Abstract
Background/Objectives
Recent data suggests that common genetic risk for metabolic disorders such as obesity may be human-specific and exert effects through the central nervous system. To overcome the limitation of human tissue access for study, we have generated induced human pluripotent stem cell (hiPSC)-derived neuronal cultures which recapture many features of hypothalamic neurons within the arcuate nucleus. Here we have comprehensively characterized this model across development, benchmarked these neurons to in vivo events, and demonstrate a link between obesity risk variants and hypothalamic development.
Methods
The dynamic transcriptome across neuronal maturation was examined using microarray and RNAseq methods at 9 time points. K-means clustering of the longitudinal data was conducted to identify co-regulation and miRNA control of biological processes. The transcriptomes were compared to those of 103 samples from 13 brain regions reported in the Genotype-Tissue Expression database (GTEx) using principal components analysis. Genes with proximity to body mass index (BMI)-associated genetic variants were mapped to the developmentally expressed genesets, and enrichment significance assessed with Fisher’s exact test.
Results
The human neuronal cultures have a transcriptional and physiological profile of NPY/AGRP ARC neurons. The neuronal transcriptomes were highly correlated with adult hypothalamus as compared to any other brain region from the GTEx. Also, roughly 25% of the transcripts showed substantial changes in expression across neuronal development and potential co-regulation of biological processes that mirror neuronal development in vivo. These developmentally expressed genes were significantly enriched for genes in proximity to BMI-associated variants.
Conclusions
We affirmed the utility of this in vitro human model to study development of key hypothalamic neurons involved in energy balance and show that genes at loci associated with body weight regulation may share a pattern of developmental regulation. These data support the need to investigate early development to elucidate human-specific CNS pathophysiology underlying obesity susceptibility.
INTRODUCTION
The hypothalamus is a major brain hub that regulates energy homeostasis.(1–5) A core component is the arcuate nucleus (ARC) which is unique in its role of sensing and integrating peripheral and nutrient signals to orchestrate appropriate whole body responses aimed at restoring energy balance, including modulation of brown adipose tissue (BAT) thermogenesis(6, 7). The neural projections from the ARC are central in this homeostatic regulation. Exciting advances have been made in understanding this hypothalamic activity; however, the genetic and molecular basis of this regulation in humans is not well understood as access to this tissue is a major barrier and available only postmortem. While rodent models are useful for a broad understanding of hypothalamic regulation, genetic risk for associated disorders such as diabetes and obesity may be human-specific and, therefore, require human models to identify the molecular mechanism. Recent meta-analyses of genome-wide association studies (GWAS) to identify genetic risk for measures of obesity in diverse populations have shown that the majority are located in noncoding and potentially regulatory regions which are not conserved between mouse and human.(8–11) Moreover, genes in proximity to genetic variants associated with body mass index (BMI), a common clinical measure of obesity, were strikingly overrepresented in the processes of hypothalamic function, neuronal transmission and development.(8–11) Because of metabolic and genetic regulatory differences between humans and rodent models, it will be important to investigate the role of these variants on hypothalamic and neuronal function in human context. Additionally, growing evidence from maternal programming paradigms has underscored the critical importance of proper hypothalamic neuronal development in maintaining energy homeostasis.(12–14) Therefore, genetic variants that impact on traits influencing energy homeostasis such as obesity may manifest their effects early in hypothalamic development, which in humans occurs largely in utero.
To overcome the barrier of limited access to tissue for study, we have developed protocols to generate human induced pluripotent stem cell (hiPSC)-derived neuronal cultures that recapitulate many of the features of hypothalamic neurons from the arcuate nucleus. We have also benchmarked this in vitro model to in vivo events that are pivotal in hypothalamic development using isoform switching information. To further affirm the utility of our model in uncovering mechanisms faithful to the hypothalamus we compared gene expression patterns of iPSC-derived hypothalamic cells to adult hypothalamus gene expression reported in the Genotype-Tissue Expression database (GTEx)(15). This benchmarking revealed that our terminally differentiated cultures manifest gene expression patterns closely mirroring that of adult hypothalamus as compared to any other brain region. Further, our human model provides a unique window into exploring the impact of genetic variants on changes in the hypothalamus in a neuronal stage-specific manner. We have performed a longitudinal analysis of expression profiles of control human iPSC-derived neurons to examine co-regulation of biological processes and potential control by miRNA across development and maturation. With these data, we have identified distinct developmental expression patterns of obesity GWAS-implicated genes and potential regulators that may be involved in neuronal influence on body mass index.
METHODS
Human iPSC generation and characterization is described in full in Supplemental Methods and Figure S1. Briefly, human newborn fibroblasts (ATCC CRL-2703) were reprogrammed using episomal vectors expressing the transcription factors C-MYC, OCT4 (POU5F1), SOX2, NANOG, and KLF4.
Neuronal Differentiation
The verified hiPSC from passage 15 were dissociated and grown as cellular aggregates or embryoid bodies (EBs) on ultra-low attachment 6-well plates in low insulin-modified hES medium without bFGF for 4 days, Neural induction medium (NIM) containing DMEM/F12, supplemented with 1× N2, 0.1 mM NEAA, 4ug/ml heparin was then supplied for 2 days. At day 6, the EB aggregates were transferred to dishes coated with a combination of poly-L-ornithine and laminin and fed fresh NIM every other day. Primitive neuroepithelial (NE) cells appeared at day 8–10 and were treated with 50 ng/ml WNT3A in NIM. On day 14–17, neural tube-like rosettes were formed and the rosettes were then isolated and transferred onto new coated dishes and grown in NIM containing WNT3A 50ng/ml for 5–10 days, dissociated with Accutase and re-plated onto coated dishes in neuronal differentiation medium (NDM) containing reduced insulin neurobasal medium(16), 1× N2 and 2X B27, 0.1 mM NEAA, and supplemented with 1uM cAMP and 10ng/ml WNT3A for 3 days. The neurons were then maintained in this medium supplemented with BDNF, GDNF and IGF1 (10ng/ml each).
Immunocytochemical characterization of human neurons
The differentiated neurons were harvested and transferred to PLO/laminin-coated glass coverslips for immunostaining with a combination of neuronal-specific markers: anti-beta III tubulin (Abcam, CAT#ab7751); anti-PAX6 (Abcam CAT#ab78545); anti-VGLUT1 (vesicular glutamate transporter1) (Abcam, CAT#ab72311); anti-GRM3 (glutamate receptor, metabotropic-3) (Abcam, CAT# ab188750), anti-MAP2 (microtubule-associated protein-2) (Abcam, CAT#ab11267), anti-AGRP (agouti-related peptide) (Abcam, CAT#113481), anti-VGAT (vesicular GABA transporter, VIAAT)(Synaptic Systems GMBH,CAT# 131 002), anti-NPY (Neuropeptide Y) (ThermoFisher Scientific, CAT#028-08-02), anti-GLP1R (glucagon-like peptide 1 receptor)(Abcam, CAT#ab39072).
Flow cytometry
Day 70 neuronal cultures were dissociated with Accutase, washed 1× in neurobasal medium and 2×10^6 cells aliquoted for each assay. Cells were washed and resuspended in 1ml protein/serum free PBS. A fixable viability dye (Ghost dye Violet 510, Tonbo Biosciences, San Diego) was used to discriminate dead cells with minor modification to vendor protocols to accommodate primary neurons. Briefly, 1ul of 1:5 diluted Ghost dye was added to each 1 mL of cell suspension, vortexed, and incubated for 10 minutes at RT protected from light. Cells were washed 1× with 2% FBS 0.05% azide in PBS and 1× with PBS, fixed with 2% paraformaldehyde 20 min, and then permeabilized in 0.1% Triton for 15min at RT. For indirect immunofluorescence staining, Fc binding sites were saturated with Fc blocking mAb (Human BD Fc clone 3070, Fisher Scientific) 30 min at 4°C in the dark. Primary mAbs were added and incubated for 30 min at 4°C. Cells were then washed twice followed by secondary mAb staining for 30 min a 4°C and analyzed by flow cytometry. Primary antibodies used were AGRP (Abcam, cat#113481) and NPY (Thermo Fisher Scientific, cat#028-08-02). Secondary antibodies were Alexa Fluor 555 conjugated anti-rabbit (Invitrogen, cat#A21428) and Alexa Fluor 488 conjugated anti-mouse IgG1 (Invitrogen, cat#A21121). Controls included unstained samples, single stained samples for compensation, and fluorescent minus one (FMO) for gating purposes. Analyses were carried out using a flow cytometer (LSRII; Becton Dickinson, San Jose, CA). Data analysis was performed using Diva software 8.0.1 (Becton Dickinson, San Jose, CA). All assays were performed on 3 independent neuronal cultures (i.e., from different iPS colonies).
Neuronal Expression Profiling
Total RNA at 9 time points from day 0 to 115 during the differentiation and maturation process was isolated using Qiagen RNeasy kit, and quality control assessed using the Agilent 2100 Bioanalyzer. The microarray experiment was conducted in duplicate using Illumina HT-12 v4 Beadchips. For small RNA sequencing, 1 ug of total RNAs were processed using the BIOO Scientific NextFlex Small RNA Sequencing Kit (Cat # 5132-01) according to the manufacturer’s instructions. Libraries were constructed using Illumina/Universal (KAPA Biosystems). A pool of 16 libraries was used for cluster generation at a concentration of 10 nM using cBot Cluster System (Illumina). Single-read 50bp sequencing was performed with an Illumina HiSeq 2000. qRTPCR verification of transcripts and Western blotting is described in Supplemental Methods and Figure S2.
Physiologic verification of neurons
Functioning adiponectin and leptin receptors were assessed by changes in NPY expression in response to adiponectin/aCrp30 and leptin treatment: ~75Day neuronal cultures were held in neuronal maintenance medium free of insulin and at 5.5mM glucose 24hours and then supplemented with either 1ug/ml recombinant human adiponectin/aCrp30 or 50 nM recombinant human leptin (R&D Systems, Minneapolis MN). Cells in 24-well plates were collected at 4 hours of exposure and RNA isolated for expression analysis using qRT-PCR as described above and using 18SRNA for normalization. Four independent neuronal cultures derived from 2 iPSC were tested in triplicate. A two-tailed t-test was used to determine p-values. The sequences for the primer pairs used are given in Table S1. Functioning glutamate receptors were assessed by patch clamp as described in Supplemental Methods Figure S5.
Statistical analyses
Differential transcriptomics
Data generated from Illumina Inc. GenomeStudio was pre-processed by log2 transformation and quantile normalization implemented in the lumi R package (17). Differential expression analysis was carried out with Limma package implemented in R(18). Sample relationship during time progression was constructed using multidimensional scaling algorithm. K-means cluster algorithm was applied to identify gene clusters that follow similar patterns during progression from Day 35–115. Transcripts with significant differential expression (adjusted p-value<.01, absolute fold change>2) between at least 2 time points were included in the analysis. Calinski and Silhouette index were evaluated to identify optimum number of clusters for k-means algorithm. Then, for each cluster, gene ontology and enrichment analysis for miRNAs and transcriptional factors were carried out using Toppgene(19), David(20) and FIRE(21). Bonferroni adjusted p-values are reported. The nearest-range as well as long-range effect of BMI-associated single nucleotide polymorphisms (SNPs) on each cluster were evaluated by using Fisher’s exact test. An additional K-means cluster analysis was performed using only RNAseq data for small RNAs.
miRNA sequencing data analysis
Sequence reads were processed using Illumina Flicker 3.0 pipeline and alignment using Illumina ELAND. Possible contamination sequence reads containing mitochondrial DNA, rRNA, and primers were removed. Remaining sequence reads were then matched against miRBase release 18 (http://www.miRBase.org) to identify mature and precursor sequences, and read counts aligned to each microRNA were reported for further analysis.
Comparison to GTEx data
Raw data (.CEL files) of all brain tissue originated samples from GEO database (GSE45878) were downloaded and pre-processed using oligo package in R(22). Probes that map to multiple genes were discarded from further analyses. Potential batch effects between our hiPSCs and GTEx samples were removed by using sva package in R(23). Sample relationship between our hiPSCs and brain tissues from GTEx project were examined using principal component analysis (PCA). Hierarchical clustering based on average linkage was also carried out using genes that met the criteria of coefficient of variation above second quantile, average gene expression above second quantile and standard deviation above second quantile.
RESULTS
The derived neuronal cultures were characterized through transcriptomics and verified using immunocytochemistry with specific neuronal markers. Total RNA for 2 samples at each of 9 time points, and from the source human fibroblast cells, were assayed with Illumina HT-12 v4 Beadchips. Following standardization steps, 34 694 unique probes representing 15 631 genes were robustly detected and their expression compared across the multiple time points. The neuronal differentiation protocol and corresponding time points for expression analyses are shown in Figure 1A. A heatmap of the expression profiles for the genes of moderate to high expression that were differentially expressed across at least one time point is shown in Figure 1B. Distinctive expression profiles were observed for the time points before and after Day 35 as expected since Day 35 marks the earliest differentiated neurons. Results from multidimensional scaling of mRNA and miRNA expression profiles between each time point are plotted in Figure 2. Similar relationships are seen for both mRNA and miRNA temporal profiles including a similar progression from Days 35–115, suggesting capture of miRNA regulation during differentiation and maturation. The GEO accessions for the mRNA and miRNA datasets are GSE78789 and GSE78812, respectively. qRT-PCR and Western blotting verification and characterization was conducted on a subset of genes as shown in Figure S2.
Figure 1.
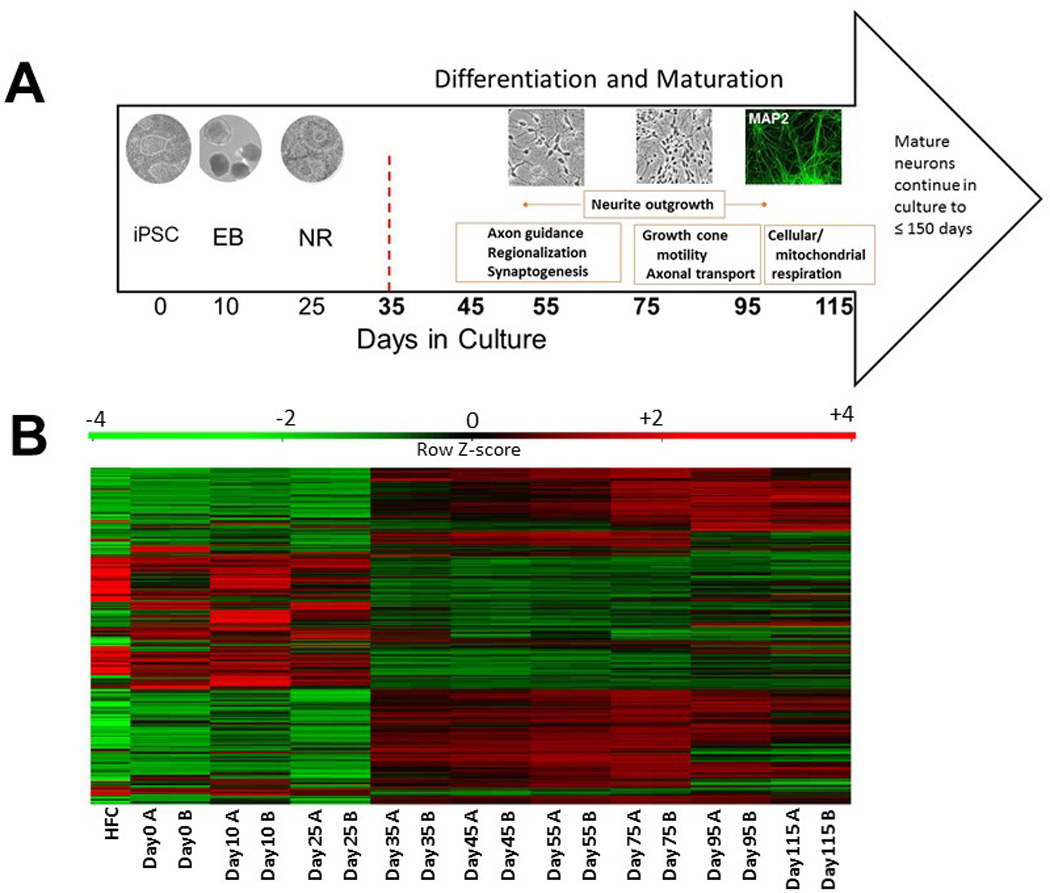
A) Neuronal differentiation timecourse, Days in culture and representative culture images are shown from left to right. Representative culture images taken at 20× are shown above corresponding time point. Day 35 is marked with a red hatched line to indicate the neuronal developmental shift at this stage in which first early neurons appear. Biological processes occurring across stages, as interpreted using GO data, are shown in orange blocks. iPSC – induced pluripotent stem cell colony; EB – embryoid body; NR – neural rosette; B) Heatmap of temporal expression profiles for iPSC and derived neurons. Heatmap plotting genes with average log2 intensity > 6 and variance across samples above 1 (~1500 genes). HFC - human fibroblast cells
Figure 2. Multidimensional scaling of temporal A) mRNA and B) miRNA expression profiles.
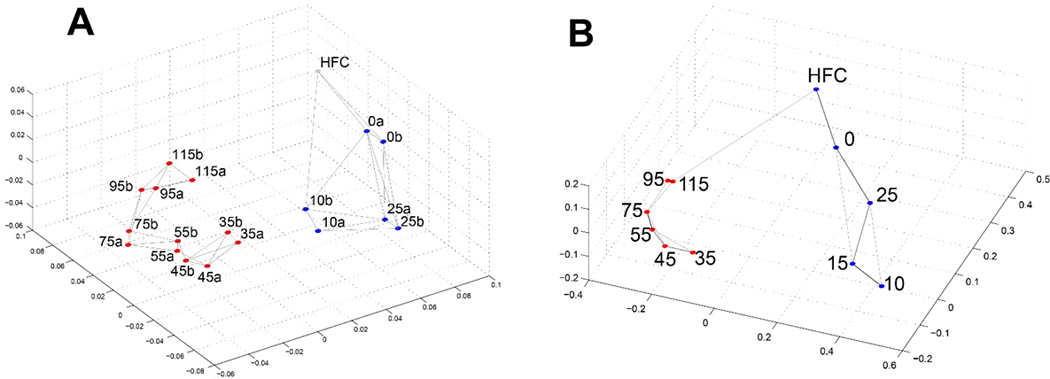
Pairwise sample Pearson correlation is mapped into distance as 1-correlation. Pre- and post-neuronal induction time points are marked as blue and red dots, respectively. HFC is human fibroblast cell before reprogramming
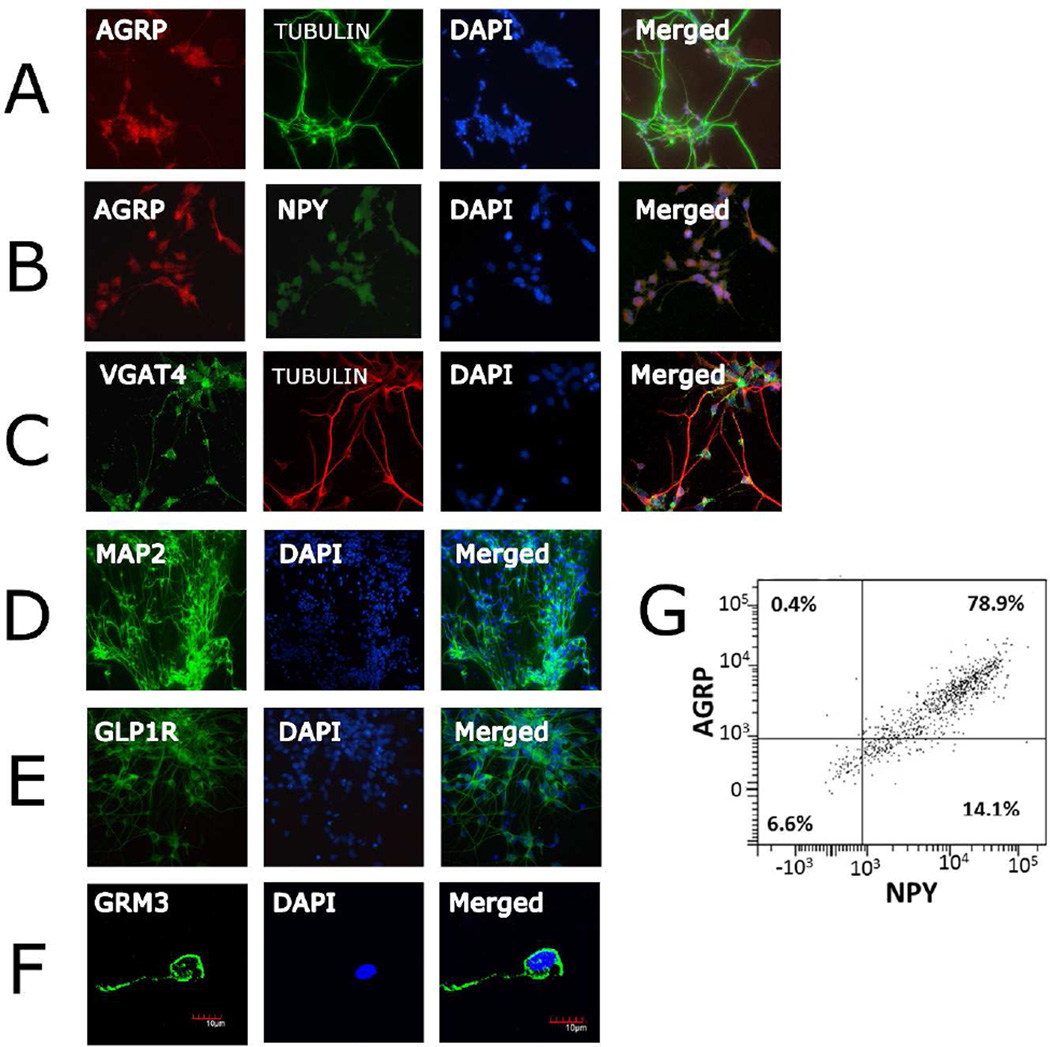
The hypothalamic ARC contains several distinct neuronal populations including the orexigenic neuropeptide Y (NPY)/agouti-related peptide (AGRP)-expressing neurons, the anorexigenic proopiomelanocortin (POMC)-expressing neurons, dopaminergic, catecholaminergic, and growth hormone-releasing hormone (GHRH) neurons. The profiles of the mature iPSC-derived neurons indicate that they are primarily NPY/AGRP neurons. In addition to NPY, the glutamate transporters SLC17A6 (VGLUT2) and SLC17A7 (VGLUT1), the metabotropic glutamate receptors GRM2 and GRM3, and the GABA vesicular transporter SLC32A1 (VGAT) and GABA B receptors(24) are among the highly expressed genes, whereas no expression is observed for the dopamine transporter gene SLC6A3 (DAT1), the norepinephrine transporter SLC6A2 (NET), the serotonin transporter SLC6A4 (SERT), the vesicular monoamine transporters SLC18A1 and SLC18A2 (VMAT1 and VMAT2), nor POMC or MC4R. The transcription factors archaete-scute complex 1 (ASCL1) and POU class 3 homeobox 2 (POU3F2) were highly expressed by early neurons. In mice, ASCL1 promotes proliferation of NPY/AGRP, GHRH and catecholaminergic neurons of the ARC.(25) Importantly, GHRH is not expressed by our iPSC-derived neurons and as stated above, nor are the catecholamines NET and DAT1. High levels of adiponectin receptors 1 and 2 (ADIPOR1 and ADIPOR2) as well as leptin receptor (LEPR) were detected in the differentiated neurons. No expression of oligodendrocyte specific markers or the astrocyte marker glial fibrillary acidic protein (GFAP) was detected (Figure S4). Immunocytochemical analyses show strong expression and co-localization of AGRP and NPY in the cultures and confirm expression of key proteins such as VGAT, glucagon-like peptide 1 receptor (GLP1R)(26) and metabotropic glutamate receptor 3 (Figure 3). Further, we conducted flow cytometric analyses of neurons using indirect immunofluorescent staining for NPY and AGRP on 3 independent neuronal cultures under standard non-fasting conditions similar to that used for transcriptomic profiling. We observed between 93–96% of the cells to be positive for NPY, and a total of 80% to be double positive for NPY and AGRP (Figure 3G and Figure S3). Less than 0.5% of cells were positive only for AGRP as expected. It is well known that AGRP expression in NPY neurons is increased by fasting conditions, and therefore it is possible that a greater number of the NPY positive neurons would also express measureable AGRP if tested under a fasting-like state. Taken together, the differentiation method we used results in neurons with a profile similar to the ARC orexigenic neurons expressing NPY and AGRP.
Figure 3. Verification of hiPSC-derived neurons with neuron-specific markers.
Immunostaining at day 95 with A) AGRP/tubulin/DAPI, 20×, B) AGRP/NPY/DAPI, 20×, C) VGAT/tubulin/DAPI, 20× D) MAP2/DAPI, 20×, E) GLP1R/DAPI, 20×; F) GRM3/ DAPI, 40×; G) Representative FACS analysis of neuronal cultures using indirect immunofluorescence for NPY and AGRP. Percentage of cells positive for NPY or AGRP, or both are shown in each quadrant.
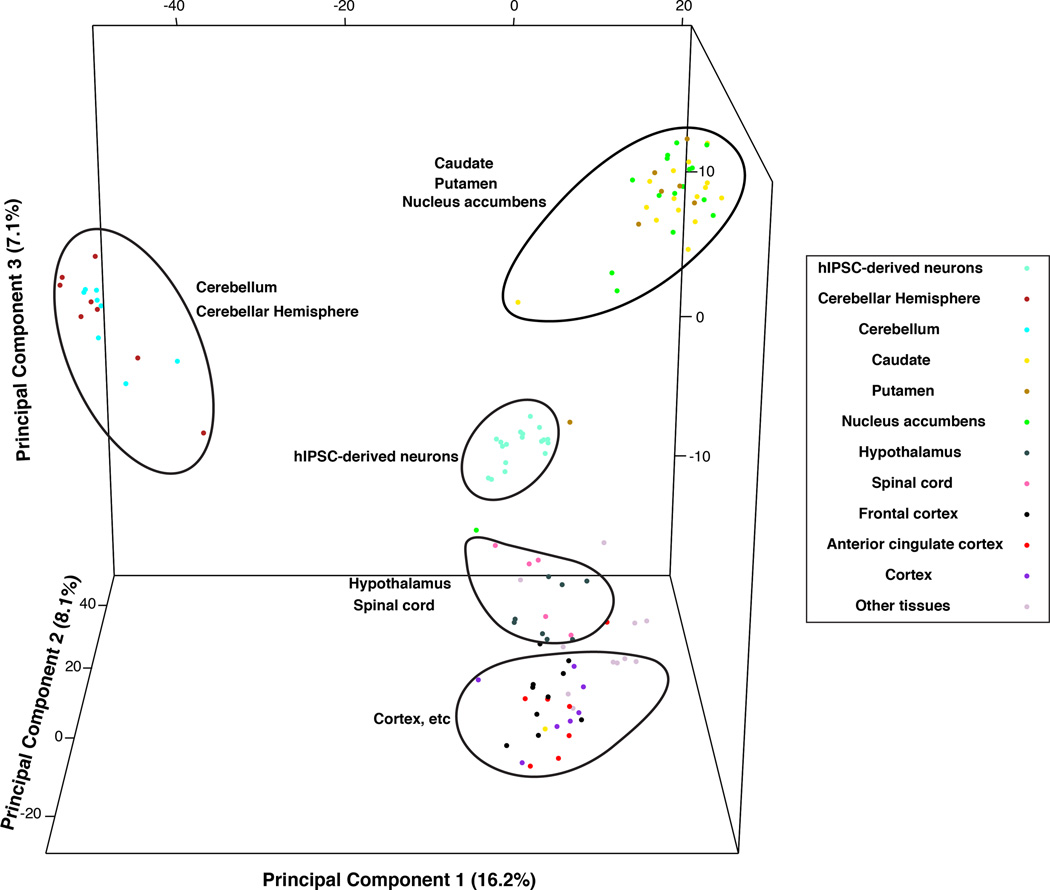
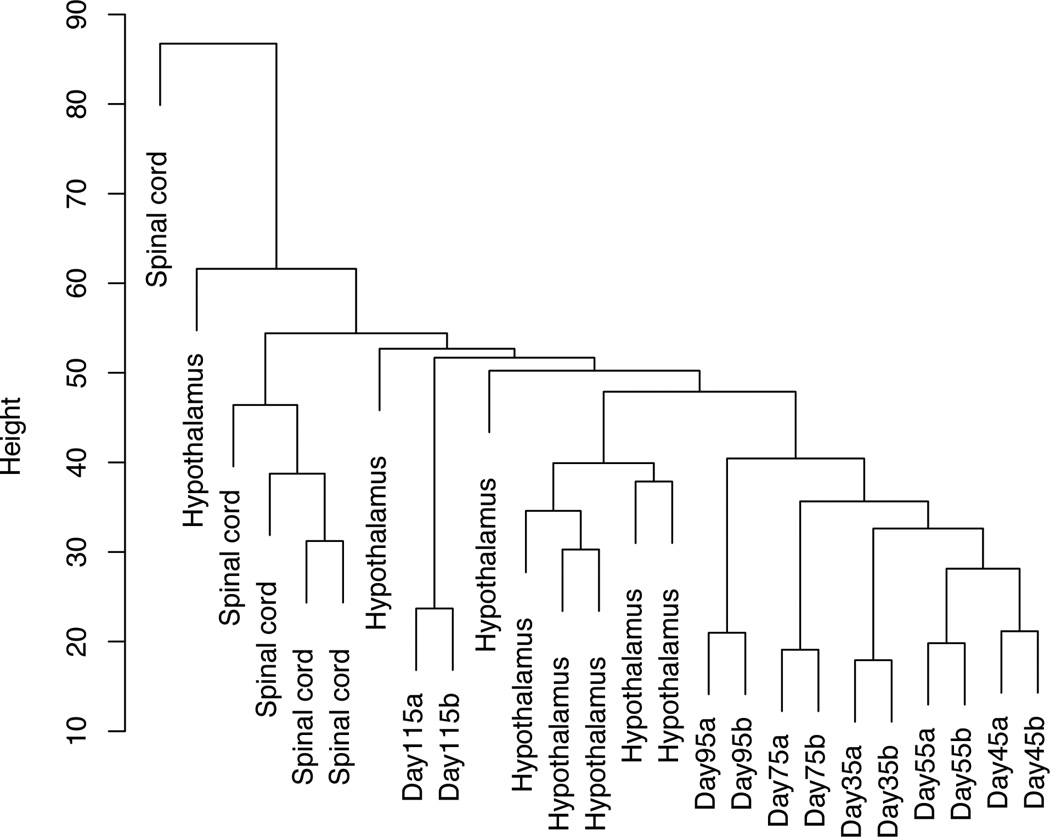
To further affirm the utility of our model in representing specific neurons of the hypothalamus, we compared gene expression patterns of iPSC-derived hypothalamic cells to adult brain-region specific gene expression reported in the GTEx. Principal components analyses of GTEx gene expression data for 103 samples from 13 brain regions as well as expression profiles from each post-neuronal induction time point for the iPSC-derived neurons indicates that the expression profiles of our derived neurons most closely correlate to data from hypothalamic samples (Figure 4A). Next, we performed hierarchical cluster analyses using all GTEX spinal cord and hypothalamic samples and all time points for the iPSC-derived neurons. The results and dendogram (Figure 4B) indicate that the neurons we generated are more similar to hypothalamic tissues as compared to spinal cord. The correlation with hypothalamic samples does not completely overlap as expected as the GTEx hypothalamic tissue samples include many non-neuronal cell types as well.
Figure 4.
A) Plot of top 3 principal components distinguishing expression profiles for GTEx brain samples and iPSC-derived neurons Days 35–115. Sample relations plotted are based on the 4354 genes with log2 expression > 5 and coefficient of variation > 0.1. B) Dendogram of hierarchical cluster analysis using GTEx hypothalamus and spinal cord samples and iPSC-derived neurons. Sample relations are based on 1763 shared genes.
Physiologic testing of neuronal cultures
The functionality of these hypothalamic-like neurons was confirmed using neuropeptide response to external hormonal stimuli and patch-clamp recordings. Several studies have demonstrated hormonal regulation of peptide expression in NPY/AGRP neurons of the arcuate nucleus in relation to obesity whereby adiponectin increases and leptin decreases NPY expression and secretion. Following treatment of our neurons with either 1ug/ml recombinant human adiponectin/aCrp30 or 50 nM recombinant human leptin in 5mM glucose-containing media, we observed a significant >2-fold increase and 1.6-fold decrease, respectively, of NPY mRNA (Figure 5). Although an indirect effect of treatment cannot be ruled out, these results suggest that these cells are expressing functioning adiponectin and leptin receptors. Next, as glutamatergic input to NPY/AGRP neurons of the ARC plays a key role in controlling energy balance(27), we used whole-cell recordings of 75-day neurons to confirm the presence of functioning glutamate receptors in the cultures as shown in Supplemental Figure S5.
Figure 5. Fold ΔNPY mRNA levels following 4-hour treatment with adiponectin/ACRP30 or leptin.
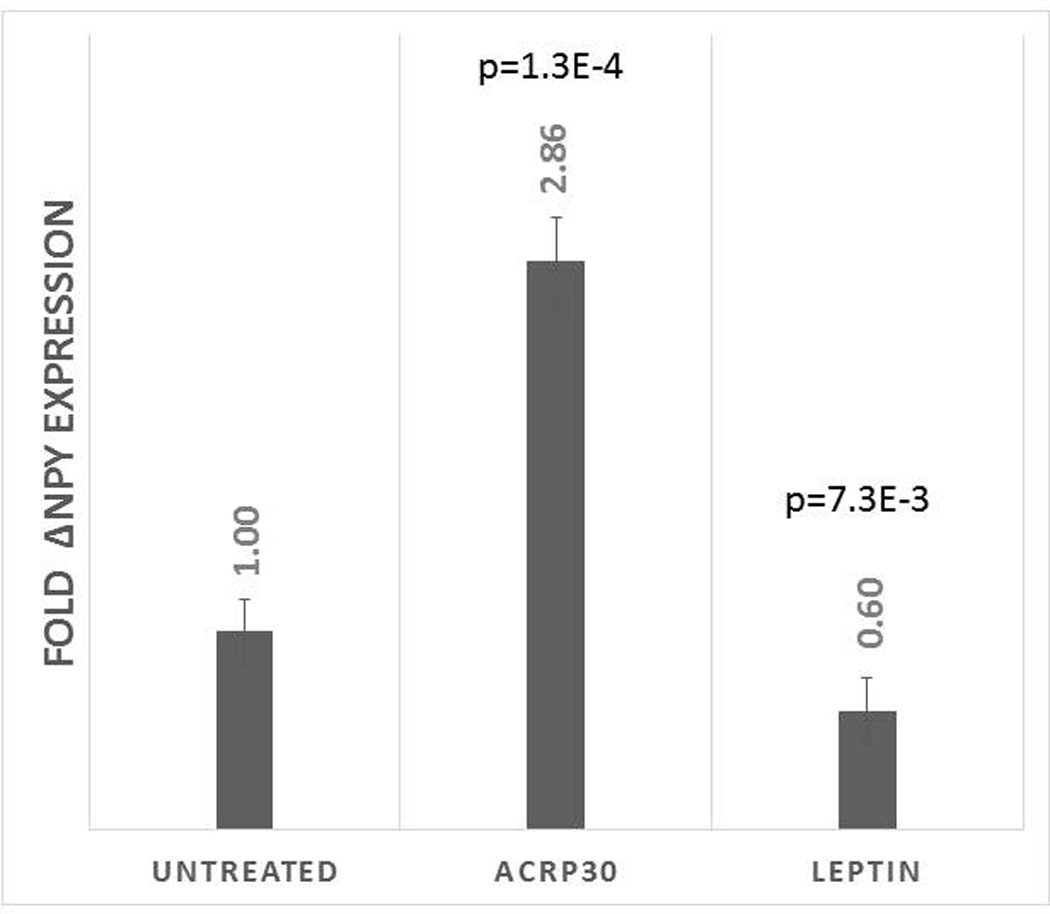
P-values are relative to NPY mRNA from untreated cells which is normalized to 1.
Co-regulation of genes, miRNAs and pathways across neuronal maturation
In order to examine potential co-regulation of genes involved in distinct biological processes during maturation of the human neurons, K-means clustering algorithm was applied to the normalized expression values for Days 35–115. Twelve clusters, or genesets, best represented the time course data across the neuronal maturation phase. Of the 15,631 genes expressed by these neurons, only 4079 showed distinct patterns of temporal expression changes (Figure 6). For each gene cluster, we examined geneset enrichment for known biological processes and gene ontology terms as well as predicted miRNA and transcription factor enrichment. Full GO results and miRNA correlation for each gene cluster are provided in Supplemental Tables S1 and S2. The results for clusters of particular note in relation to metabolism are described below. We also assessed potential co-regulation of miRNAs across neuronal maturation by applying K-means clustering algorithm separately to the normalized expression values of miRNA measured with RNAseq for Days 35–115. Of the 1206 miRNA expressed by these neurons at any time point, 199 met the criteria of being significantly differentially expressed between at least two time points, and six clusters best represented the time course data for these potentially developmentally expressed miRNAs across the neuronal maturation phase. (Supplemental Figure S6). Detailed data for each miRNA cluster and predicted gene targets are provided in Supplemental Tables S3 and S4.
Figure 6. K-means cluster analysis of expression profiles across neuronal maturation Days 35–115.
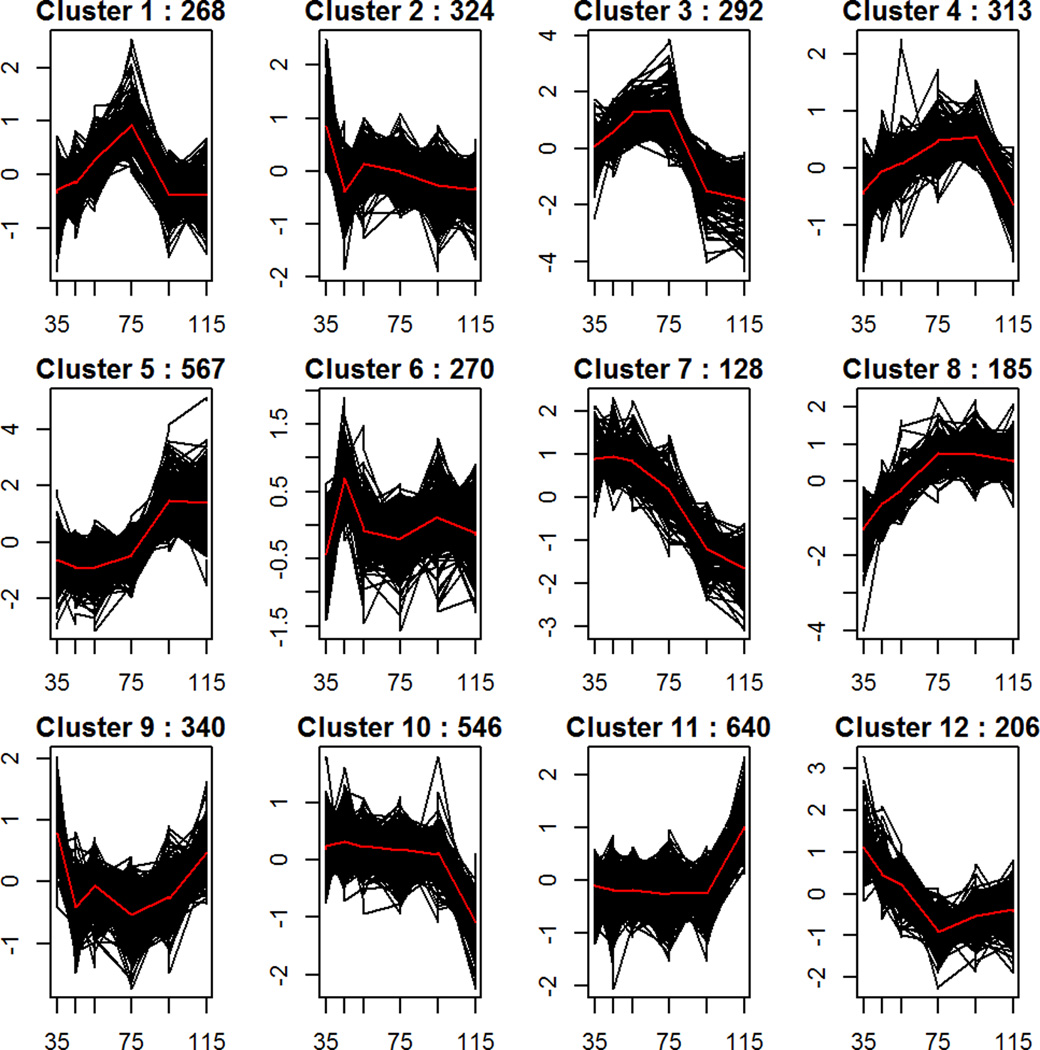
Calinski and Silhouette index evaluation indicated 12 clusters as optimum to distinguish the 4079 genes that displayed significant differential expression (adjusted p-value<.01, absolute fold change>2) between at least two time points. The number of genes falling into each cluster are shown at the top of each plot. The y-axis plots the expression of each transcript centered and scaled so that each gene will have mean 0 and sd 1. The x axis plots days in culture. Cluster 1 showed statistically significant enrichment for BMI GWAS-implicated genes.
Cluster 1 contains genes whose normalized expression sharply increases across early development peaking at Day 75 and then rapidly declines to that of early neurons. Key GO terms enriched in this cluster are “neuron projection”, “synapse” and “transmission of nerve impulse” (p= 3.9E-8, 1.1E-7, and 7.7E-3). Many genes in this cluster are targets of the repressor Polycomb protein SUZ12 in human embryonic stem cells (p=1.54E-10), and a significant number of genes have MAM (meprin/A5-protein/PTPmu) protein domains (p=6E-5), including MDGA2 which interacts with neuroligin 2 to regulate inhibitory synapse development(28), neuropilin 1 (NRP1) which regulates dendritic growth and branching, and the ALK receptor tyrosine kinase which induces neurite outgrowth(29). Predicted binding sites for miR96-5p, a miRNA with diurnal activity and evidence for involvement with circadian rhythm regulation(30, 31), was the most significantly enriched in our data (p=7.4E-5) and showed negative co-expression with its putative targets (r=−0.76, p=0.01). Significant enrichment and positive correlation with expression of miR-9 and negative correlation for miR-181a (enrichment p=0.009 and 0.006; correlation r = 0.97 and −0.93; p=4E-6 and 1E-4, respectively) was also found, reflecting the biological process of specific synapse development for this cluster. Whereas, mir-9 targets the 3’UTR of dopamine receptor D2 (DRD2) and results in its down-regulation, mir-181a promotes the generation of neurons of dopaminergic fate (32, 33).
Cluster 3 genes are similar in temporal profile to cluster 1, but show much stronger differential expression between early and late stages of neuronal development and are dominated by those containing a homeobox domain (2.6E-28) such as IRX3. The obesity-associated intronic interval of the fat mass and obesity associated (FTO) gene harbors an enhancer for IRX3 in adipocytes(34) and the embryonic mouse hypothalamus(35). Further, IRX3 deficient mice show a significant reduction in body weight through increase in basal metabolic rate and browning of white adipose tissue, and knockout of IRX3 specific to either the hypothalamus or adipocytes recapitulates a significant portion of the phenotype, indicating potential cell autonomous roles for adipocytes and hypothalamic neurons which together influence obesity. Roles involved in regionalization and pattern specific processes are enriched in this cluster of genes including those from the ROBO and SLIT families as well as netrin 1 (NTN1) which are central to directional guidance of axons during establishment of neural circuitry. In mice, the outgrowth of NPY neurites from the ARC was observed to be impaired in offspring of obese mothers as compared to those from non-obese mothers, and genes from the ROBO and SLIT families as well as receptors for NTN1 had altered expression in the ARC at midgestation in the affected offspring(13). Thus, this cluster may provide information regarding the critical developmental window in this in vitro model that relates to understanding how the in utero environment might affect fetal programming and susceptibility to metabolic disorders through neuronal development. Several miRNAs are predicted to target genes of this cluster and also showed significant correlation of expression values with their targets in our human neurons (Supplemental Table S2); however, their function in hypothalamic development is still unknown.
In contrast, cluster 5 genes, whose expression sharply increased across late maturation stages, are enriched for “actin-myosin filament sliding”, and “axonogenesis” processes, the GO terms “extracellular matrix” and “cell adhesion”, as well as targets of MEF2A (p=2.1E-7), a transcription factor that affects dendrite development and plasticity during CNS neural circuit formation(36). Motor proteins have been shown to have various neuronal functions including cell migration, growth cone motility and axonal transport. It is interesting to note that 3 (MYL1, MYLPF, TNNC2) of the 5 myosin cytoskeletal protein genes shown to have increased hypothalamic expression following glucocorticoid treatment in mice(37) are contained in cluster 5. Whether these changes play a role in glucocorticoid-induced hypothalamic leptin resistance and weight gain is not known. Binding sites for miR132 and miR219, two microRNAs involved in circadian clock modulation(38), were significantly overrepresented in cluster 5 (p<0.05); however, expression of these microRNAs was not significantly negatively correlated with expression of their predicted targets in our data, perhaps due to their diurnal nature.
Clusters 4 and 8 are very similar in that their genes are highly expressed and show a steady increase in expression across neuronal maturation. Cluster 4 genes, however, peak at Day 95 and exhibit a sharp decrease in expression levels at Day 115 whereas those of cluster 8 remain high. Cluster 4 is enriched for genes related to “calmodulin binding” (5.0E-04), “genes involved in NCAM signaling for neurite outgrowth” (3.7E-04) and “regulation of vesicle-mediated transport” (p=5.3E-04). As calcium signaling is critical for proper dendritic arborization(39) and neurotransmitter release, coordinated temporal expression of calcium ion channels and molecules involved in crosstalk between calmodulin and cAMP pathways may be needed for appropriate neuronal development and function.
Cluster 8 genes comprise those involved in “multicellular organismal signaling” (p=9E-8) and “dendrite” (p=4E-11). Several genes encoding GABA receptors, glutamate receptors and those found in neurite growth cones belong to this cluster. Predicted targets of miR218-5p are enriched in cluster 8 (p=8E-3), and expression of this miRNA was significantly positively correlated with that of genes of this cluster (r=0.94; p=6E-6). This microRNA has been shown to localize to dendrites and to negatively regulate ROBO1 and ROBO2 expression yet be positively correlated with NRXN1 suggesting a role in refinement of synaptic connections(40). The positive correlation of mIR218-5p expression with genes of this cluster may be surprising; however, miR218 is reported to be one of a group of miRNAs that are co-expressed with their target genes and either spatially or temporally modulate their protein levels following miRNA inhibition release via extracellular stimuli such as BDNF(41).
Geneset enrichment analyses for genes comprising cluster 7 which are highly expressed by early neurons across Days 35–55 and then consistently decreased in expression across each consecutive time point, indicated significant enrichment for the biological processes “tube development” and “negative regulation of cell differentiation” (p-value range 1.1E-08 to 7.4E-07). FIRE results showed genes in this cluster to be enriched for targets of MZF1 (p<0.05), a transcription factor known to induce cell proliferation(42). This gene cluster appears to be regulated by the miRNA let7b as shown by binding site enrichment (p=3.9E-6) as well as a significant negative correlation of expression of let7b with that of this cluster (r=−0.99; p-value= 1.1E-08). Surprisingly, the GO term “abnormal fetal growth/weight/body size” was significantly represented in the cluster by 12 genes (p=2.6E-6) including several imprinted genes such as delta-like 1 homologue (DLK1) known to co-localize with AGRP/NPY and be involved in hypothalamic development(43).
Cluster 11 genes are distinct in that their transcript levels rise steeply in the mature neuronal cultures at Day 115. Genes coding for components of the ribonucleoprotein complex are enriched in this cluster (p=3.1E-10), and their increase in expression in the late stage neurons may correspond to packaging and transport of mRNAs to the dendrites and synapses. Genes involved in cellular respiration and translation, such as many encoding mitochondrial ribosomal proteins, also belong to this cluster. Sensing of nutrients by the hypothalamus may involve mitochondrial respiration(44), and studies suggest that lipid metabolism and mitochondrial free radical production in NPY/AGRP neurons of the ARC are key factors in the regulation of appetite. Ghrelin-induced food intake was shown to require uncoupling protein 2 (UCP2)-dependent mitochondrial proliferation and activation of these specific hypothalamic neurons(45). Thus, the late-stage iPS-derived neuronal cultures may be a good model for study of mitochondria-mediated effects of signaling proteins on appetite and behavior in human context.
Benchmarking in vitro neuronal development to in vivo development
In order to determine which in vivo developmental time points are represented by the neuronal culture maturation time points, we used isoform switching information for 3 key genes. The gene transcription factor 7-like 2 (TCF7L2) undergoes brain developmental alternative splicing. In mouse late embryonic thalamus, 2 predominant isoforms are observed at 35 and 58kD and an isoform at 78kD is also detected; however, expression of the 35kD form is greatly reduced at postnatal day 10 and barely detectable at day 60.(46) Similar to mouse, we see 35kD, 58kD and weak 75kD TCF7L2 isoforms early in the human neuronal cultures. From Day 75–115, only 58kD and 75kD are expressed (Figure S2). Further, overall expression of TCF7L2 is much lower in the mature neurons at Days 95–115 which is consistent with data reported for postnatal mouse. In addition, the vesicular glutamate transporter isoforms VGLUT1 and VGLUT2 are developmentally regulated. In rat, VGlut1 and VGlut2 are moderately expressed across gestation, but after birth, there is a marked increase in VGlut1 and a decrease in VGlut2 expression.(47) Likewise, glycine receptor isoforms switch from the neonatal alpha2 subunit to the adult alpha1 and beta subunits by the 2nd postnatal week in mice.(48) Consistent with this, we observe striking switch in expression of these developmentally regulated genes in the neuronal cultures beginning at Day95 (Figure S7). Taken together, these data suggest that the neuronal culture time points of Days 95 and 115 may represent in vivo postnatal stage neurons in mice and rat.
Temporal expression changes of obesity GWAS-implicated genes
Using GWAS data from Locke et al.(8), we sought to determine whether genes implicated by GWAS to be associated with BMI shared temporal expression patterns during hypothalamic development. First we identified the genes nearest the GWAS-reported SNPs (n=289) from Locke et al.(8) that reached BMI association significance of P<E-5. A total of 58 of these GWAS-implicated genes that were expressed by GTEx hypothalamus (out of 203) show distinct temporal expression patterns represented by our 12 clusters giving significant evidence of enrichment with p=3.7E-7. We also tested whether this set of genes was enriched in any particular cluster. Cluster 1 described above and in Figure 6 was significantly enriched for GWAS-implicated genes (p=3.3E-7, >5-fold enrichment). Additionally, six of the 14 GWAS-implicated genes that are present in cluster 1 are predicted by both PicTar and TargetScan to be targets of mir-96 (Table 1), which may be involved in circadian rhythm modulation(30). Nominal enrichment was seen for clusters 4 and 5 (p=0.02 and 0.03, respectively). Given that trait-associated genetic variants may exert long-range regulatory effects, we also conducted an enrichment test by considering all genes located within 250kb of the index SNPs. Of the 1122 hypothalamic-expressed genes in this interval, 231 belong to our gene clusters with significant enrichment evidence of p=3E-6. Cluster 1 remained enriched but with attenuated significance (p=0.010). Detailed results are given in supplementary Table S5.
Table 1.
Hypothalamic genes proximal to BMI GWAS variants and with Cluster 1 temporal expression.
| ENTREZ ID |
Gene Symbol |
Gene name | GWAS SNP | GWAS p-value |
|---|---|---|---|---|
| 253559 | CADM2 | cell adhesion molecule 2 | rs13078960 | 1.74E-14 |
| 158038 | LINGO2 | leucine rich repeat and Ig domain containing 2 | rs10968576 | 2.34E-14 |
| 54566 | EPB41L4B* | erythrocyte membrane protein band 4.1 like 4B | rs6477694 | 1.71E-08 |
| 138046 | RALYL | RALY RNA binding protein-like | rs2033732 | 4.89E-08 |
| 65125 | WNK1 | WNK lysine deficient protein kinase 1 | rs11611246 | 1.70E-07 |
| 29951 | PDZRN4* | PDZ domain containing ring finger 4 | rs285575 | 6.28E-07 |
| 10500 | SEMA6C | semaphorin 6C | rs4357530 | 1.64E-06 |
| 114788 | CSMD3 | CUB and Sushi multiple domains 3 | rs4389974 | 1.71E-06 |
| 375567 | VWC2 | von Willebrand factor C domain containing 2 | rs10269783 | 1.72E-06 |
| 54715 | RBFOX1 | RNA binding protein, fox-1 homolog 1 | rs7189501 | 3.37E-06 |
| 214 | ALCAM* | activated leukocyte cell adhesion molecule | rs1436351 | 3.81E-06 |
| 93986 | FOXP2* | forkhead box P2 | rs12705981 | 4.99E-06 |
| 596 | BCL2* | B-cell CLL/lymphoma 2 | rs12454712 | 6.04E-06 |
| 7534 | YWHAZ | tyrosine 3-/tryptophan 5-monooxygenase activation rotein, zeta |
rs3134353 | 7.22E-06 |
| 238 | ALK* | anaplastic lymphoma receptor tyrosine kinase | rs7578465 | 8.14E-06 |
Predicted target of mir-96
For comparison we conducted a similar analyses using GWAS data from Shungin D et al.(49) for waist hip ratio (WHR) as a proxy for body fat distribution. Associated loci for this trait have been shown to be enriched for genes expressed in adipose tissue and the processes of adipogenesis, angiogenesis and insulin resistance. None of the 12 clusters were significantly enriched for genes proximal to the 61 loci with reported GWAS significance of P<E-5 for WHR (cluster enrichment p≥0.6). In addition, no clusters were significantly enriched for genes within 250kb of the WHR loci. Nominal enrichment (p=0.013) was seen for cluster 3; however, 6 of the 7 genes in this cluster were members of the homeobox A cluster near a single GWAS locus on chromosome 7 which confounds the enrichment.
Discussion
The importance of hypothalamic neural signaling in the regulation of energy homeostasis has been established through animal studies as well as rare and severe, monogenic disorders of human obesity. The orexigenic AGRP/NPY neurons of the ARC regulate food intake and the behaviors associated with this mission. Several rodent studies have linked the release of AGRP, NPY and GABA by these neurons with changes in energy balance. Study of these key neurons in humans, however, is extremely limited by tissue access. We have developed a protocol to generate cultures of neurons from human iPSC that recapitulate many of the features of these important hypothalamic neurons. Our protocol, which uses low-insulin modified hES media without bFGF, incorporates the findings from Wataya et al.(16) which showed that minimization of exogenous signals in ES media induces rostral hypothalamic differentiation. Unlike Wataya et al.(16), our protocol includes treatment of the resulting self-patterned purified neural precursors with Wnt. The early neural progenitors then express high levels of a number of proneural genes critical for hypothalamic development and known to be repressed by the Notch signaling pathway including the transcription factor ASCL1 which is key to ARC development(50, 51). Two other groups have developed mixed cultures of hypothalamic neurons expressing POMC, NPY, AGRP, somatostatin, dopamine, melanin concentrating hormone, and vasopressin using a directed differentiation protocol through early activation of SHH signaling and dual SMAD inhibition followed by inhibition of Notch signaling(52, 53). Thus, our protocol which combines aspects of these self-patterning and directed approaches is unique in that the derived cultures are less heterogeneous and consist mainly of NPY/AGRP neurons.
Recent data also suggests that common genetic variation influencing body weight regulation may exert their effects through the CNS and especially the hypothalamus. Neither the cellular effects of these risk genetic variants nor the developmental timing of these events is yet known. Using developmental expression profiles from human hypothalamic-like neuronal cultures that we have generated from iPSC, we have defined sets of potentially co-regulated genes that exhibit distinct, and intrinsic dynamic expression patterns across neuronal stages. One distinct set of temporally expressed genes consisting of less than 2% of the total neuronal genes is strikingly enriched for genes that have been implicated by GWAS to be associated with body weight regulation and obesity. This set of genes which by GO analysis are involved in “neuron projection”, “synapse” and “transmission of nerve impulse” exhibit a temporal expression pattern that sharply peaks in mid-stage neurons and then plateaus at a much reduced level in the mature neurons which correspond to those in mice at approximately one month of age. Many of these genes are predicted targets of miR-96, a microRNA with a significant inverse expression pattern to them and one with growing evidence for regulation of and by circadian factors. This hints that these GWAS obesity candidates might be influenced by circadian rhythm; however, these in silico miRNA predictions need to be confirmed experimentally. This relationship is an intriguing finding as the fat mass and obesity associated gene FTO has been shown to modulate circadian rhythms(54), and variation in genes related to the circadian rhythm of food intake and hypothalamic signaling have been associated with extreme obesity(55).
The developmental expression pattern observed by these GWAS-implicated genes in our hypothalamic neuronal cultures suggests that their influences on body weight regulation may occur during early development. This is consistent with findings reported by both Smemo et al.(35) and Claussnitzer et al.(34) with regard to FTO intronic variants. The chromatin looping between the FTO intronic region and IRX3, and the effect of associated SNPs on expression of IRX3 and IRX5 was only observed in embryonic brain and differentiating adipocytes, respectively, and not in the mature tissue. In our model, high levels of gene expression of IRX3 and IRX5 are observed only in the early neurons with little to no expression detected in the later stage neurons. Whether the obesity causal risk variants exert effects through IRX3/IRX5 in the hypothalamus in humans is not known. A recent study has described allelic-based differential expression of FTO and the proximal gene retinitis pigmentosa GTPase regulator-interacting protein-1 like (RPGRIP1L), but not IRX3 or IRX5, in early neurons derived from human iPS cells using a dual-SMAD inhibition protocol(56). Their results suggest that the molecular mechanism for the risk variants involves RPGRIP1L and FTO rather than the IRX gene family in the hypothalamus; however, it is important to note that the early neurons studied were not representative of the hypothalamus. Moreover, expression of IRX3 and IRX5 was very low at the time point studied, and thus the pertinent time point may have been missed. Further study in hypothalamic-specific models may be needed to resolve these findings. Results from these and similar studies highlight a need to study obesity GWAS variants during relevant cellular or tissue development as their effects may not be observed in adult tissue biopsies or post-mortem samples, for example.
As hypothalamic developmental processes occur in utero in humans, this accessible in vitro human model will be useful to begin to understand how alterations of these genes affect neuronal development and/or plasticity and ultimately pathophysiology underlying obesity susceptibility. Additionally, these neurodevelopmental processes may occur in a similar fashion during adult hypothalamic neurogenesis which is necessary for response of energy-balance circuits to environmental challenges yet declines with age. Elucidating the genetic and molecular events of this highly controlled process in these key hypothalamic neurons may help to uncover drivers that may serve as targets for interventions on body weight and appetite regulation.
Our temporal profiling results also point to a critical developmental window in this in vitro human model that can be exploited for study of in utero environmental effects on fetal programming and susceptibility to metabolic disorders through alterations in neuronal development. In animal models of maternal obesity, the outgrowth of NPY/AGRP neurites from the ARC to the PVN is impaired, and this may be a consequence of altered expression of genes involved in directional guidance of axons during development.(13) We show that these genes are highly expressed in early neurons, show peak expression at or near a time point that correlates with the end of gestation in rodents and then show minimal expression in late-stage neurons. Whether similar changes in gene expression and hypothalamic neurite connections from the ARC occur in humans in response to adverse intrauterine environments is not known. This model can be useful for such studies.
In conclusion, we have developed an in vitro human system for study of NPY/AGRP neurons of the arcuate nucleus. Although complex neural network interactions cannot be studied using this in vitro system, this human model provides a unique tool to examine molecular and cellular effects of genetic variation in a neuronal-stage specific manner in neurons directly involved in energy homeostasis.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Thomas Drawdy, Deborah Holstein, and Iriscilla Ayala for their technical assistance in this project and the UTHSCSA and Cancer Therapy and Research Center (CTRC) shared resources and staff that assisted on this project, including the Optical Imaging Core, the Flow Cytometry Core and the Next Generation Sequencing and Bioinformatics Core at the Greehey Children’s Cancer Research Institute, as well as the IIMS Clinical and Molecular Cytogenetics Lab. This work was funded in part by NIH DK47482, P30CA054174, and National Center for Advancing Translational Sciences Grant UL1 TR001120,
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Supplementary information is available at the Journal of Neuroendocrinology website.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Arble DM, Sandoval DA. CNS control of glucose metabolism: response to environmental challenges. Front Neurosci. 2013;7:20. doi: 10.3389/fnins.2013.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grayson BE, Seeley RJ, Sandoval DA. Wired on sugar: the role of the CNS in the regulation of glucose homeostasis. Nature reviews Neuroscience. 2013;14(1):24–37. doi: 10.1038/nrn3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Konner AC, Klockener T, Bruning JC. Control of energy homeostasis by insulin and leptin: targeting the arcuate nucleus and beyond. Physiology & behavior. 2009;97(5):632–638. doi: 10.1016/j.physbeh.2009.03.027. [DOI] [PubMed] [Google Scholar]
- 4.Sandoval D, Cota D, Seeley RJ. The integrative role of CNS fuel-sensing mechanisms in energy balance and glucose regulation. Annual review of physiology. 2008;70:513–535. doi: 10.1146/annurev.physiol.70.120806.095256. [DOI] [PubMed] [Google Scholar]
- 5.Sandoval DA, Obici S, Seeley RJ. Targeting the CNS to treat type 2 diabetes. Nature reviews Drug discovery. 2009;8(5):386–398. doi: 10.1038/nrd2874. [DOI] [PubMed] [Google Scholar]
- 6.Vogt MC, Bruning JC. CNS insulin signaling in the control of energy homeostasis and glucose metabolism - from embryo to old age. Trends in endocrinology and metabolism: TEM. 2013;24(2):76–84. doi: 10.1016/j.tem.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Zhang W, Bi S. Hypothalamic Regulation of Brown Adipose Tissue Thermogenesis and Energy Homeostasis. Front Endocrinol (Lausanne) 2015;6:136. doi: 10.3389/fendo.2015.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Speakman JR. Functional analysis of seven genes linked to body mass index and adiposity by genome-wide association studies: a review. Human heredity. 2013;75(2–4):57–79. doi: 10.1159/000353585. [DOI] [PubMed] [Google Scholar]
- 10.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature genetics. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nature genetics. 2009;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Begum G, Davies A, Stevens A, Oliver M, Jaquiery A, Challis J, et al. Maternal undernutrition programs tissue-specific epigenetic changes in the glucocorticoid receptor in adult offspring. Endocrinology. 2013;154(12):4560–4569. doi: 10.1210/en.2013-1693. [DOI] [PubMed] [Google Scholar]
- 13.Sanders TR, Kim DW, Glendining KA, Jasoni CL. Maternal obesity and IL-6 lead to aberrant developmental gene expression and deregulated neurite growth in the fetal arcuate nucleus. Endocrinology. 2014;155(7):2566–2577. doi: 10.1210/en.2013-1968. [DOI] [PubMed] [Google Scholar]
- 14.Zhang C, Xu D, Luo H, Lu J, Liu L, Ping J, et al. Prenatal xenobiotic exposure and intrauterine hypothalamus-pituitary-adrenal axis programming alteration. Toxicology. 2014;325:74–84. doi: 10.1016/j.tox.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 15.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wataya T, Ando S, Muguruma K, Ikeda H, Watanabe K, Eiraku M, et al. Minimization of exogenous signals in ES cell culture induces rostral hypothalamic differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(33):11796–11801. doi: 10.1073/pnas.0803078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 18.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic acids research. 2009;37(Web Server issue):W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome biology. 2007;8(9):R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell. 2007;28(2):337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26(19):2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith AW, Bosch MA, Wagner EJ, Ronnekleiv OK, Kelly MJ. The membrane estrogen receptor ligand STX rapidly enhances GABAergic signaling in NPY/AgRP neurons: role in mediating the anorexigenic effects of 17beta-estradiol. American journal of physiology Endocrinology and metabolism. 2013;305(5):E632–E640. doi: 10.1152/ajpendo.00281.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jo YH, Chua S., Jr Transcription factors in the development of medial hypothalamic structures. American journal of physiology Endocrinology and metabolism. 2009;297(3):E563–E567. doi: 10.1152/ajpendo.00064.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ten Kulve JS, van Bloemendaal L, Balesar R, RG IJ, Swaab DF, Diamant M, et al. Decreased Hypothalamic Glucagon-Like Peptide-1 Receptor Expression in Type 2 Diabetes Patients. J Clin Endocrinol Metab. 2016;101(5):2122–2129. doi: 10.1210/jc.2015-3291. [DOI] [PubMed] [Google Scholar]
- 27.Liu T, Kong D, Shah BP, Ye C, Koda S, Saunders A, et al. Fasting activation of AgRP neurons requires NMDA receptors and involves spinogenesis and increased excitatory tone. Neuron. 2012;73(3):511–522. doi: 10.1016/j.neuron.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee K, Kim Y, Lee SJ, Qiang Y, Lee D, Lee HW, et al. MDGAs interact selectively with neuroligin-2 but not other neuroligins to regulate inhibitory synapse development. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(1):336–341. doi: 10.1073/pnas.1219987110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Motegi A, Fujimoto J, Kotani M, Sakuraba H, Yamamoto T. ALK receptor tyrosine kinase promotes cell growth and neurite outgrowth. J Cell Sci. 2004;117(Pt 15):3319–3329. doi: 10.1242/jcs.01183. [DOI] [PubMed] [Google Scholar]
- 30.Hansen KF, Sakamoto K, Obrietan K. MicroRNAs: a potential interface between the circadian clock and human health. Genome Med. 2011;3(2):10. doi: 10.1186/gm224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinoshita C, Aoyama K, Matsumura N, Kikuchi-Utsumi K, Watabe M, Nakaki T. Rhythmic oscillations of the microRNA miR-96-5p play a neuroprotective role by indirectly regulating glutathione levels. Nat Commun. 2014;5:3823. doi: 10.1038/ncomms4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi S, Leites C, He D, Schwartz D, Moy W, Shi J, et al. MicroRNA-9 and microRNA-326 regulate human dopamine D2 receptor expression, and the microRNA-mediated expression regulation is altered by a genetic variant. J Biol Chem. 2014;289(19):13434–13444. doi: 10.1074/jbc.M113.535203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stappert L, Borghese L, Roese-Koerner B, Weinhold S, Koch P, Terstegge S, et al. MicroRNA-based promotion of human neuronal differentiation and subtype specification. PloS one. 2013;8(3):e59011. doi: 10.1371/journal.pone.0059011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med. 2015;373(10):895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez-Marin C, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507(7492):371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen SX, Cherry A, Tari PK, Podgorski K, Kwong YK, Haas K. The transcription factor MEF2 directs developmental visually driven functional and structural metaplasticity. Cell. 2012;151(1):41–55. doi: 10.1016/j.cell.2012.08.028. [DOI] [PubMed] [Google Scholar]
- 37.Nishida Y, Yoshioka M, St-Amand J. Regulation of hypothalamic gene expression by glucocorticoid: implications for energy homeostasis. Physiological genomics. 2006;25(1):96–104. doi: 10.1152/physiolgenomics.00232.2005. [DOI] [PubMed] [Google Scholar]
- 38.Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54(5):813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenberg SS, Spitzer NC. Calcium signaling in neuronal development. Cold Spring Harb Perspect Biol. 2011;3(10):a004259. doi: 10.1101/cshperspect.a004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Small EM, Sutherland LB, Rajagopalan KN, Wang S, Olson EN. MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circ Res. 2010;107(11):1336–1344. doi: 10.1161/CIRCRESAHA.110.227926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bicker S, Lackinger M, Weiss K, Schratt G. MicroRNA-132, 134, and -138: a microRNA troika rules in neuronal dendrites. Cell Mol Life Sci. 2014;71(20):3987–4005. doi: 10.1007/s00018-014-1671-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaboli M, Kotsi PA, Gurrieri C, Cattoretti G, Ronchetti S, Cordon-Cardo C, et al. Mzf1 controls cell proliferation and tumorigenesis. Genes & development. 2001;15(13):1625–1630. doi: 10.1101/gad.902301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Persson-Augner D, Lee YW, Tovar S, Dieguez C, Meister B. Delta-like 1 homologue (DLK1) protein in neurons of the arcuate nucleus that control weight homeostasis and effect of fasting on hypothalamic DLK1 mRNA. Neuroendocrinology. 2014;100(2–3):209–220. doi: 10.1159/000369069. [DOI] [PubMed] [Google Scholar]
- 44.Benani A, Troy S, Carmona MC, Fioramonti X, Lorsignol A, Leloup C, et al. Role for mitochondrial reactive oxygen species in brain lipid sensing: redox regulation of food intake. Diabetes. 2007;56(1):152–160. doi: 10.2337/db06-0440. [DOI] [PubMed] [Google Scholar]
- 45.Andrews ZB, Liu ZW, Walllingford N, Erion DM, Borok E, Friedman JM, et al. UCP2 mediates ghrelin's action on NPY/AgRP neurons by lowering free radicals. Nature. 2008;454(7206):846–851. doi: 10.1038/nature07181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagalski A, Irimia M, Szewczyk L, Ferran JL, Misztal K, Kuznicki J, et al. Postnatal isoform switch and protein localization of LEF1 and TCF7L2 transcription factors in cortical, thalamic, and mesencephalic regions of the adult mouse brain. Brain structure & function. 2013;218(6):1531–1549. doi: 10.1007/s00429-012-0474-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bean AJ. Protein trafficking in neurons. Amsterdam; Boston: Elsevier/Academic Press; 2007. p. xv.p. 448. [Google Scholar]
- 48.Bock G, Goode J. Sodium channels and neuronal hyperexcitability. Chichester, England New York: Wiley; 2002. p. viii.p. 244. [Google Scholar]
- 49.Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Magi R, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518(7538):187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ratie L, Ware M, Barloy-Hubler F, Rome H, Gicquel I, Dubourg C, et al. Novel genes upregulated when NOTCH signalling is disrupted during hypothalamic development. Neural Dev. 2013;8:25. doi: 10.1186/1749-8104-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ware M, Hamdi-Roze H, Dupe V. Notch signaling and proneural genes work together to control the neural building blocks for the initial scaffold in the hypothalamus. Front Neuroanat. 2014;8:140. doi: 10.3389/fnana.2014.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merkle FT, Maroof A, Wataya T, Sasai Y, Studer L, Eggan K, et al. Generation of neuropeptidergic hypothalamic neurons from human pluripotent stem cells. Development. 2015;142(4):633–643. doi: 10.1242/dev.117978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L, Meece K, Williams DJ, Lo KA, Zimmer M, Heinrich G, et al. Differentiation of hypothalamic-like neurons from human pluripotent stem cells. J Clin Invest. 2015;125(2):796–808. doi: 10.1172/JCI79220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang CY, Shie SS, Hsieh IC, Tsai ML, Wen MS. FTO modulates circadian rhythms and inhibits the CLOCK-BMAL1-induced transcription. Biochem Biophys Res Commun. 2015;464(3):826–832. doi: 10.1016/j.bbrc.2015.07.046. [DOI] [PubMed] [Google Scholar]
- 55.Mariman EC, Bouwman FG, Aller EE, van Baak MA, Wang P. Extreme obesity is associated with variation in genes related to the circadian rhythm of food intake and hypothalamic signaling. Physiological genomics. 2015;47(6):225–231. doi: 10.1152/physiolgenomics.00006.2015. [DOI] [PubMed] [Google Scholar]
- 56.Stratigopoulos G, Burnett LC, Rausch R, Gill R, Penn DB, Skowronski AA, et al. Hypomorphism of Fto and Rpgrip1l causes obesity in mice. J Clin Invest. 2016;126(5):1897–1910. doi: 10.1172/JCI85526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.