higher percentage of Lactobacillus in females compared to males (p=0.01). Alternatively, there was statistically higher amount of Streptococcus in males (p=0.006).
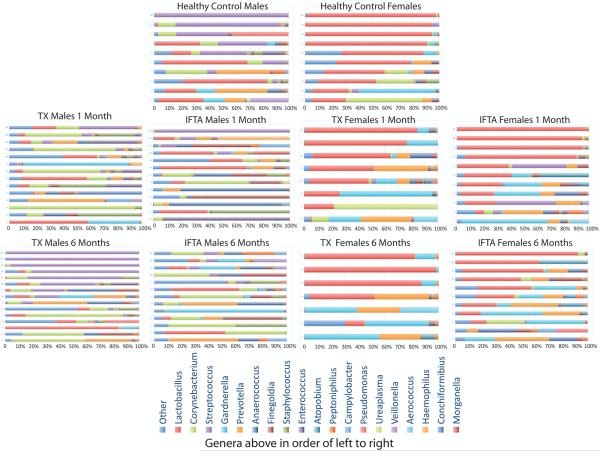
It is important to note that we observed a high variability in bacterial genera composition between individuals. This was true for healthy controls and all the transplant patient microbiomes (Figure 2). As mentioned, Streptococcus was collectively the most abundant genera in healthy males, yet it was the dominant genera in only 50% of samples. Lactobacillus was collectively the most abundant genera in healthy females, and was the dominant genera in 80% of samples.
Figure 2. Bacteria genera composition (percent) for each individual.
Most of the male healthy controls (HC) (top row, left) have dominance by Streptococcus (purple) and/or Lactobacillus (red). Most of the female HCs (top row, right) are dominated by Lactobacillus (red) genera. In the transplant patients, there is a decrease in the dominant bacteria at 1-month post-transplant (second row). At 6-months post-transplant (third row), there is a general restoration of dominant bacteria in the treatment group with excellent functioning kidney (TX) participants. In contrast, there is a further decrease in normally dominant genera in interstitial fibrosis and tubular atrophy (IFTA) participants from 1 month to 6 months post-transplant. Despite these generalized findings, this image makes it apparent that there exists significant variation in bacteria genera composition in every phenotype.
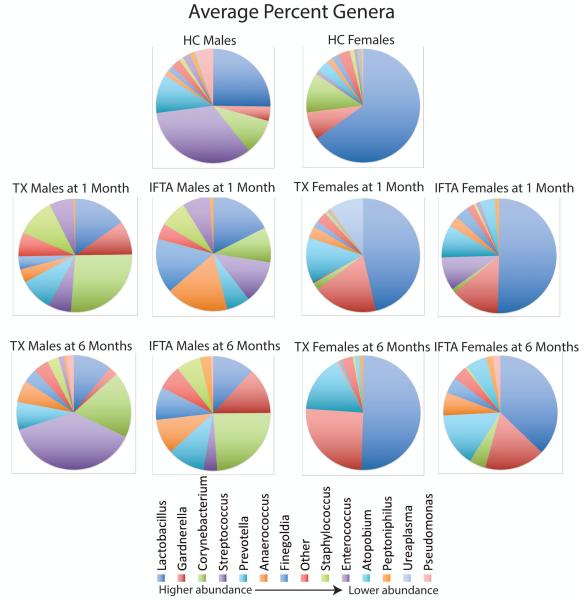
Microbiome at 1 month post-transplant for IFTA and TX samples
There were differences at the genera level in UMBs from IFTA and TX samples at 1 month post-transplant in comparison to healthy controls (Figure 3). The considerable variability observed between individuals within the same phenotype made it difficult to statistically detect differences in collective microbiome populations.
Figure 3. Average bacteria genera composition (percent) for each phenotype.
Streptococcus (purple) and Lactobacillus (dark blue) have the highest average abundance in healthy control (HC) males (top row, left). Lactobacillus (dark blue) has the highest average abundance in HC females (top row, right). At 1 month post-transplant (2nd row), both treatment group with excellent functioning kidney (TX) and interstitial fibrosis and tubular atrophy (IFTA) males and females demonstrate a decrease in Lactobacillus and Streptococcus genera, with an increase in non-dominant species. At 6 months (3rd row), there is restoration of dominant bacteria (i.e. Lactobacillus and Streptococcus) in TX, but a further loss of these two genera in the IFTA patients.
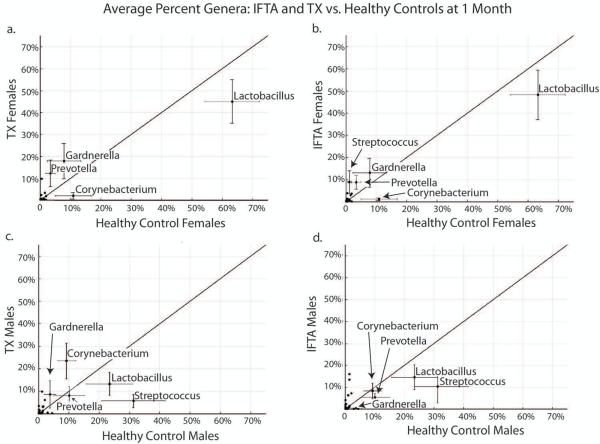
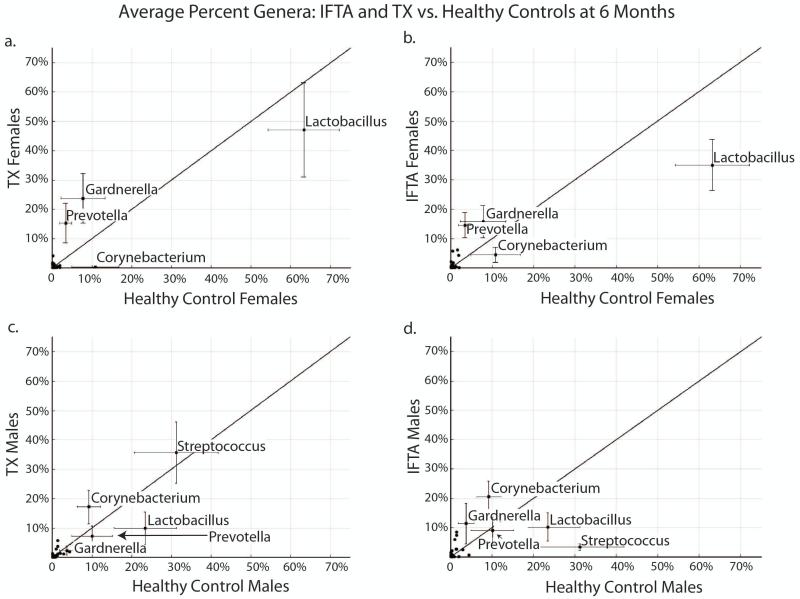
In females, Lactobacillus was non-significantly lower in IFTA (48%) and TX (45%) compared to healthy controls (63%; p=0.20 and p=0.24, respectively) (Figure 4a, b). There was also a non-significant increase in Gardnerella from TX females compared to healthy control females (13% vs. 8%, p=0.14). In males, Streptococcus was significantly lower in IFTA (11%) and TX (6%) patients compared to healthy controls (32%; p<0.058 and p=0.05, respectively) (Figure 4c, d). The percent composition of Lactobacillus was lower in TX (10%) compared to healthy controls (24%) but was not significant, again due to individual variation.
Figure 4. IFTA and TX vs. healthy controls at 1 month post-transplant.
In females (a. and b.), Lactobacillus was non-significantly lower in treatment group with excellent functioning kidney (TX) (a.) and interstitial fibrosis and tubular atrophy (IFTA) (b.) compared to healthy controls. In males (c. and d.), Streptococcus was significantly lower in TX (c.) and TX (d.) patients compared to healthy controls (p<0.058, p=0.05, respectively).
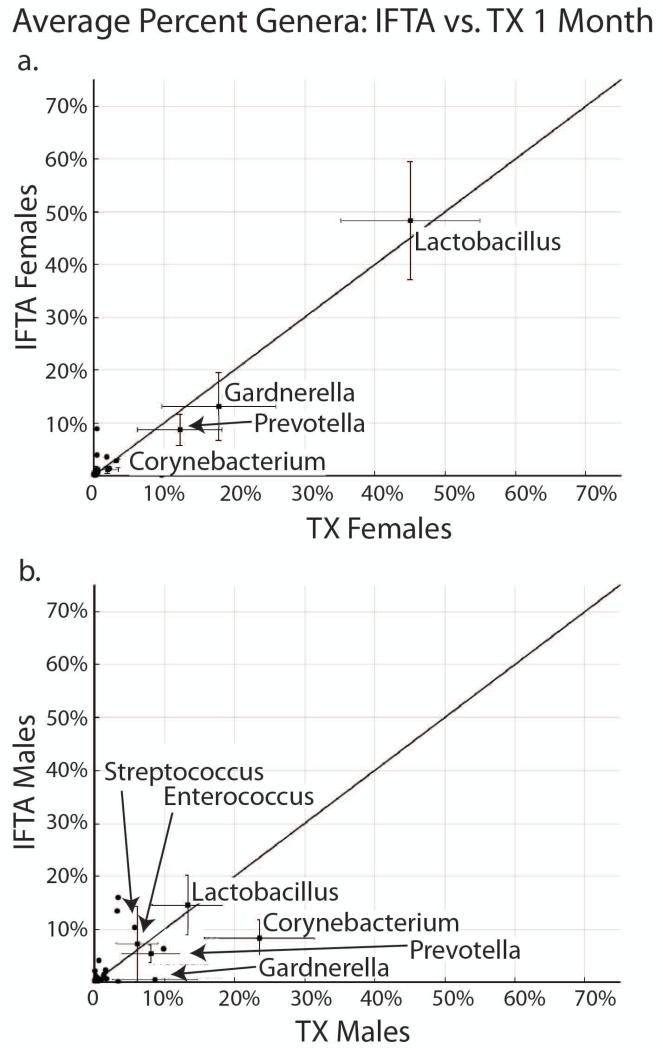
In comparison of female IFTA vs. TX at 1 month, there were no statistically significant differences in UMBs. Lactobacillus was the dominant genus in both IFTA (49%) and TX (45%) (Figure 5a), and there was a similar percent of Gardnerella, Prevotella and Corynebacterium. There were also no statistically significant differences in comparison of male IFTA vs. TX at 1 month, differences in UMBs (Figure 5b). There was no dominant genus in either IFTA or TX. Corynebacterium had the highest abundance of any genus, comprising 24% of TX males and 9% of IFTA males.
Figure 5. IFTA vs. TX at 1 month post-transplant.
a. In females, there were no differences in urinary microbiome composition between treatment group with excellent functioning kidney (TX) and interstitial fibrosis and tubular atrophy (IFTA) at 1 month. Lactobacillus was the dominant genus in both, followed by Gardnerella and Prevotella. b. In males, there were also no differences in comparison of IFTA vs TX at 1 month. There was no dominant genus in either IFTA or TX. Corynebacterium had the highest abundance of any genus, comprising 24% of TX males.
Microbiome at 6-8 months post-transplant for IFTA and TX samples
In TX females, Lactobacillus was relatively non-changed (45% to 47%) from 1 month to 6-8 months, and non-significantly lower than normal control females (63%) (Figure 6a). In contrast, in IFTA females, Lactobacillus decreased further from 48% to 36%, which was also lower than normal controls (63%; p=0.06) (Figure 6b). In male TX patients, Streptococcus increased from 6% at 1 month to 36% 6-8 months post-transplant, returning to the level of healthy controls (32%, p=0.98) (Figure 6c). Meanwhile, in male IFTA patients, Streptococcus decreased from 11% at 1 month to 3.4% at 6-8 months post-transplant, and was significantly lower than healthy control males (p=0.03) (Figure 6d).
Figure 6. IFTA and TX vs. healthy controls at 6-8 months post-transplant.
a. In females with excellent functioning kidney (TX), Lactobacillus remained stable from 1 month to 6 months, and non-significantly lower than healthy control (HC) females. b. In females with interstitial fibrosis and tubular atrophy (IFTA), Lactobacillus decreased from 6 months to 1 month, and was lower than HCs at borderline significance (p=0.06). c. In male patients with TX, Streptococcus increased from 1 month to 6 months, and was higher than HCs. d. In male patients with IFTA, Streptococcus decreased from 1 to 6 months to 3%, significantly lower than HCs (p=0.03).
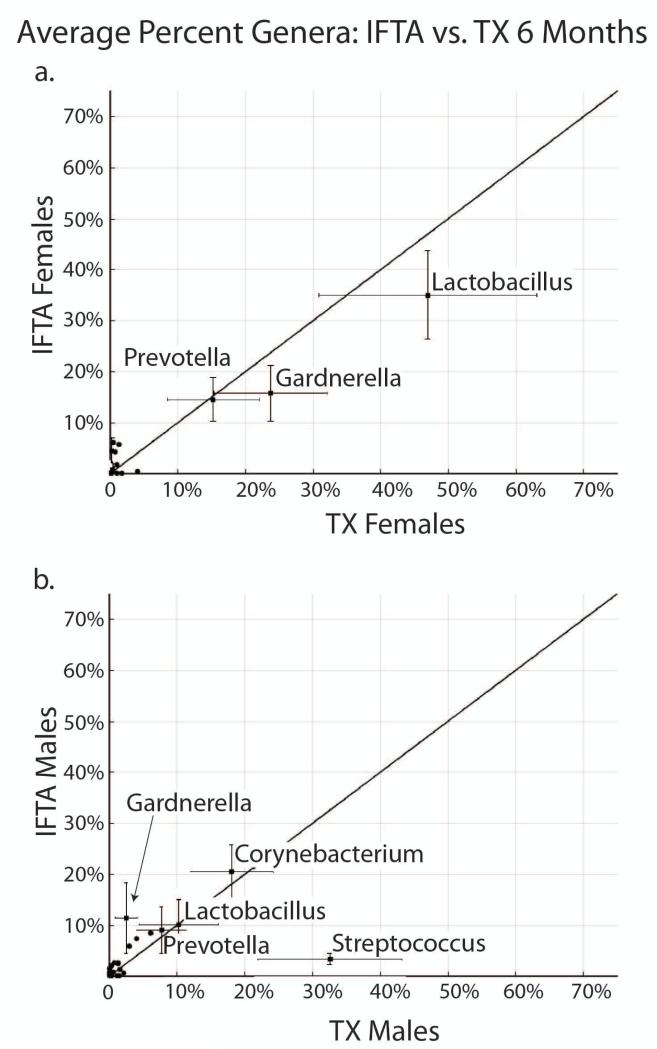
In comparison of female IFTA vs. TX at 6-8 months, there were no statistically significant differences in UMBs. Lactobacillus was the dominant genus in both IFTA (36%) and TX (47%), followed by Gardnerella and Prevotella (Figure 7a). Corynebacterium, which was present in both IFTA and TX females at 1 month, was not found at significant abundance at 6-8 months.
Figure 7. IFTA vs. TX at 6-8 months post-transplant.
a. In females, Lactobacillus was non-significantly higher in treatment group with excellent functioning kidney (TX) compared to interstitial fibrosis and tubular atrophy (IFTA) at 6 months. Lactobacillus was the dominant genus in both, followed by Gardnerella and Prevotella. b. In males, Streptococcus was significantly higher in IFTA compared to TX at 6 months (p=0.03).
In comparison of male IFTA vs TX at 6-8 months, there was a significantly higher abundance of Streptococcus in TX (mean: 36% vs 3%, p=0.03) (Figure 7b). Of note, the range and 25th-75th quartile of Streptococcus in TX was 0-99% and 1-74%, respectively, compared to 0-12% and 0-6% in IFTA (Figure S1). There was an increase in Gardnerella in IFTA compared to TX of borderline statistical significance (12% vs 2%; p=0.06). Otherwise, there was no difference in Corynebacterium, Lactobacillus and Prevotella.
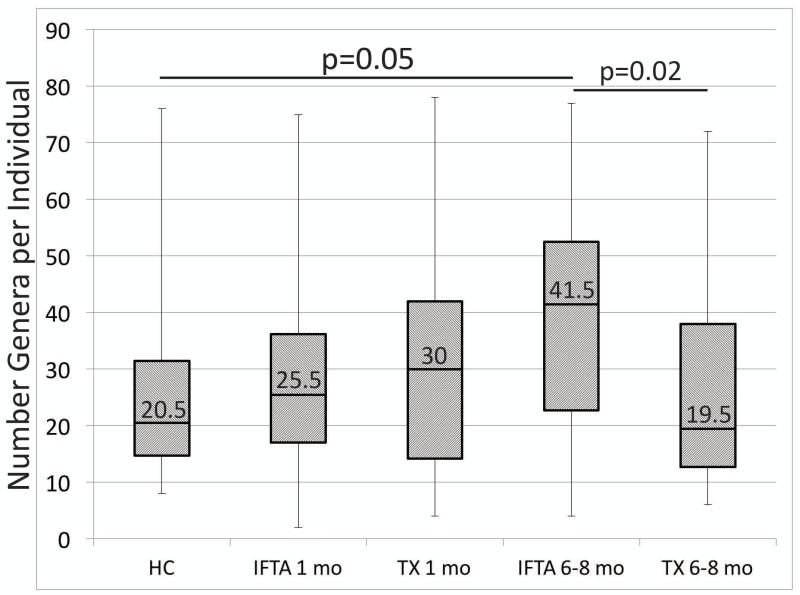
Number of genera per sample is increased in IFTA samples
At 1 month post-transplant, there was no significant difference in the number of genera per individual as a function of IFTA vs. TX patients and IFTA vs. healthy controls (Figure 8). In contrast, by 6-8 months, IFTA patients demonstrated a significant increase in the number of genera per individual (median=41.5) in comparison to healthy controls (median=20.5; p-value=0.05) and TX at 6-8 months (median=19.5; p-value=0.02).
Figure 8. Number of bacteria genera per individual.
At 6-8 months post-transplant, interstitial fibrosis and tubular atrophy (IFTA) patients demonstrated a significantly higher number of genera per individual in comparison to healthy controls (HCs) and treatment group with excellent functioning kidney (TX). Box plots and whiskers indicate 25-75th percentiles and ranges, respectively.
Discussion
In this study, we determined urinary microbiome compositions of 48 transplant patients at two time points (1 and 6-8 months post-transplant) and 20 healthy controls to determine microbiome changes associated with the development of IFTA. We found that healthy control female microbiomes were dominated by the Lactobacillus genus, whereas healthy control males were comprised mostly of Streptococcus genus. In comparison to healthy controls, all transplant The clinical significance of this study is that a future prospective trial may demonstrate that monitoring urinary microbiome changes in individual patients as a function of time and selective treatment may reduce susceptibility to rejection, including chronic rejection and development of IFTA, and ultimately improve long-term clinical outcomes.change from normal may reflect the common use of antibiotics, including prophylaxis, in the early post-transplant period. It is also possible that UMBs are impacted by the first exposure to immunosuppressive drugs and a major surgical procedure. Finally, there could be disturbance of the normal urinary microbiome prior to transplantation due to the presence of chronic kidney disease. Thus, transplantation and the use of immunosuppressant and antibiotic medications may be further worsening an already dysregulated UMB.
At 6-8 months post-transplant, Lactobacillus stabilized in females with healthy grafts (TX) but decreased in females with histological evidence of IFTA. Similarly, Streptococcus actually increased to normal levels in TX males from 1 month to 6-8 months post-transplant, but decreased to represent only 3.4% in the IFTA group. Concurrently, we found that the number of genera per sample was significantly higher in IFTA patients at 6-8 months compared to all transplant patients at 1 month, TX patients at 6-8 months and all healthy controls. In addition to these collective differences in microbiome composition, it is important to note that we also observed a high variability between individual samples, even within the same phenotypic group. Therefore, we conclude that this approach is not appropriate for developing a biomarker strategy based simply on 16S microbial ribosomal gene profiling signatures.
Bacterial DNA sequences taken from the urine of our 10 healthy females were predominantly found in the following phyla: Firmicutes (73%), Actinobacteria (19%), Bacteroidetes (3%) and Proteobacteria (3% females). In a prior study that evaluated the urinary microbiomes in 8 healthy females by also using 16S sequencing methods, the authors reported UMB compositions that were very similar to our female patients: Firmicutes (65%), Bacteroidetes (18%), Actinobacteria (12%), Fusobacteria (3%) and Proteobacteria (2%) (30). The phyla composition of the male healthy controls in our study were also very similar to our female subjects and the previous study: Firmicutes (66%), Actinobacteria (15%), Bacteroidetes (10%) and Proteobacteria (8%). Finally, a similarly designed study featuring both adult men (n=6) and women (n=10), three quarters of the samples had Firmicutes (31) as the predominant phylum. All of these findings suggest that Firmicutes is the dominant and relatively stable phylum in the UMB of both healthy men and women, followed by three other less prevalent bacterial phyla: Actinobacteria, Bacteroidetes and Proteobacteria.
Notwithstanding strong similarities at the phyla level, healthy males and females differed substantially at the genus level in our study. The urine of our female healthy controls included the following genera: Lactobacillus (63%), Corynebacterium (11%), Gardnerella (8%), Prevotella (3%) and Bacillus (2%). In contrast, the urine of our male controls included the following genera: Streptococcus (31%), Lactobacillus (23%), Prevotella (10%), Corynebacterium (9%), and Pseudomonas (4%). Supportively, in the study of 8 urinary microbiomes from healthy females mentioned previously, the authors found the following three genera to be of highest abundance: Lactobacillus (57%), Prevotella (20%) and Gardnerella (10%). Corynebacterium was not found in healthy UMBs in the published study, although this may be due to differences in sequencing techniques and read mapping. For example, the researchers sequenced 3 regions of the rRNA (V1, V2 and V6), whereas our technique sequenced 7 regions of the rRNA (V2, V3, V4, V5, V6, V7, V8 and V9). We also mapped gene sequences to later versions of microbial databases.
In another published study, the authors used 16S sequencing to evaluate UMBs of 26 healthy controls and 27 subjects at risk for asymptomatic bacteriuria due to spinal cord injury-related neurogenic bladder (32). In agreement with our results, Lactobacillus was higher and the predominant genera in healthy women, while Streptococcus was higher in healthy males. The study found that Lactobacillus, Corynebacterium, Gardnerella, Prevotella and Enterococcus defined gender differences. Finally, the authors concluded that there was decreasing abundance of Lactobacillus in relation to increasing duration of a neurogenic bladder. Concurrently, Enterobacteriales also increased in abundance in relation to neurogenic bladder duration. The authors concluded that the loss of Lactobacillus dominance and increase of Enterobacteriales likely predisposed these individuals to urinary tract infections. This parallels our results in female transplant recipients with IFTA in which levels of Lactobacillus decrease by 6-8 months.
It is worth noting that the Streptococcus and Lactobacillus genera identified in our study are actually from same bacteria order (Lactobacillales) and phylum (Firmicutes). That explains why there were no differences in healthy control male vs. females at the phyla level. The evolutionary closeness between these two genera suggests that these bacteria have evolved in parallel and adapted to the role as primary human commensal organisms in the genitourinary space. Within the genera of Streptococcus, we identified 2 known commensal species commonly found in healthy individuals: S. anginosus and S. intermedius. Likewise, within the Lactobacillus genera in females, we found the following 3 commensal species: L. iners, L. jensenii and L. vaginalis. Combining Streptococcus and Lactobacillus, we found that these two genera comprise the vast majority of urinary microbiomes in all healthy men and women.
While cognizant of the large variability in genera between individuals, we found that the collective abundance of Streptococcus was significantly lower in all male transplants at 1 month post-transplant in comparison to healthy controls. We also found that Lactobacillus was non-significantly lower in all transplant patients compared to healthy controls at the same time point. An important finding of our study is that Streptococcus further decreased to only 3.4% in males with biopsy-proven IFTA, but actually increased to normal levels in male transplants with biopsy-documented normal histology and normal renal function (TX). Similarly, Lactobacillus decreased to 36% in females with IFTA, but stabilized in the TX females at 47%, although this did not reach statistical significance. Concomitantly, there was a significant increase in the number of genera or microbial complexity per sample in the male and female IFTA samples at 6 months in comparison to all the TX samples and healthy controls. The increase in the number of genera per sample represents a replacement of the dominant bacteria (i.e. Lactobacillus and Streptococcus) with a multitude of non-resident bacteria, many of which are known to be pathogenic (Table 2). Studies that provide evidence that these non-resident bacteria species can cause infection in transplant patients, or elicit an immune response, are also referenced in Table 2.
Table 2.
Non-resident pathogenic bacteria at high abundance in IFTA samples at 6-8 months post-transplant. Literature citations are provided for bacteria species that have demonstrated the ability to produce infection or cause immune stimulation.
| Males | Females | ||
|---|---|---|---|
| Sample | Bacteria Species (% composition) | Sample | Bacteria Species (% composition) |
| IIFTA |
Propionibacterium acne (17%), Cupriavidus NA (11%) (37) |
14IFTA |
Gardnerella vaginalis (18%) (38), Prevotella timonensis (15%) (39) |
| 2IFTA | Gardnerella vaginalis (90%) (38) | 15IFTA |
Gardnerella NA (47%), Prevotella melaninogenica (28%) (40) |
| 3IFTA |
Prevotella disiens (51%)(41), Campylobacter ureolyticus (14%) (42) |
16IFTA |
Gardnerella NA (45%), Corynebacterium NA (29%) |
| 4IFTA |
Gardnerella vaginalis (22%) (38), Veillonella NA (25%) |
17IFTA | NA |
| 5IFTA |
Corynebacterium sundsvallense & C.
tuberculostearicum (25%) (43), Peptoniphilus NA (23%) |
18IFTA |
Anaerococcus NA (29%), Pseudomonas NA (23%), Campylobacter ureolyticus (15%) (42) |
| 6IFTA |
Corynebacterium tuberculostearicum (51%) (43), Staphylococcus NA (39%) |
19IFTA |
Prevotella timonensis (15%) (39), Anaerococcus NA (11%) |
| 7IFTA |
Corynebacterium pyruviciproducens (46%) (43, 44) |
20IFTA | Atopobium vaginae (37%) (41) |
| 8IFTA |
Corynebacterium pyruviciproducens & C.
tuberculostearicum (50%) (43), Finegoldia magna (15%) (41, 45) |
21IFTA |
Finegoldia magna (22%) (45), Prevotella bivia & P. buccalis (41) (13%) |
| 9IFTA |
Corynebacterium pyruviciproducens C.
tuberculostearicum (41%) (43), Finegoldia magna (34%) (41), Anaerococcus prevoti) (18%) (46) |
22IFTA |
Prevotella timonensis (41%) (39), Gardnerella vaginalis (22%) (38), Anaerococcus lactolyticus (20%) (41) |
| 10IFTA |
Corynebacterium pyruviciproducens & C.
sundsvallense (14%) (43, 44) |
23IFTA |
Prevotella timonensis (39) & P. disiens
(36%) (41), Atopobium NA (16%) |
| 11IFTA |
Finegoldia magna (45%) (45), Corynebacterium pyruviciproducens (24%) (44) |
24IFTA | Gardnerella vaginalis (35%) (38) |
| 12IFTA |
Gardnerella NA (28%), Anaerococcus NA (20%) |
||
| 13IFTA |
Prevotella bivia (40%) (47), Anaerococcus lactolyticus & A. tetradius (35%) (41) |
||
Note: No specie (NA) is given in cases where the majority of the reads are unable to be mapped to a bacteria specie (i.e. ‘slash reads’).
In the months after kidney transplant surgery, there are alterations in the host microbiome in relation to exposure to hospital microbes, administration of perioperative and prophylactic antibiotics, and initiation of immunosuppressive medications (14). These published changes in the host microbiome as a function of time post-transplant, including our new data on the urinary microbiome in patients developing IFTA, suggest that the replacement of dominant bacteria with non-resident bacteria are likely influencing the immunobiology of the transplant but may also impact the overall clinical health of the transplant recipient. Our recent publication demonstrated that molecular profiling of kidney transplant biopsies with IFTA share a majority of both the genes and immune pathways found in biopsies of patients with clinical acute rejection (18). We concluded that IFTA is part of the arc of the same immune-mediated rejection process; in other words, IFTA is a histological feature of chronic rejection. Therefore, given the new data in this paper on the UMB, we propose that chronic, immune-mediated rejection is influenced directly and/or indirectly by the changing microbial populations in the genito-urinary tract.
In a study that characterized the vaginal microbiota of 396 asymptomatic, sexually active women using 16S sequencing, the most abundant vaginal bacteria genera were Lactobacillus, Prevotella, Megasphaera, Sneathia, Atopobium and Streptococcus (33). In all the female participants of our study, Lactobacillus, Prevotella and Streptococcus were among the top 5 most abundant UMB bacteria genera. Thus, given the known propensity for urine collection to be contaminated by vaginal flora, there remains a possibility that some of our UMB findings are a reflection of normal vaginal flora. Notwithstanding, these 3 bacteria genera were also among the top 5 abundant bacteria in healthy control males and transplanted male patients.
Alternatively, it is likely that maintenance of the Lactobacillus genus, whether as the dominant bacteria in UMB or vagina flora, has a correlation with overall health of the host. Lactobacillus is a facultative anaerobe that dominates the vaginal environment in women of reproductive age, and is important in maintaining an overall acidic environment (33). The maintenance of Lactobacillus as the dominant species has been consistently associated with healthy pregnancy outcomes and prevention of infections, including sexually transmitted pathogens like HIV (34), and a condition known as bacterial vaginosis. Bacterial vaginosis is a type of vaginal inflammation in which atypical bacteria such as Gardnerella replace Lactobacillus and other dominant vaginal resident bacteria. In a study of 231 vaginal samples, the molecular quantification of Atopobium vaginae and Gardnerella vaginalis by DNA levels had the highest predictive value for the diagnosis of bacterial vaginosis with a 99% sensitivity (35). Bacterial vaginosis consistently associates with preterm labor complications and worse pregnancy outcomes (25). In addition to protection from the influence of potential pathogenic bacteria, Lactobacillus likely plays an important role in tempering the immune response. Indeed, in a mouse model of graft-versus-host disease (GVHD), replenishment of the dominant Lactobacillus species in the gut significantly protected mice from disease (36).
A number of other limitations must be acknowledged. First, the high level of inter-individual variation we document in the urinary microbiomes obviates thinking about this work in simple biomarker or molecular diagnostic terms. Perhaps, monitoring individual changes and/or identifying specific classes of microbiota associated with increased risk for chronic rejection might eventually prove valuable. Second, the current early state of microbiome research makes any kind of mechanistic conclusions with the profiling results of human patient samples impossible. Are the changes we observe due to changes in transplant function with chronic rejection? Or is there a link between urinary microbiome changes with altered and/or activated immune effector cell populations? Third, since urine collection in females can often be contaminated with vaginal flora, the UMBs of healthy and transplant females may have been biased to demonstrate an increased prevalence of vagina flora and making it more difficult to identify IFTA-specific microbial changes.
In conclusion, unbalanced microbial communities have been associated with dysregulated immune responses (2, 3) and the development of inflammatory diseases such ulcerative colitis, Crohn’s disease and asthma. Interestingly, several studies including our own recent work (17), now demonstrate an association of cellular immune rejection with the development of IFTA (15, 19-26), suggesting that dysregulated immune responses may also be responsible for chronic rejection. Our theory is that the loss of protective, typically predominant resident bacteria and replacement with pathogenic non-resident bacteria in the UMB is acting to increase the immune response towards the transplanted kidney. This up-regulation of the immune system is then leading to non-specific graft injury and exacerbating the specific immune response against the allograft in a feed forward activation mechanism. The clinical significance of this study is that a future prospective trial may demonstrate that monitoring urinary microbiome changes in individual patients as a function of time and selective treatment may reduce susceptibility to rejection, including chronic rejection and development of IFTA, and ultimately improve long-term clinical outcomes. Finally, there is much to learn about how the microbiome in different tissues influences the immune repertoire and with that understanding should emerge additional and novel opportunities to prevent or treat human disease.
Supplementary Material
Figure S1: IFTA vs. TX at 6-8 months post-transplant. The range and 25th-75th quartile of Streptococcus in TX was 0-99% and 1-74%, respectively, compared to 0-12% and 0-6% in IFTA.
Acknowledgments
Funding for this work was provided by NIH NIAID grant U19 AI063603 (DRS, FH) and CTSA KL2 TR001112 (BDM).
Abbreviations
- ANOVA
analysis of variance
- IFTA
Interstitial fibrosis and tubular atrophy
- IRB
institutional review board
- NIH
National Institutes of Health
- SE
standard error
- TGCG
Transplant Genomic Collaborative Group
- TX
Treatment group with excellent functioning kidney
- UMB
urinary microbiome
Footnotes
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- 1.Proctor LM. The Human Microbiome Project in 2011 and beyond. Cell Host Microbe. 2011;10(4):287–91. doi: 10.1016/j.chom.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474(7351):307–17. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gevers D, Kugathasan S, Denson Lee A, Vázquez-Baeza Y, Van Treuren W, Ren B, et al. The Treatment-Naive Microbiome in New-Onset Crohn’s Disease. Cell Host & Microbe. 2014;15(3):382–92. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9(10):599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Blaser MJ. Helicobacter pylori colonization is inversely associated with childhood asthma. J Infect Dis. 2008;198(4):553–60. doi: 10.1086/590158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnold IC, Dehzad N, Reuter S, Martin H, Becher B, Taube C, et al. Helicobacter pylori infection prevents allergic asthma in mouse models through the induction of regulatory T cells. J Clin Invest. 2011;121(8):3088–93. doi: 10.1172/JCI45041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7(307) doi: 10.1126/scitranslmed.aab2271. 307ra152. [DOI] [PubMed] [Google Scholar]
- 9.Alegre ML, Mannon RB, Mannon PJ. The microbiota, the immune system and the allograft. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2014;14(6):1236–48. doi: 10.1111/ajt.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6(237) doi: 10.1126/scitranslmed.3008599. 237ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan S, Rahman HN, Okamoto T, Matsunaga T, Fujiwara Y, Sawa T, et al. Promotion of atherosclerosis by Helicobacter cinaedi infection that involves macrophage-driven proinflammatory responses. Sci Rep. 2014;4:4680. doi: 10.1038/srep04680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balmer ML, Slack E, de Gottardi A, Lawson MA, Hapfelmeier S, Miele L, et al. The liver may act as a firewall mediating mutualism between the host and its gut commensal microbiota. Sci Transl Med. 2014;6(237) doi: 10.1126/scitranslmed.3008618. 237ra66. [DOI] [PubMed] [Google Scholar]
- 13.Whiteside SA, Razvi H, Dave S, Reid G, Burton JP. The microbiome of the urinary tract[mdash]a role beyond infection. Nat Rev Urol. 2015;12(2):81–90. doi: 10.1038/nrurol.2014.361. [DOI] [PubMed] [Google Scholar]
- 14.Fricke WF, Maddox C, Song Y, Bromberg JS. Human Microbiota Characterization in the Course of Renal Transplantation. American Journal of Transplantation. 2014;14(2):416–27. doi: 10.1111/ajt.12588. [DOI] [PubMed] [Google Scholar]
- 15.Mannon RB, Matas AJ, Grande J, Leduc R, Connett J, Kasiske B, et al. Inflammation in areas of tubular atrophy in kidney allograft biopsies: a potent predictor of allograft failure. American Journal of Transplantation. 2010;10(9):2066–73. doi: 10.1111/j.1600-6143.2010.03240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park WD, Griffin MD, Cornell LD, Cosio FG, Stegall MD. Fibrosis with Inflammation at One Year Predicts Transplant Functional Decline. Journal of the American Society of Nephrology : JASN. 2010;21(11):1987–97. doi: 10.1681/ASN.2010010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cosio FG, Grande JP, Wadei H, Larson TS, Griffin MD, Stegall MD. Predicting Subsequent Decline in Kidney Allograft Function from Early Surveillance Biopsies. American Journal of Transplantation. 2005;5(10):2464–72. doi: 10.1111/j.1600-6143.2005.01050.x. [DOI] [PubMed] [Google Scholar]
- 18.Modena BD, Kurian SM, Gaber LW, Waalen J, Su AI, Gelbart T, et al. Gene Expression in Biopsies of Acute Rejection and Interstitial Fibrosis/Tubular Atrophy Reveals Highly Shared Mechanisms That Correlate With Worse Long-Term Outcomes. American Journal of Transplantation. 2016 doi: 10.1111/ajt.13728. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cosio FG, Grande JP, Wadei H, Larson TS, Griffin MD, Stegall MD. Predicting subsequent decline in kidney allograft function from early surveillance biopsies. Am J Transplant. 2005;5(10):2464–72. doi: 10.1111/j.1600-6143.2005.01050.x. [DOI] [PubMed] [Google Scholar]
- 20.El Ters M, Grande JP, Keddis MT, Rodrigo E, Chopra B, Dean PG, et al. Kidney Allograft Survival After Acute Rejection, the Value of Follow-Up Biopsies. American Journal of Transplantation. 2013;13(9):2334–41. doi: 10.1111/ajt.12370. [DOI] [PubMed] [Google Scholar]
- 21.Nickerson PW, Rush DN. Rejection: an integrated response. Am J Transplant. 2013;13(9):2239–40. doi: 10.1111/ajt.12365. [DOI] [PubMed] [Google Scholar]
- 22.Nankivell BJ, Chapman JR. The significance of subclinical rejection and the value of protocol biopsies. Am J Transplant. 2006;6(9):2006–12. doi: 10.1111/j.1600-6143.2006.01436.x. [DOI] [PubMed] [Google Scholar]
- 23.Legendre C, Thervet E, Skhiri H, Mamzer-Bruneel MF, Cantarovich F, Noel LH, et al. Histologic features of chronic allograft nephropathy revealed by protocol biopsies in kidney transplant recipients. Transplantation. 1998;65(11):1506–9. doi: 10.1097/00007890-199806150-00020. [DOI] [PubMed] [Google Scholar]
- 24.Seron D, Moreso F. Protocol biopsies in renal transplantation: prognostic value of structural monitoring. Kidney Int. 2007;72(6):690–7. doi: 10.1038/sj.ki.5002396. [DOI] [PubMed] [Google Scholar]
- 25.Heilman RL, Devarapalli Y, Chakkera HA, Mekeel KL, Moss AA, Mulligan DC, et al. Impact of subclinical inflammation on the development of interstitial fibrosis and tubular atrophy in kidney transplant recipients. Am J Transplant. 2010;10(3):563–70. doi: 10.1111/j.1600-6143.2009.02966.x. [DOI] [PubMed] [Google Scholar]
- 26.Rush DN, Henry SF, Jeffery JR, Schroeder TJ, Gough J. Histological findings in early routine biopsies of stable renal allograft recipients. Transplantation. 1994;57(2):208–11. doi: 10.1097/00007890-199401001-00009. [DOI] [PubMed] [Google Scholar]
- 27.Cheng J, Torkamani A, Grover RK, Jones TM, Ruiz DI, Schork NJ, et al. Ectopic B-cell clusters that infiltrate transplanted human kidneys are clonal. Proc Natl Acad Sci U S A. 2011;108(14):5560–5. doi: 10.1073/pnas.1101148108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grover RK, Cheng J, Peng Y, Jones TM, Ruiz DI, Ulevitch RJ, et al. The costimulatory immunogen LPS induces the B-Cell clones that infiltrate transplanted human kidneys. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(16):6036–41. doi: 10.1073/pnas.1202214109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alneberg J, Johan Bengtsson-Palme, Ino de Bruijn, Luisa Hugerth, Mikael Huss, Thomas Svensson. Normalization of count data from the metagenomic data sets GitHub2014. [Available from: http://2014-5-metagenomics-workshop.readthedocs.org/en/latest/annotation/normalization.html. [Google Scholar]
- 30.Siddiqui H, Nederbragt AJ, Lagesen K, Jeansson SL, Jakobsen KS. Assessing diversity of the female urine microbiota by high throughput sequencing of 16S rDNA amplicons. BMC Microbiology. 2011;11:244. doi: 10.1186/1471-2180-11-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis DA, Brown R, Williams J, White P, Jacobson SK, Marchesi JR, et al. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Frontiers in Cellular and Infection Microbiology. 2013;3:41. doi: 10.3389/fcimb.2013.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fouts DE, Pieper R, Szpakowski S, Pohl H, Knoblach S, Suh M-J, et al. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. Journal of Translational Medicine. 2012;10:174. doi: 10.1186/1479-5876-10-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4680–7. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marrazzo JM. Interpreting the epidemiology and natural history of bacterial vaginosis: Are we still confused? Anaerobe. 2011;17(4):186–90. doi: 10.1016/j.anaerobe.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menard J-P, Fenollar F, Henry M, Bretelle F, Raoult D. Molecular Quantification of Gardnerella vaginalis and Atopobium vaginae Loads to Predict Bacterial Vaginosis. Clinical Infectious Diseases. 2008;47(1):33–43. doi: 10.1086/588661. [DOI] [PubMed] [Google Scholar]
- 36.Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, Dudakov JA, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. The Journal of Experimental Medicine. 2012;209(5):903–11. doi: 10.1084/jem.20112408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tena D, Losa C, Medina MJ, Saez-Nieto JA. Muscular abscess caused by Cupriavidus gilardii in a renal transplant recipient. Diagnostic microbiology and infectious disease. 2014;79(1):108–10. doi: 10.1016/j.diagmicrobio.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 38.Sivadon-Tardy V, Roux A-L, Piriou P, Herrmann J-L, Gaillard J-L, Rottman M. Gardnerella vaginalis Acute Hip Arthritis in a Renal Transplant Recipient. Journal of Clinical Microbiology. 2009;47(1):264–5. doi: 10.1128/JCM.01854-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glazunova OO, Launay T, Raoult D, Roux V. Prevotella timonensis sp. nov., isolated from a human breast abscess. International journal of systematic and evolutionary microbiology. 2007;57:883–6. doi: 10.1099/ijs.0.64609-0. Pt 4. [DOI] [PubMed] [Google Scholar]
- 40.Kierzkowska M, Majewska A, Sawicka-Grzelak A, Mlynarczyk A, Chmura A, Durlik M, et al. Specific character of anaerobic bacterial infections in patients treated in transplantation wards at one of the clinical hospitals in Warsaw. Transplantation proceedings. 2014;46(8):2586–8. doi: 10.1016/j.transproceed.2014.08.033. [DOI] [PubMed] [Google Scholar]
- 41.Imirzalioglu C, Hain T, Chakraborty T, Domann E. Hidden pathogens uncovered: metagenomic analysis of urinary tract infections. Andrologia. 2008;40(2):66–71. doi: 10.1111/j.1439-0272.2007.00830.x. [DOI] [PubMed] [Google Scholar]
- 42.O'Donovan D, Corcoran GD, Lucey B, Sleator RD. Campylobacter ureolyticus: a portrait of the pathogen. Virulence. 2014;5(4):498–506. doi: 10.4161/viru.28776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernard K. The genus corynebacterium and other medically relevant coryneform-like bacteria. J Clin Microbiol. 2012;50(10):3152–8. doi: 10.1128/JCM.00796-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tong J, Han Q, Wang S, Su Z, Zheng D, Shen P, et al. Corynebacterium pyruviciproducens, as an immune modulator, can promote the activity of macrophages and up-regulate antibody response to particulate antigen. Experimental biology and medicine (Maywood, NJ) 2012;237(11):1322–30. doi: 10.1258/ebm.2012.012181. [DOI] [PubMed] [Google Scholar]
- 45.Levy PY, Fenollar F, Stein A, Borrione F, Raoult D. Finegoldia magna: a forgotten pathogen in prosthetic joint infection rediscovered by molecular biology. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2009;49(8):1244–7. doi: 10.1086/605672. [DOI] [PubMed] [Google Scholar]
- 46.Murphy EC, Frick I-M. Gram-positive anaerobic cocci – commensals and opportunistic pathogens. FEMS Microbiology Reviews. 2013;37(4):520–53. doi: 10.1111/1574-6976.12005. [DOI] [PubMed] [Google Scholar]
- 47.Brook I. Microbiology of polymicrobial abscesses and implications for therapy. Journal of Antimicrobial Chemotherapy. 2002;50(6):805–10. doi: 10.1093/jac/dkg009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: IFTA vs. TX at 6-8 months post-transplant. The range and 25th-75th quartile of Streptococcus in TX was 0-99% and 1-74%, respectively, compared to 0-12% and 0-6% in IFTA.