Abstract
Cronobacter sakazakii is an opportunistic nosocomial and foodborne pathogen that causes severe infections with high morbidity and mortality rates in neonates, the elderly, and immunocompromised individuals. Little is known about the pathogenesis mechanism of this pathogen and if there are any consequences of C. sakazakii colonization in healthy individuals. In this study, we characterized the mechanisms of autoaggregation in C. sakazakii ATCC 29544 (CS29544). Autoaggregation in CS29544 occurred rapidly, within 30 min, and proceeded to a maximum of 70%. Frameshift mutations in two flagellum proteins (FlhA and FliG) were identified in two nonautoaggregating CS29544 clonal variant isolates. Strategic gene knockouts were generated to determine if structurally intact and functional flagella were required for autoaggregation in CS29544. All structural knockouts (ΔflhA, ΔfliG, and ΔfliC) abolished autoaggregation, whereas the functional knockout (ΔmotAB) did not prevent autoaggregation. Complementation with FliC (ΔfliC/cfliC) restored autoaggregation. Autoaggregation was also disrupted by the addition of exogenous wild-type CS29544 filaments in a dose-dependent manner. Finally, filament supercoils tethering neighboring wild-type CS29544 cells together were observed by transmission electron microscopy. In silico analyses suggest that direct interactions of neighboring CS29544 FliC filaments proceed by hydrophobic bonding between the externally exposed hypervariable regions of the CS29544 FliC flagellin protein. Further research is needed to confirm if flagella-mediated autoaggregation plays a prominent role in C. sakazakii pathogenesis.
Keywords: autoaggregation, Cronobacter sakazakii, flagella, FliC, protein-protein interactions
Introduction
Cronobacter spp. are motile, biofilm-forming, facultative anaerobic Gram-negative bacilli. Cronobacter sakazakii, formerly known as Enterobacter sakazakii (Iversen et al., 2008), the most prominent species, is an opportunistic pathogen associated with fatal infections in neonates and immunocompromised children and adults (Lai, 2001). Most notably, C. sakazakii infections in neonates have been linked epidemiologically to the consumption of powdered infant formula (PIF) (Biering et al., 1989; Simmons et al., 1989; van Acker et al., 2001). Furthermore, C. sakazakii withstands desiccation in PIF and thrives in reconstituted PIF, especially when PIF is temperature-abused (Breeuwer et al., 2003; Riedel and Lehner, 2007; Osaili et al., 2009). In response, medical and health professionals had been cautioned regarding the use of PIF; however, C. sakazakii infections in neonatal units are not solely due to consumption of contaminated PIF (Jason, 2012). For example, C. sakazakii has been reported in infants exclusively breastfed (Hurrell et al., 2009b; Broge and Lee, 2013; Ravisankar et al., 2014). Another concern is the frequency with which nasogastric tubes are used to deliver enteral nutrition in premature neonates (Axelrod et al., 2006). A surveillance study reported that several species of Enterobacteriaceae, including a single C. sakazakii isolate, were recovered from used nasogastric enteral feeding tubes (Hurrell et al., 2009b). These researchers cautioned that microbial biofilms on nasogastric enteral feeding tubes might serve as a continuous inoculum during bolus feedings while the tube is in place. A simple solution may be to switch from indwelling nasogastric tubes to insertion of a nasogastric tube at each feeding; however, the comfort of the neonate and associated economic costs must be considered (Symington et al., 1995). A multifactorial approach to protecting neonates from microbial infections associated with feedings is needed, including identification of the mechanisms C. sakazakii uses during biofilm formation and gastrointestinal colonization.
Many bacteria, especially pathogens, have developed elaborate mechanisms to permit attachment to and formation of dense sessile mono- or polymicrobial aggregates on biotic and abiotic surfaces (Costerton et al., 1987; An and Friedman, 1998; Schluter et al., 2015). Following this initial attachment, bacterial aggregates can cooperatively form biofilms, thereby increasing their chance of survival. Herein, the formation of monospecies aggregates is referred to as autoaggregation. Autoaggregation is common in the Enterobacteriaceae family, including Escherichia coli (Girón et al., 1991; Czeczulin et al., 1997; Prigent-Combaret et al., 2000; Schembri et al., 2001; Sherlock et al., 2005; Girard et al., 2010; Nakao et al., 2012), Salmonella spp. (Collinson et al., 1993), Klebsiella pneumoniae (Favre-Bonte et al., 1995), Edwardsiella tarda (Gao et al., 2015), Citrobacter freundii (Bai et al., 2012), Yersina pestis (Vadyvaloo et al., 2015), and Proteus mirabilis (Rocha et al., 2007; Alamuri et al., 2010), and often occurs via self-recognizing cell-surface appendages. Autoaggregation is mediated by adhesins (Sherlock et al., 2005; Alamuri et al., 2010; Girard et al., 2010; Abdel-Nour et al., 2014; Arenas et al., 2015; Wang et al., 2015) and other cell-surface molecules, such as surface-associated proteins (Prigent-Combaret et al., 2000; Gao et al., 2015), pili (Girón et al., 1991), fimbriae (Nataro et al., 1992; Collinson et al., 1993; Czeczulin et al., 1997; Schembri et al., 2001), flagella (Sjoblad et al., 1985; Näther et al., 2006), and lipopolysaccharides (Nakao et al., 2012; Wang et al., 2012). Using microscopy, supercoiling between neighboring microorganisms promoted by pili in Escherichia coli was observed (Girón et al., 1991). Additionally, autoaggregation was mediated by flagella in Pseudomonas marina (Sjoblad et al., 1985) and Pyrococcus furiosus (Näther et al., 2006). The gastrointestinal colonization of two Escherichia coli pathotypes occurs via different fully characterized mechanisms of autoaggregation. Bundle-forming fimbriae (AAF/I and AAF/II) in enteroaggregative E. coli promote autoaggregation and biofilm formation along the intestinal surface (Nataro et al., 1992; Czeczulin et al., 1997), whereas enteropathogenic E. coli adheres to the intestinal surface via interactions between Intimin and Tir (Donnenberg and Kaper, 1991) and establishes three-dimensional microcolonies using bundle-forming pili (Girón et al., 1991). Although autoaggregation was observed in some C. sakazakii strains (Lehner et al., 2005; Wang et al., 2012; Hu et al., 2015), the extracellular factor mediating autoaggregation in C. sakazakii and its biological function were not described.
The role of bacterial flagella in motility and bacterial chemotaxis is well characterized (Sourjik and Wingreen, 2012), but motility is not its sole biological function. Bacterial flagella contribute to the virulence of bacterial pathogens, including adhesion, microcolony formation, invasion, and biofilm formation, as reviewed by others (Haiko and Westerlund-Wikström, 2013). Unlike other Enterobacteriaceae, the contribution of C. sakazakii's flagellum to its virulence has received little attention. The flagella of C. sakazakii ES5 are required for adhesion to Caco-2 monolayers and biofilm formation to microtiter plates (Hartmann et al., 2010). Herein, we describe the role played by the bacterial flagella in the autoaggregation of C. sakazakii ATCC 29544 (CS29544). A collection of gene knockout and complementation strains revealed that structurally intact FliC containing filaments were required for autoaggregation. Additionally, we provide evidence to suggest that direct interactions between neighboring filaments promote autoaggregation of liquid CS29544 cultures.
Materials and methods
Bacterial strains and growth conditions
CS29544 was cultured in brain heart infusion (BHI) broth (Becton Dickinson), pH 7.38 at 37°C overnight aerobically with agitation (250 rpm) unless specified. CS29544 was enumerated and spread-plated on BHI agar plates following serial dilution in 1 × phosphate buffered saline (PBS; Dulbecco's Formula), pH 7.4. Escherichia coli was cultured in lysogeny broth (LB, Miller's formula) at 37°C overnight aerobically with agitation (250 rpm) unless specified. When necessary, ampicillin or chloramphenicol were added to BHI or LB at a final concentration of 100 and 35 μg/mL, respectively. To test for motility, CS29544 were grown on 0.4% agar composed of 3 g/L beef extract, 10 g/L Bacto peptone, 5 g/L sodium chloride (BPN) supplemented with 1% of 2, 3, 5-triphenyltetrazolium chloride (redox indicator) or observed microscopically by wet mount.
Autoaggregation assays
Stationary phase CS29544 cultures, grown in 10 mL BHI at 37°C, were held statically at room temperature (~25°C) for 6 h to allow autoaggregation. The change in optical density at 600 nm was gently measured at 30 min intervals for 2 h followed by 1 h intervals until 6 h. Autoaggregation was reported as the maximum percent autoaggregation. Typically the endpoint was used, and calculated by Equation (1).
| (1) |
Several additional autoaggregation assays were conducted with modifications after the growth of CS29544 in 10 mL BHI, including static incubation at different temperatures (4 and 37°C), the addition of 50 mM EDTA or PBS, and before and after blending at “whip” speed for 30 s (BL113SG; Black and Decker). Furthermore, autoaggregation assays were completed with CS29544 following growth in 10 mL BHI at different pH values (pH = 4, 5, 6, 7.38, and 8) or incubated anaerobically (90% N2, 5% CO2, 5% H2). Finally, autoaggregation assays were run with CS29544 following growth (10 mL) in different media, including Miller and Lennox LB formulations (LB10 and LB5, respectively), tryptic soy broth (TSB), and BPN broth.
Stationary phase CS29544 and nonautoaggregating clonal variant (CV) cultures (described below) grown in BHI were mounted and held statically at room temperature for 1 h to allow autoaggregation. Still images were taken every 10 s for a total of 1 h by a stationary DSLR camera (Rebel T2i; Canon) with an intervalometer. Images (360 frames) were stitched together to create a video file with 24 frames per second. An additional time lapse video was constructed as previously described with the CS29544 and flagellum competition assays (described below) with still images taken every 20 s for a total of 6 h. Images (1,080 frames) were stitched together to create a video file with 72 frames per second.
Isolation of nonautoaggregating CS29544 clonal variants
Stationary phase CS29544 cultures autoaggregated for 2 h. Then, two separate 100 μL (1%; v/v) aliquots, one from the top fraction of the CS29544 culture and one from the bottom fraction (autoaggregating control) were passed into two fresh tubes of 10 mL BHI broth and incubated as described above. Successive passages following autoaggregation continued until autoaggregation was arrested. Two independent nonautoaggregating variants were isolated and characterized.
DNA extraction and whole-genome sequencing
Genomic DNA was isolated from CS29544 and CVs using the UltraClean® Microbial DNA Isolation Kit (MoBio Laboratories) according to manufacturer's instructions. High-quality genomic DNA libraries were prepared and sequenced using the Illumina platform by the DNA Services group affiliated with the Roy J. Carver Biotechnology Center at the University of Illinois-Urbana Champaign. Paired-end libraries of CS29544 and CVs were prepared with the TruSeq Genomic DNA Sample Prep Kit (length: 200–600 bp) and sequenced on a HiSeq2500 with the TruSeq SBS Sequencing Kit v1 producing a mean read length of 160 nt. An additional mate-pair library of CS29544 was prepared with the Nextera Mate Pair Library Sample Prep Kit (length: 3–8 kbp) and sequenced on a MiSeq V3 with the MiSeq 600-cycle Sequencing Kit v3 producing a mean read length of 300 nt. Paired-end reads were imported into CLC Genomics Workbench v7.5, and quality and adapter trimmed using default settings. Mate-pair reads were quality and adapter trimmed using Cutadapt (Martin, 2011) and an in-house Perl script provided by the Roy J. Carver Biotechnology Center. Processed mate-pair reads were imported into CLC Genomics Workbench and de novo assembled using default parameters, and only contigs larger than 1,000 bp were kept. The paired-end reads were mapped to the CS29544 de novo assembly and putative single nucleotide polymorphisms were identified (>90% frequency) using the Basic Variant Detection tool with default parameters confirmed by targeted Sanger sequencing using an ABI 3730XL capillary sequencer (Life Technologies).
Construction of CS29544 gene knockout strains and complementation vector
Targeted gene disruptions (flhA, fliG, motAB, fliC, and flaA) were constructed in the wild-type CS29544 using the lambda Red recombinase system (Cherepanov and Wackernagel, 1995; Datsenko and Wanner, 2000). All bacterial strains, plasmids, and primers used in this study are listed in Tables 1, 2. Briefly, linear DNA fragments were amplified by PCR with pKD3 DNA using the target gene specific primer set (60 bp) and appropriate experimental conditions. CS29544 containing the pKD46 plasmid were grown in 10 mL of LB containing 10 μg/mL of ampicillin and 10 mM L-arabinose at 30°C overnight aerobically with agitation (250 rpm). CS29544 pKD46 electrocompetent cells were transformed with 500 ng of the purified linear DNA fragment. The FRT-Cmr-FRT cassette in the recombinant mutants was cured by transformation and subsequent removal of the temperature-sensitive flippase (FLP) recombinase helper plasmid (pCP20). The double gene knockout (fliC and flaA) was constructed as described above for the CS29544 ΔflaA strain. Gene disruptions were confirmed by junction fragment PCR using the appropriate primer sets and experimental conditions.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristics or purpose | Internal number | Source or references |
|---|---|---|---|
| C. SAKAZAKII STRAINS | |||
| CS29544 | Cronobacter sakazakii ATCC 29544, wild-type | MJM187 | ATCC |
| CS29544 pKD46 | CS29544 harboring pKD46 | MJM396 | This study |
| flhA_CV (2.10) | CS29544 flhA nonautoaggregating variant | MJM387 | This study |
| fliG_CV (3.6) | CS29544 fliG nonautoaggregating variant | MJM388 | This study |
| ΔflhA | CS29544 flhA | MJM425 | This study |
| ΔfliG | CS29544 fliG | MJM426 | This study |
| ΔmotAB | CS29544 motAB | MJM476 | This study |
| ΔfliC | CS29544 fliC | MJM477 | This study |
| ΔflaA | CS29544 flaA | MJM478 | This study |
| ΔflaA pKD46 | CS29544 flaA harboring pKD46 | MJM480 | This study |
| ΔflaAΔfliC | CS29544 flaAfliC | MJM484 | This study |
| ΔfliC/cfliC | CS29544 fliC/fliC complementation vector | MJM481 | This study |
| ΔflaAΔfliC/cfliC | CS29544 flaAfliC fliC complementation vector | MJM483 | This study |
| E. COLI STRAINS | |||
| DH5α | pKD3 and pCP20 host strain | MJM363, MJM364 | Gift from J. M. Slauch |
| MG1655 | pKD46 host strain | MJM365 | Gift from J. M. Slauch |
| Top10 | cfliC host strain | MJM423 | Invitrogen |
| PLASMIDS | |||
| pCP20 | bla cat cI857 λPRflp | Cherepanov and Wackernagel, 1995 | |
| pKD3 | bla FRT cat FRT | Datsenko and Wanner, 2000 | |
| pKD46 | bla PBAD gam bet exo | Datsenko and Wanner, 2000 | |
| cfliC | 1 kb fragment fliC native promoter and ORF cloned into pET-11a | pMJM49 | This study |
Table 2.
Primer sequences used to create and confirm gene knockouts.
| Locus | Primers | Sequence (5′–3′) |
|---|---|---|
| cat | c2_F | GATCTTCCGTCACAGGTAGG |
| c1_R | TTATACGCAAGGCGACAAGG | |
| flhA | flhA_ko_F | ATCACCAAGGGTGCGGGGCGTATCGCGGAAGTGGGCGCGC |
| flhA_ko_R | GATAGGCTTCACCGCTGCCGATTTCCACGCCTTTCATCAG | |
| flhA_flank_F | GAAATCGGTCAGCAGATCC | |
| flhA_flank_R | CAGACCACGACCGTCATCAC | |
| fliG | fliG_ko_F | TGACCATCGGTGAAGACCGCGCGGCGGAGGTGTTCAAACA |
| fliG_ko_R | ATAGCTTTCTGTTCGTTTTCCACCTGAGACAGACGCACCG | |
| fliG_flank_F | AGCGTAAAGAAGTGGAAGAG | |
| fliG_flank_R | CGTCGACTTCTTCGCTTTC | |
| motAB | motAB_ko_F | TGAACATCCTCGCCATAGCCAACAGCGGAAGGATGATGTC |
| motAB_ko_F | AAATGTCTGATAAAAATCGCTAATATCCATACTCACGCTA | |
| motAB_flank_F | CGTAAACTTTCGCGAGATGCTG | |
| motAB_flank_R | CGCGGAATATGGCATTTAGCTG | |
| fliC | fliC_ko_F | CAACCTGAACAAATCTCAGTCTGCTCTGGGCACTGCTATC |
| fliC_ko_F | GAAAGCTTGAGCGTCTATTAACGCAGCAGGGACAGCATGG | |
| fliC_flank_F | ATGCAGACGCAGGCTATTGAG | |
| fliC_flank_R | TCCGGCTATATCTGTCGCAAC | |
| flaA | flaA_ko_F | GCCACGCCGGTTGAGTCAGCGTCCGGGCAGGCGCGTGACG |
| flaA_ko_F | GCCTTGTTAACGACTCTCCGATGTGCGAAACGGCAACCCT | |
| flaA_flank_F | CTTCCCGCTGACCATTTC | |
| flaA_flank_R | AACGTGCCGTCCATATCC |
A fliC complementation vector was constructed by GenScript. Briefly, a 1,011 bp sequence, containing the fliC coding sequence and native promoter, was obtained from the publically available CS29544 genome (NCBI Reference Sequence: NZ_CP011047.1). The entire DNA fragment was synthesized and cloned into the pET-11a vector with the restriction enzymes BglII and BamHI. The cfliC vector was electroporated into E. coli Top10 and subsequently electroporated into the CS29544 ΔfliC and ΔflaAΔfliC strains using LB broth. Putative complements were grown in BHI or on motility agar plates supplemented with 50 μg/mL ampicillin. Restoration of wild-type function was assessed by autoaggregation assays, motility assays, microscopy, and flagella harvest as detailed above and below.
Flagella staining and microscopy
The presence of extracellular flagella of CS29544, gene knockout, and complementation strains were determined by a combination of imaging techniques. Log or stationary phase CS29544, gene knockout, and complementation cultures were stained using a crystal violet-based flagella stain (Hardy Diagnostics) according to manufacturer's instructions. Stains were visualized using a light microscope at 1,000 × total magnification (BA210; Motic). Images were captured with a 2-megapixel Motic camera.
Several overnight colonies of CS29544, gene knockout, and complementation strains were gently lifted from BHI agar plates and suspended in phosphate buffered Karnovsky's fixative containing 2% glutaraldehyde and 2.5% paraformaldehyde. Transmission electron microscopy (TEM) was completed by the Beckman Institute's Microscopy Suite at the University of Illinois-Urbana Champaign. Briefly, the samples were stained with 2% uranyl acetate for 1 min and visualized using a CM200 LaB6 transmission electron microscope (FEI Co.). TEM was conducted at 120 kV and images were captured with a 2 k × 2 k digital camera (Tietz; Gauting; Germany). Several locations on the grids were examined, and the pictures were representative of the whole sample.
Flagella harvest and filament protein identification, sequencing, and In silico analysis
The extracellular protein fraction of CS29544, gene knockout, and complementation strains was harvested by differential centrifugation (DePamphilis and Adler, 1971). Bacteria were cultured in two baffled flasks each containing 500 mL of BHI and incubated overnight at 37°C with agitation (250 rpm). Stationary phase cultures (1 L total) were centrifuged at 3,220 × g for 10 min at 4°C. Bacterial pellets were resuspended in a total of 250 mL 0.1 M Tris-HCl, pH 7.8, and blended at room temperature for 30 s at “whip” speed. Blended suspensions were centrifuged at 12,000 × g for 10 min at 4°C. The supernatant was further ultracentrifuged at 55,000 × g for 1 h at 4°C. Protein pellets were resuspended in a total of 1 mL 0.1 M Tris-HCl, pH 7.8, containing 50% glycerol (v/v, protein storage buffer) and stored at −20°C. Total protein was quantified with the Bradford Assay (BioRad Laboratories) and visualized with SDS-polyacrylamide gel electrophoresis. Typical flagellum protein recovery was 0.5–0.7 mg/mL from 1 L of cell mass (~1012 cells).
The putative FliC (28.9 kDa) band from CS29544 flagellum preparation was gel-excised and treated in-gel with trypsin (G-Bioscience) by the DNA Services group affiliated with the Roy J. Carver Biotechnology Center at the University of Illinois-Urbana Champaign. Protein was digested at a ratio of 1:20 (trypsin:protein) in 25 mM ammonium bicarbonate at 55°C for 30 min. Following lyophilization, peptides were analyzed by liquid chromatography-mass spectrometry. A total of 1–2 μg of digested peptides were loaded into a Dionex Ultimate 3000 RSLCnano connected directly to a Thermo LTQ-Velos-ETD Pro Mass Spectrometer (Thermo Fisher Scientific). Peptide were run on an Acclaim 300 C18 nano column (Thermo Fisher Scientific) using a gradient of 100% A (water + 0.1% formic acid) to 60% B (acetonitrile + 0.1% formic acid) at a flow rate of 300 nL/min. Raw data were collected by Xcalibur (Thermo Fisher Scientific) and processed with an in-house Mascot Distiller and Mascot Server (Matrix Science) and identified with the NCBInr database.
The secondary structure of the CS29544 FliC protein was predicted from the amino acid coding sequence (NCBI Reference Sequence: NZ_CP011047.1) using the Iterative Threading Assembly Refinement (I-TASSER) method (Yang and Zhang, 2015) with default parameters. The I-TASSER method is publically available at http://zhanglab.ccmb.med.umich.edu/I-TASSER/, accessed 10/15/2015. The theoretical secondary structures of FliC were visualized and modified using UCSF Chimera v.1.10.2 (Pettersen et al., 2004), publically available at https://www.cgl.ucsf.edu/chimera/. The hydrophobicity index of the primary FliC amino acid sequence was determined using the ProtScale tool from the ExPASy Bioinformatics Resource Portal (Gasteiger et al., 2005), publically available at http://www.expasy.org/, accessed 06/01/2016. The hydrophobicity index was calculated using the Kyte and Doolittle amino acid scale (Kyte and Doolittle, 1982) with a window size of 15 amino acids.
CS29544 and flagellum competition assays
A 3 mL aliquot of stationary phase CS29544 culture was mixed with detached flagellar pieces at a concentration of 0.1, 1, 5, 10, or 20 μg/mL of total flagellum protein and autoaggregated for 6 h. Controls included adding 20 μg/mL of bovine serum albumin (BSA) or equal volume of protein storage buffer (no protein).
CS29544 biofilm formation to polyvinyl chloride tubing
Polyvinyl chloride tubing (PVC; 0.318 cm outer diameter; 0.159 cm inner diameter; U.S. Plastic Corporation) was cut into 5 cm long pieces (external surface area ~5.15 cm2) with a sterile blade. The PVC tube pieces were disinfected and submerged in 70% ethanol for 10 min and aseptically dried. Two PVC tube pieces were aseptically transferred to a 15 mL centrifuge tube containing 10 mL of BHI supplemented with 50 μg/mL ampicillin for the ΔfliC/cfliC strain. Each experimental vessel was inoculated with 1% (v/v) stationary phase CS29544, ΔmotAB, ΔfliC, and ΔfliC/cfliC cells and incubated vertically at 37°C aerobically with agitation (250 rpm) for 24 h. After incubation, each PVC tube piece was transferred with sterile forceps and washed three times in 5 mL PBS, pH 7.4. After washing, each PVC tube piece was placed into 30 mL PBS, pH 7.4, containing 3 g of autoclaved 0.1 mm diameter glass beads (Research Products International). Biofilms were subsequently disrupted by vortex at maximum speed for 1 min. Bacterial biofilm populations were enumerated by serial dilution (10−1–10−3) in 0.1% peptone water, spread-plated (0.1 mL in triplicate) on BHI agar plates, and incubated at 37°C overnight aerobically.
Statistical analysis
Autoaggregation and biofilm formation assay results were from a minimum of three independent replicates. The differences between the mean maximum percent autoaggregation due to temperature, redox balance, pH, blending, and various media; the differences between biofilm formation due to the presence of FliC were determined with SAS® (Version 9.4; SAS Institute) using the generalized linear model. When statistical significance was observed (P < 0.05), a post-hoc mean separation was run using Tukey's Honest Significance Difference test which controlled for unequal sample sizes. All data satisfied the assumptions of normality and homogeneity of variance.
Results
Characterization of autoaggregation in CS29544
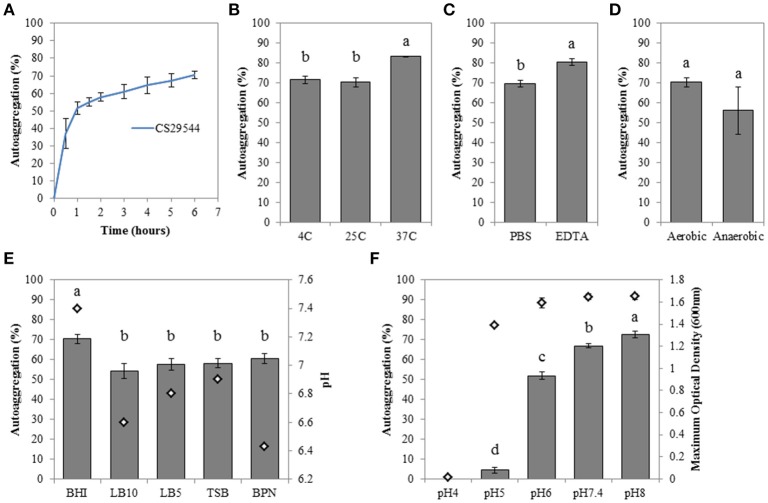
Stationary phase BHI cultures of CS29544 autoaggregated at 25°C within 30–60 min (Figure 1A). Following 6 h static incubation, the maximum mean autoaggregation percentage was 70.3 ± 2.2%. Various growth media and physiological conditions were tested to better understand CS29544 autoaggregation. Autoaggregation of BHI-grown cells at 37°C was significantly higher when statically incubated at 37°C than at 4 or 25°C (P = 0.0022; Figure 1B) or in the presence of EDTA (P = 0.0092; Figure 1C) compared to the PBS-added control. Although the maximum mean autoaggregation percentage was lower for cells grown in a reduced environment, the difference was not significant (P = 0.3003, Figure 1D). Autoaggregation following overnight growth in BHI was significantly higher than overnight growth in LB10, LB5; TSB, and BPN (P = 0.0203; Figure 1E). The difference in autoaggregation was not readily explained by the presence of salt, extracts, protein sources, or inclusion of phosphates but rather the initial pH of the media. Therefore, CS29544 was grown in BHI with varying initial pH values (range pH 4–8). CS29544 did not grow at pH 4, but its maximum growth was not affected by BHI from pH 5–8 (Figure 1F). As predicted, the maximum mean autoaggregation percentage decreased with decreasing pH (P < 0.0001; Figure 1F). The maximum mean autoaggregation percentage in BHI at an initial pH 6 was 51.9 ± 1.6%, which was slightly lower than the maximum mean autoaggregation percentage observed in the various media tested. During this initial characterization, an abolishment of autoaggregation was only achieved when grown in BHI at pH 5; however, this observation did not clearly point to a specific mechanism. Since the maximum mean autoaggregation percentage never reached 100%, we hypothesized that there might be a nonautoaggregating subpopulation of CS29544 mediated by an identifiable genetic variation.
Figure 1.
Autoaggregation of stationary phase CS29544 cultures. (A) Time course of autoaggregation in BHI broth over 6 h. Maximum percent autoaggregation (B) in BHI broth during static incubation at different temperatures; (C) in BHI broth in the presence of 50 mM EDTA or PBS; (D) in BHI broth following growth aerobically or anaerobically; (E) in a variety of growth media; LB10 = LB Miller's formula (10 g/L NaCL), LB5 = LB Lennox's formula (5 g/L NaCL), TSB = tryptic soy broth and BPN = motility broth, initial pH of growth media is reported for reference; (F) in BHI at different initial pH values. Maximum optical density (600 nm; open diamonds) is reported for reference. All experiments are mean ± standard error of three independent replicates. Values with no letters in common are significantly different (P < 0.05).
Nonautoaggregating CS29544 are a stable genetically distinct subpopulation
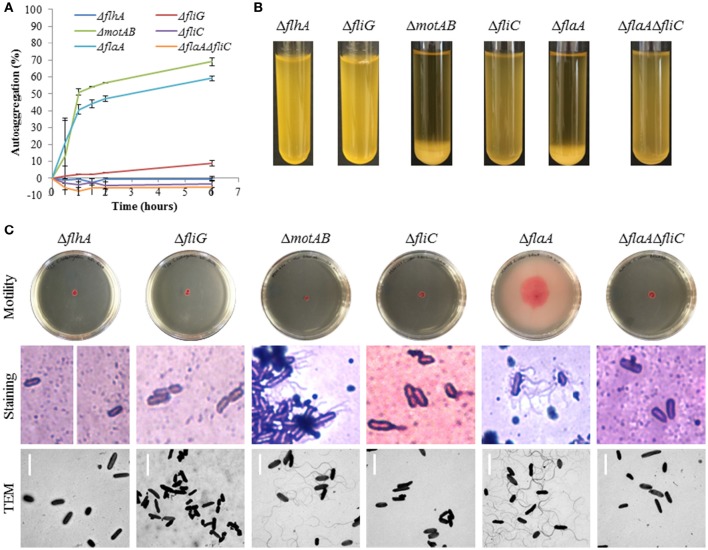
Following five successive passages selecting against autoaggregating CS29544, we were able to isolate two independent nonautoaggregating CVs of CS29544 (Figures 2A,B; see Movie S1 in the Supplementary Material). De novo assembly of the mate-pair library preparation from wild-type CS29544 was used as the reference genome for comparative genomic analysis with the nonautoaggregating CVs (2.10 and 3.6) assemblies. One unique single nucleotide polymorphism was detected in each of the nonautoaggregating CV. Strain 2.10 contained a putative deletion of two consecutive base pairs (GC), while strain 3.6 contained a putative deletion of a single base pair (C). Further corroboration by Sanger sequencing revealed the two nonautoaggregating CVs contained frameshift mutations, and NCBI BLAST analysis revealed that these mutations were located in the open reading frames of two flagellum proteins, FlhA (2.10) and FliG (3.6). Comparison with the full length wild-type FlhA and FliG proteins (692 and 340 amino acids, respectively) revealed that the frameshift mutations results in truncated proteins (157 and 183 amino acids, respectively). Strains carrying these variant mutations are referred to as flhA_CV (2.10) and fliG_CV (3.6), respectively. Accordingly, we assessed the flagellation of the wild-type CS29544, flhA_CV, and fliG_CV with motility assays and microscopy (Figure 2C). Both the flhA_CV and fliG_CV strains were nonmotile and aflagellate by staining and TEM (Figure 2C). As a result, we constructed a variety of gene knockouts to determine if structurally intact and functional flagella were required for autoaggregation in CS29544.
Figure 2.
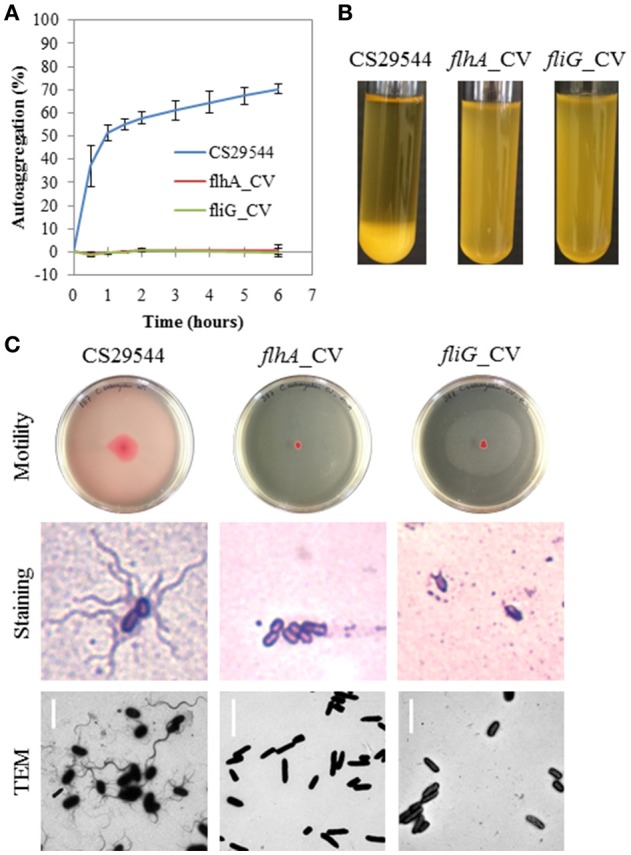
Autoaggregation, motility and flagellation of nonautoaggregating CS29544. (A) Time course of autoaggregation in BHI broth over 6 h. CS29544 (included for reference) and clonal variants: flhA_CV and fliG_CV. Experiment is mean ± standard error of three independent replicates. (B) Autoaggregation tube assays in BHI broth after static incubation for 6 h. (C) Motility agar plates centrally inoculated and imaged after 24 h, flagellation as detected by crystal violet staining at 1,000 × total magnification and TEM, bar = 4 μm.
Flagellum structure, not function, is required for autoaggregation in CS29544
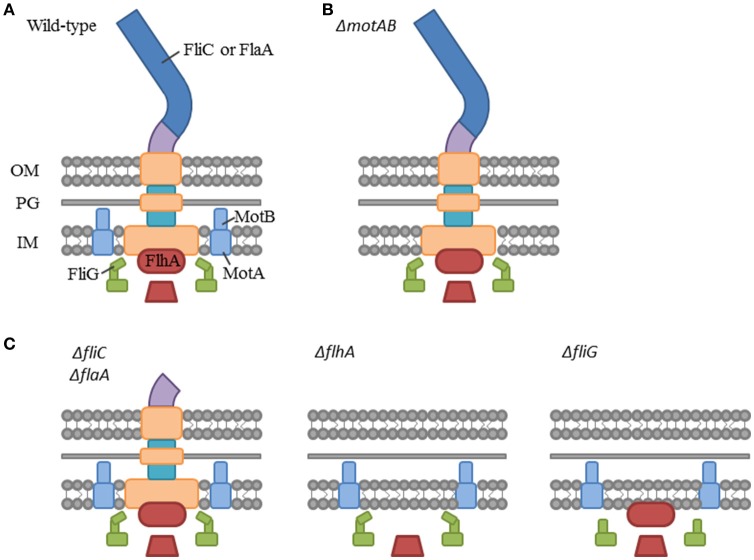
Published C. sakazakii genomes, available from NCBI, have annotated some 40 genes related to their flagellum's structure, function, and regulation. Therefore, gene knockout strains (Table 1) were constructed to disrupt the structure and function of the CS29544 flagella (see Figures 3A–C for a simplified diagram of the bacterial flagella outlining our knockout strategy). Two basal body proteins, FlhA and FliG (same genes as the nonautoaggregating CVs) were targeted. Previously, FlhA truncation mutants in Campylobacter jejuni resulted in aflagellate cells lacking flagellar components past the inner membrane (Abrusci et al., 2013). FliG, along with FliN and FliM, forms the C ring of the basal body (Zhao et al., 2014). Proton-driven conformational changes in the MotA and MotB stator (motor) proteins (Kojima and Blair, 2001) directly interact with the C terminus of the FliG protein of the C ring to provide rotation to the flagella (Irikura et al., 1993; Lloyd et al., 1996). Although FliG is integral for the flagellum function, its necessity for assembly is disputed (Irikura et al., 1993; Lloyd et al., 1996). It was hypothesized that the disruption of motAB would render the cells nonmotile while retaining the structural components. Additionally, the disruption of flhA and fliG would block the early assembly of the flagella and the cells would be aflagellate. Based on the published CS29544 genome, CS29544 has redundant filament proteins; therefore, both FliC and FlaA single and double gene knockouts were constructed. Finally, the function of the flagella was disrupted by targeting the two motor proteins, MotA and MotB.
Figure 3.
Simplified diagram of the bacterial flagellum highlighting our gene knockout strategy. (A) Bacterial flagellum labeled with gene knockout targets. Filament proteins: FliC and FlaA, Stator (motor) proteins: MotA and MotB, C-ring protein: FliG, and Exporter apparatus protein: FlhA. (B) Hypothesized flagellum structure in the functional mutant: ΔmotAB. (C) Hypothesized flagellum structure in the structural mutants: ΔfliC and ΔflaA, ΔflhA, and ΔfliG.
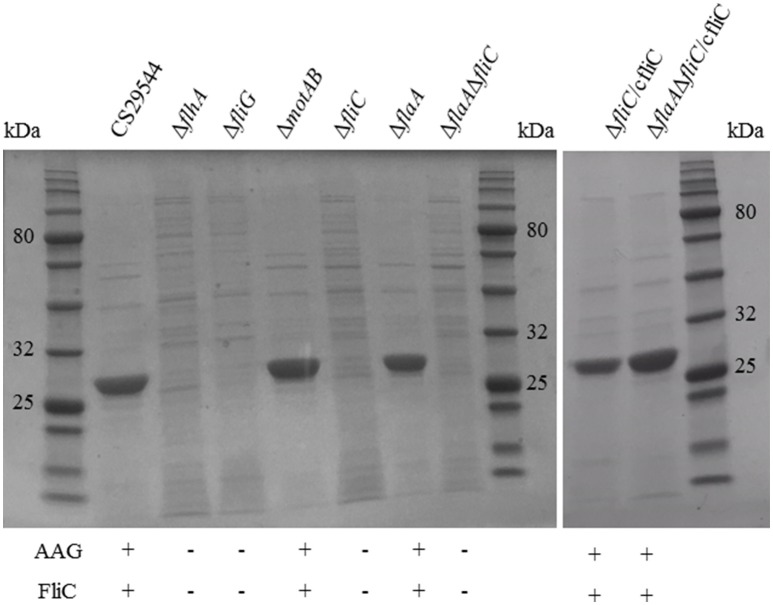
The ΔflhA, ΔfliG, ΔfliC, and ΔflaAΔfliC strains did not autoaggregate and were aflagellate by staining and TEM and nonmotile (Figures 4A–C). Conversely, the ΔmotAB and ΔflaA strains remained autoaggregative and had visible flagella by staining and TEM (Figures 4A–C). These two strains differed in their motility: ΔflaA was motile while ΔmotAB was not motile. Therefore, we concluded that motility was not required for autoaggregation in CS29544. Based on the phenotypes of the structural gene knockouts, we hypothesized that filaments composed of FliC, not FlaA, were required for autoaggregation. Upon mechanical removal of the filaments, only the autoaggregating CS29544, ΔmotAB, and ΔflaA strains possessed a dense 28.9 kDa protein band (Figure 5). The band was confirmed as FliC by a total of five peptides with individual ion scores of 44 and a Mascot ion score of 1,191 (data not shown). Furthermore, the ΔfliC strain did not contain a 50.1 kDa band, which is the predicted size for FlaA. Autoaggregation phenotype, motility, and flagellation were restored in the ΔfliC/cfliC and ΔflaAΔfliC/cfliC complementation strains (Figures 6A–C). Finally, we confirmed a loss of autoaggregation in CS29544 after mechanical removal of flagella from 79.4 ± 4.4% to 2.2 ± 2.2% with no reduction in cellular viability (data not shown).
Figure 4.
Autoaggregation, motility, and flagellation of CS29544 and gene knockout strains. (A) Time course of autoaggregation in BHI broth over 6 h. Gene knockouts: ΔflhA, ΔfliG, ΔmotAB, ΔfliC, ΔflaA, ΔflaAΔfliC. Experiment is mean ± standard error of three independent replicates. (B) Autoaggregation tube assays in BHI broth after static incubation for 6 h. (C) Motility agar plates centrally inoculated and imaged after 24 h, flagellation as detected by crystal violet staining at 1,000 × total magnification and TEM, bar = 4 μm.
Figure 5.
Harvested filaments from of CS29544, gene knockout, and complementation strains. Gene knockouts: ΔflhA, ΔfliG, ΔmotAB, ΔfliC, ΔflaA, ΔflaAΔfliC, and FliC complements: ΔfliC/cfliC and ΔflaAΔfliC/cfliC. Each lane was loaded with ~2 μg of total protein. FliC expected size 28.9 kDa.
Figure 6.
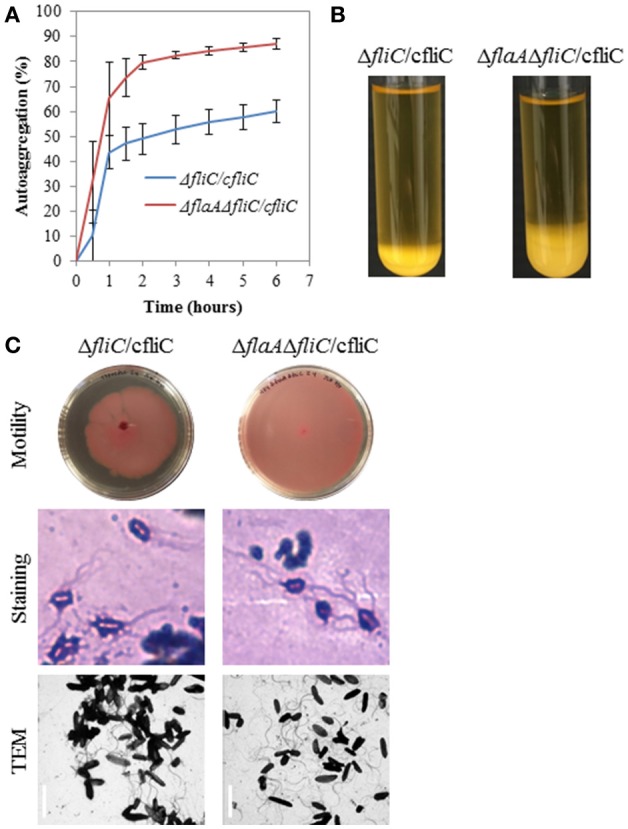
Autoaggregation, motility, and flagellation of FliC complementation strains. (A) Time course of autoaggregation in BHI broth over 6 h. FliC complements: ΔfliC/cfliC and ΔflaAΔfliC/cfliC. Experiment is mean ± standard error of three independent replicates. (B) Autoaggregation tube assays in BHI broth after static incubation for 6 h. (C) Motility agar plates centrally inoculated and imaged after 24 h, flagellation as detected by crystal violet staining at 1,000 × total magnification and TEM, bar = 4 μm.
Flagella-mediated autoaggregation occurs by flagellum-flagellum interactions
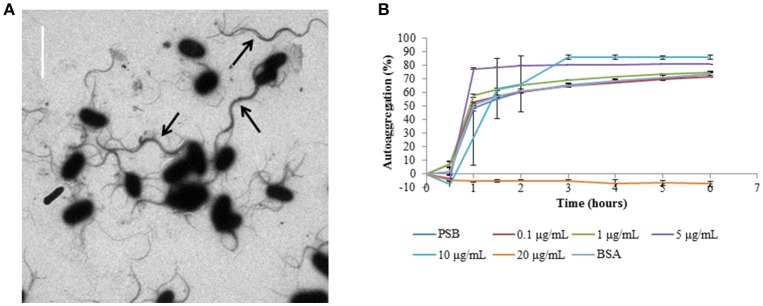
Visual analysis of the CS29544 TEM image revealed supercoiled flagella linking several bacteria together in a cluster (Figure 7A). Therefore, it was hypothesized that autoaggregation in CS29544 proceeds as flagella from one cell becomes entangled with the flagella from neighboring cells. To test this hypothesis, mechanically detached flagella pieces were added to stationary phase CS29544 cultures. Autoaggregation of CS29544 was not affected by adding 0.1 or 1.0 μg/mL of detached flagella, 20 μg/mL BSA, or the no-protein control (PSB; Figure 7B). Autoaggregation was prevented when 20 μg/mL of detached flagella were added. The addition of detached flagella altered the manner by which the CS29544 autoaggregated. The controls autoaggregated as before by forming small cell flocs that settled to the bottom of the tube. In the presence of 5 and 10 μg/mL of detached flagellum flocs did not form, rather a large mass of cells settled gradually to the bottom of the tube (see Movie S2 in the Supplementary Material).
Figure 7.
Autoaggregation competition assays with harvested filaments from CS29544. (A) CS29544 wild-type flagella TEM image, bar = 4 μm. Black arrows indicate bundles of flagella. (B) Time course of autoaggregation in BHI broth over 6 h. Stationary phase CS29544 cultures combined with harvested FliC filaments (0.1–20 μg/mL), bovine serum albumin (BSA, 20 μg/mL) and PSB (no protein control). Experiment is mean ± standard error of three independent replicates.
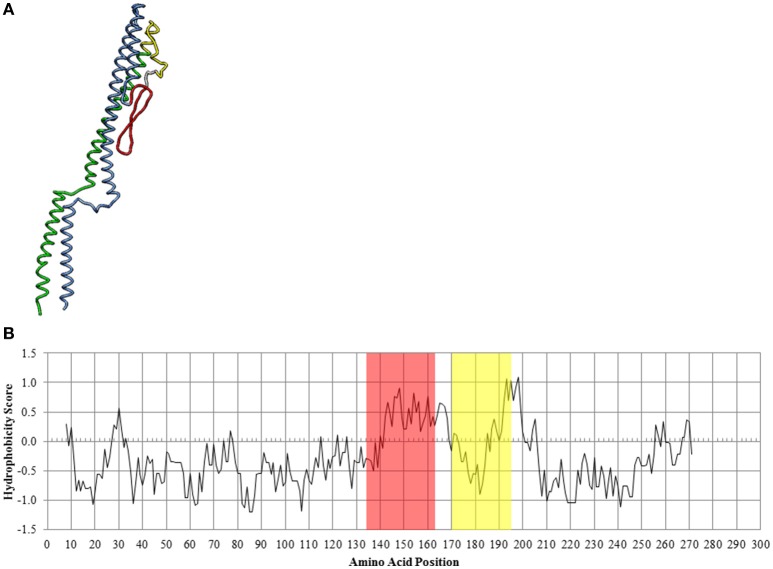
The predicted secondary structure of the CS29544 FliC protein was analogous to homologous flagellins with a highly conserved flagellin N and C termini linked by a hypervariable region (Figure 8A). Analysis of the hydrophobicity scores of the linear amino acid sequence revealed a peak followed by a valley in hydrophobicity (Figure 8B) located in the hypervariable region of the CS29544 FliC. The bacterial filament is composed of several thousand flagellin proteins with the conserved regions stacked laterally. The hypervariable region is externally exposed and able to interact with its environment. We hypothesize that the hypervariable hydrophobic peaks and valleys of neighboring cells interact laterally to promote autoaggregation in CS29544. Further work will be required to test this hypothesis.
Figure 8.
In silico analysis of the CS29544 FliC protein. (A) Theoretical secondary structure of FliC from I-TASSER. Conserved regions: N terminus in blue, C terminus in green. Hypervariable region: hydrophobic peak in red, hydrophobic valley in yellow. (B) Hydrophobicity index along the linear amino acid sequence of FliC. Hydrophobic peak highlighted in red, hydrophobic valley highlighted in yellow.
Biofilm formation on PVC is not mediated by flagella
The total cellular population of wild-type CS29544 biofilms on PVC was 1.5 × 104 ± 3.0 × 103 CFU/cm2. The total cellular populations of ΔmotAB, ΔfliC, and ΔfliC/cfliC on PVC biofilms were 4.8 × 103 ± 2.2 × 103, 2.4 × 104 ± 7.0 × 103, and 1.0 × 104 ± 6.5 × 102 CFU/cm2, respectively. CS29544 biofilm formation on PVC was not mediated by FliC under the tested conditions (P = 0.0928; Figure 9).
Figure 9.
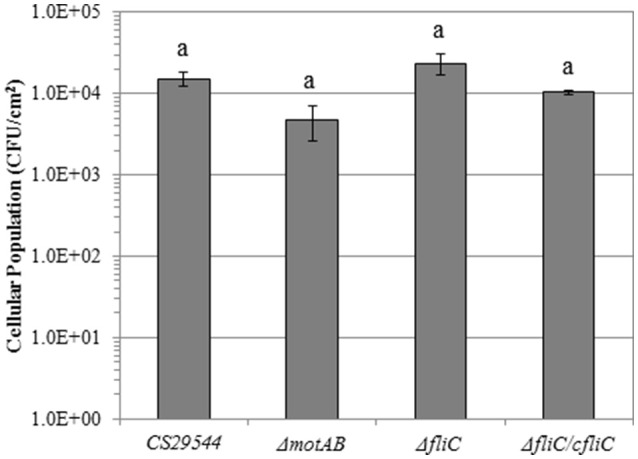
Biofilm formation on polyvinyl chloride of CS29544 and select gene knockout and complementation strains. Experiment is mean ± standard error of three independent replicates. Gene knockouts: ΔmotAB and ΔfliC and FliC complement: ΔfliC /fliC. Values with no letters in common are significantly different (P < 0.05).
Discussion
Although autoaggregation was demonstrated in C. sakazakii ATCC BAA_894 (Wang et al., 2012; Hu et al., 2015), the molecular basis for autoaggregation was not described. To understand the genetic determinants of autoaggregation in CS29544, a set of structural and functional flagellar mutants were constructed. These flagellar mutants revealed the requirement of FliC containing filaments in the autoaggregation of CS29544. Additionally, these results suggest an additional biological function for the CS29544 flagellum beyond motility.
Protein-protein interactions, such as flagella-mediated autoaggregation, may require specific environmental conditions. Previously, protein-protein aggregation in bacteria was influenced by altering the growth media (Girón et al., 1991), sodium chloride concentrations (Girard et al., 2010), pH (Sherlock et al., 2005; Alamuri et al., 2010), or the presence of divalent cations (Sjoblad et al., 1985; Abdel-Nour et al., 2014). In the present study, autoaggregation assays were conducted to identify nutritional dependencies and provide insights into potential mechanisms. Different growth media containing various nutrient extracts, protein sources, salts (NaCl and phosphates), and carbohydrates (dextrose) were tested. Additionally, autoaggregation was observed under a variety of temperatures, redox potentials, and pH values. Wild-type CS29544 was flagellated and highly motile under all tested growth conditions, except growth in BHI at pH 5. In hindsight, it is not surprising that autoaggregation in CS29544 had only minimal nutritional or conditional dependencies, even though flagellar expression is a tightly regulated system that quickly responds to changes in the surrounding bacterium environment (Osterman et al., 2015). Under favorable environmental conditions, such as nutrient-dense media, motility may be arrested following the downregulation of flagellar genes. However, nonmotile bacteria do not immediately shed their structurally intact flagella and these flagella can participate in other biological functions. Although abolishment of autoaggregation in CS29544 was observed in structural mutants (ΔflhA, ΔfliG, ΔfliC, and ΔflaAΔfliC), autoaggregation was not affected in the functional mutant (ΔmotAB) which retained the structural components. These results suggest that autoaggregation in CS29544 can serve as an additional biological function for the CS29544 flagellum under environmental conditions that favor the downregulation of motility but not the loss of structure. Of note, many other examples of autoaggregation in Enterobacteriaceae involve pili and fimbriae (Girón et al., 1991; Nataro et al., 1992; Collinson et al., 1993; Czeczulin et al., 1997; Schembri et al., 2001). While CS29544 has putative pilus and fimbrial genes, the present study did not identify a role for these structures in the autoaggregation of CS29544. Future studies are needed to investigate the role of pili and fimbriae in CS29544's pathogenicity.
The CS29544 genome encodes > 40 genes that are required for the assembly, function, and regulation of its flagellum. In this study, autoaggregation was only mediated by the loss of structural proteins, specifically, the lack of the FliC containing filament. Four structural mutants, two direct (ΔfliC and ΔflaAΔfliC) and two indirect (ΔflhA and ΔfliG), resulted in aflagellate nonautoaggregating CS29544 cells. Since the extracellular filament, comprised of several thousand FliC monomers, extends several microns from the cell, it is physically able to promote cell-cell interactions. Upon close examination of wild-type CS29544 cells by TEM, neighboring cells appeared tethered by their filaments. Similar bundles were observed in Escherichia coli (Girón et al., 1991), Pseudomonas marina (Sjoblad et al., 1985), and Pyrococcus furiosus (Näther et al., 2006). Furthermore, flagella-mediated autoaggregation was disrupted in a dose-dependent manner by the addition of exogenous wild-type FliC filaments. Protein-protein interactions can be mediated by several factors, including ionic and hydrophobic bonds. As discussed above, only growth in BHI at pH 5 abolished flagella-mediated autoaggregation in CS29544 and no other nutritional or conditional dependencies were observed. Previously, TibA-mediated autoaggregation in an enterotoxigenic E. coli was affected by changes in pH (Sherlock et al., 2005). The authors speculated that TibA-mediated autoaggregation might be promoted by pH-mediated ionic bonds between charged amino acid side chains. It is tempting to conclude that flagella-mediated autoaggregation involves ionic bonding due to abolishment at pH 5; however, our observations do not support this conclusion. CS29544 cells grown in BHI at pH 5 were growth-impaired, had no visible flagella by staining, and were nonmotile by wet mount.
The CS29544 genome encodes two different flagellin proteins, fliC and flaA; however, only FliC monomers were incorporated into the harvested CS29544 filaments under the tested conditions. Consistent with this study, FliC is the sole C. sakazakii flagellin protein reported in the literature (Proudy et al., 2008; Cruz-Córdova et al., 2012). FliC flagellin proteins and their homologs have highly conserved N and C termini connected by a hypervariable region. The conserved domains of several flagellin proteins self-assemble and form the internal channel of the filament during elongation. The exposure of conserved domains to the bacterium's environment is limited and should not contribute to flagella-mediated autoaggregation. Conversely, the hypervariable region is invariably externally exposed and likely interacts with components of the bacterium's surroundings. As a result, our in silico methods were centered on the secondary structure and hydrophobicity of the hypervariable region. The entire CS29544 FliC flagellin protein is composed of 278 amino acids, of which 50 amino acids comprise the hypervariable region. Of note, the hypervariable region in the C. sakazakii FliC flagellin protein is far shorter than FliC flagellin proteins of related Enterobacteriaceae (Proudy et al., 2008). As seen in the predicted secondary structure, the hypervariable region is relaxed and spatially aligned with the conserved regions. Unfortunately, the predicted FliC secondary structure did not reveal any obvious structural contributions to flagella-mediated autoaggregation. Rather, alterations in hydrophobicity along the amino acid sequence illustrated the potential of hydrophobic interactions. It is hypothesized that there are, along the length of every filament, thousands of FliC monomers with alternating hydrophobic peaks and valleys (colored in red and yellow, respectively, in Figure 8A). The following hypothesis can be best exemplified by the dimerization of proteins by the leucine zipper motif. When filaments are in close proximity, it is hypothesized that these hydrophobic peaks and valleys interact to allow rapid and reversible supercoiling. Once a sufficient number of CS29544 cells are tethered together by their FliC filaments, autoaggregation by this mechanism proceeds. Further work is needed to test these hypotheses and to more precisely define which amino acids interact during flagella-mediated autoaggregation in CS29544.
C. sakazakii strains form biofilms on a variety of abiotic surfaces, including stainless steel (Iversen et al., 2004; Kim et al., 2006; Jung et al., 2013), silicon (Iversen et al., 2004), latex (Iversen et al., 2004), PVC (Lehner et al., 2005; Kim et al., 2006; Hurrell et al., 2009a), and polyurethane (Hurrell et al., 2009a). The latter two plastics are used for enteral feeding tubes and formation of C. sakazakii biofilms on these plastics is of concern. Hurrell et al. (2009b) isolated C. sakazakii, along with other pathogenic Enterobacteriaceae from used enteral feeding tubes. Biofilm formation on enteral feeding tubes is problematic for several reasons. First, enteral feeding tubes typically reside within an infant at body temperature (37°C) for several days (Mehall et al., 2002). Secondly, infant feeds are nutrient-dense and provide sufficient growth substrate for bacteria. Lastly, with every feeding, bacteria might dislodge from the biofilm and continuously inoculate the neonate (Mehall et al., 2002; Hurrell et al., 2009b). To determine the impact of flagella-mediated autoaggregation on C. sakazakii biofilm formation, the biofilm formation by the wild-type CS29544 was compared to the ΔmotAB, ΔfliC, and ΔfliC/cfliC strains. To model C. sakazakii biofilm formation on neonatal enteral feeding tubes, flagella-mediated biofilm formation was tested in a nutrient-dense environment (BHI broth) at 37°C using PVC tubing. In the present study, the total cellular biofilm population on PVC tubing ranged from 3.7-log CFU/cm2 in the ΔmotAB strain to 4.4-log CFU/cm2 in the ΔfliC strain. There was no significant difference in biofilm formation between the wild-type CS29544 and the ΔmotAB, ΔfliC, and ΔfliC/cfliC strains under the tested conditions. The observed C. sakazakii population density was consistent with the mean biofilm population of 4.0-log CFU/cm2 on PVC tubing of five C. sakazakii strains grown in TSB at 12°C reported by Kim et al. (2006). Additionally, that study reported an approximate 1.5-log increase in the mean biofilm population (5.7-log CFU/cm2) on PVC tubing when C. sakazakii strains were grown in TSB at 25°C. Given that 27°C is the optimal temperature for C. sakazakii exopolysaccharide production, this result is not surprising. Admittedly, C. sakazakii biofilm formation due to differences in flagella-mediated autoaggregation phenotype was not robustly tested. To date, a single study has demonstrated the importance of C. sakazakii strain ES5 flagellum in biofilm formation and adhesion to microtiter plates (Hartmann et al., 2010). The data presented in the present study demonstrates that additional research into C. sakazakii flagella-mediated autoaggregation, biofilm formation, and gastrointestinal colonization is critically needed.
A significant shortcoming of this study is its limited scope. A single strain of C. sakazakii was characterized, and generalization to all C. sakazakii strains should be avoided. Currently, our collective understanding of C. sakazakii pathogenesis is insufficient. Several decades of work were completed to characterize the diverse pathotypes in E. coli, and it is tempting to speculate that C. sakazakii may have definable pathotypes of which flagella-mediated autoaggregation is important. Future studies should be designed to characterize flagella-mediated autoaggregation contributions to C. sakazakii pathogenesis in vivo with suitable animal models. Concurrently, autoaggregation, not necessarily flagella-mediated, should be characterized in several clinical, environmental, and laboratory C. sakazakii strains. The present study contributes much-needed knowledge to the C. sakazakii literature.
Author contributions
JH conceived of the project, contributed to the design of the experimental methods, led the acquisition, analysis, and interpretation of the data other than experiments completed by the acknowledged collaborators, wrote initial and revised drafts of the manuscript, and approved the final manuscript submission. MM contributed to project design, selection of experimental methods, interpretation of data throughout the project, contributed to drafting and revising of the manuscript, approved the final manuscript submission. JH and MM agree to be accountable for the work detailed in the final manuscript submission.
Funding
JH was supported by the Agnes and Bill Brown Fellowship in Microbiology from the University of Illinois-Urbana Champaign. This research received no direct financial support from any funding agency in the public, commercial, or not-for-profit sectors.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the assistance of staff of the DNA and Protein Services Lab at the Roy J. Carver Biotechnology Center and the Beckman Institute Microscopy Suite, particularly Catherine Wallace. We thank Dr. James Slauch for the Lambda Red Recombinase System and Daniel Hoeflinger for compiling the time lapse photos into videos.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00301/full#supplementary-material
References
- Abdel-Nour M., Duncan C., Prashar A., Rao C., Ginevra C., Jarraud S., et al. (2014). The Legionella pneumophila collagen-like protein mediates sedimentation, autoaggregation, and pathogen-phagocyte interactions. Appl. Environ. Microbiol. 80, 1441–1454. 10.1128/AEM.03254-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrusci P., Vergara-Irigaray M., Johnson S., Beeby M. D., Hendrixson D. R., Roversi P., et al. (2013). Architecture of the major component of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 20, 99–104. 10.1038/nsmb.2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamuri P., Löwer M., Hiss J. A., Himpsl S. D., Schneider G., Mobley H. L. T. (2010). Adhesion, invasion, and agglutination mediated by two trimeric autotransporters in the human uropathogen Proteus mirabilis. Infect. Immun. 78, 4882–4894. 10.1128/IAI.00718-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y. H., Friedman R. J. (1998). Concise review of mechanisms of bacterial adhesion to biomaterial surfaces. J. Biomed. Mater. Res. 43, 338–348. [DOI] [PubMed] [Google Scholar]
- Arenas J., Cano S., Nijland R., van Dongen V., Rutten L., van der Ende A., et al. (2015). The meningococcal autotransporter AutA is implicated in autoaggregation and biofilm formation. Environ. Microbiol. 17, 1321–1337. 10.1111/1462-2920.12581 [DOI] [PubMed] [Google Scholar]
- Axelrod D., Kazmerski K., Iyer K. (2006). Pediatric enteral nutrition. JPEN J. Parenter. Enteral Nutr. 30, S21–S26. 10.1177/01486071060300S1S21 [DOI] [PubMed] [Google Scholar]
- Bai L., Xia S., Lan R., Liu L., Ye C., Wang Y., et al. (2012). Isolation and characterization of cytotoxic, aggregative Citrobacter freundii. PLoS ONE 7:e33054. 10.1371/journal.pone.0033054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biering G., Karlsson S., Clark N. C., Jónsdóttir K. E., Lúdvígsson P., Steingrímsson O. (1989). Three cases of neonatal meningitis caused by Enterobacter sakazakii in powdered milk. J. Clin. Microbiol. 27, 2054–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeuwer P., Lardeau A., Peterz M., Joosten H. M. (2003). Desiccation and heat tolerance of Enterobacter sakazakii. J. Appl. Microbiol. 95, 967–973. 10.1046/j.1365-2672.2003.02067.x [DOI] [PubMed] [Google Scholar]
- Broge T., Lee A. (2013). A case of cronobacter (Enterobacter sakazakii) bacteremia in a breastfed infant. J. Pediatr. Infect. Dis. Soc. 2, e1–e2. 10.1093/jpids/pit021 [DOI] [PubMed] [Google Scholar]
- Cherepanov P. P., Wackernagel W. (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14. 10.1016/0378-1119(95)00193-A [DOI] [PubMed] [Google Scholar]
- Collinson S. K., Doig P. C., Doran J. L., Clouthier S., Trust T. J., Kay W. W. (1993). Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J. Bacteriol. 175, 12–18. 10.1128/jb.175.1.12-18.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Cheng K. J., Geesey G. G., Ladd T. I., Nickel J. C., Dasgupta M., et al. (1987). Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41, 435–464. 10.1146/annurev.mi.41.100187.002251 [DOI] [PubMed] [Google Scholar]
- Cruz-Córdova A., Rocha-Ramírez L. M., Ochoa S. A., González-Pedrajo B., Gónzalez-Pedrajo B., Espinosa N., et al. (2012). Flagella from five Cronobacter species induce pro-inflammatory cytokines in macrophage derivatives from human monocytes. PLoS ONE 7:e52091. 10.1371/journal.pone.0052091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeczulin J. R., Balepur S., Hicks S., Phillips A., Hall R., Kothary M. H., et al. (1997). Aggregative adherence fimbria II, a second fimbrial antigen mediating aggregative adherence in enteroaggregative Escherichia coli. Infect. Immun. 65, 4135–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M. L., Adler J. (1971). Purification of intact flagella from Escherichia coli and Bacillus subtilis. J. Bacteriol. 105, 376–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg M. S., Kaper J. B. (1991). Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59, 4310–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre-Bonte S., Darfeuille-Michaud A., Forestier C. (1995). Aggregative adherence of Klebsiella pneumoniae to human intestine-407 cells. Infect. Immun. 63, 1318–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z. P., Nie P., Lu J. F., Liu L. Y., Xiao T. Y., Liu W., et al. (2015). Type III secretion system translocon component eseb forms filaments on and mediates autoaggregation of and biofilm formation by Edwardsiella tarda. Appl. Environ. Microbiol. 81, 6078–6087. 10.1128/AEM.01254-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M., Appel R., et al. (2005). Protein identification and analysis tools on the ExPASy server, in The Proteomics Protocols Handbook, ed Walker J. (Totowa, NJ: Humana Press; ), 571–607. 10.1385/1-59259-890-0:571 [Accessed November 20, 2016]. [DOI] [Google Scholar]
- Girard V., Côté J.-P., Charbonneau M.-E., Campos M., Berthiaume F., Hancock M. A., et al. (2010). Conformation change in a self-recognizing autotransporter modulates bacterial cell-cell interaction. J. Biol. Chem. 285, 10616–10626. 10.1074/jbc.M109.069070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girón J. A., Ho A. S., Schoolnik G. K. (1991). An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254, 710–713. 10.1126/science.1683004 [DOI] [PubMed] [Google Scholar]
- Haiko J., Westerlund-Wikström B. (2013). The role of the bacterial flagellum in adhesion and virulence. Biology 2, 1242–1267. 10.3390/biology2041242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann I., Carranza P., Lehner A., Stephan R., Eberl L., Riedel K. (2010). Genes involved in Cronobacter sakazakii biofilm formation. Appl. Environ. Microbiol. 76, 2251–2261. 10.1128/AEM.00930-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Grim C. J., Franco A. A., Jarvis K. G., Sathyamoorthy V., Kothary M. H., et al. (2015). Analysis of the cellulose synthase operon genes, bcsA, bcsB, and bcsC in Cronobacter species: prevalence among species and their roles in biofilm formation and cell-cell aggregation. Food Microbiol. 52, 97–105. 10.1016/j.fm.2015.07.007 [DOI] [PubMed] [Google Scholar]
- Hurrell E., Kucerova E., Loughlin M., Caubilla-Barron J., Forsythe S. J. (2009a). Biofilm formation on enteral feeding tubes by Cronobacter sakazakii, Salmonella serovars and other Enterobacteriaceae. Int. J. Food Microbiol. 136, 227–231. 10.1016/j.ijfoodmicro.2009.08.007 [DOI] [PubMed] [Google Scholar]
- Hurrell E., Kucerova E., Loughlin M., Caubilla-Barron J., Hilton A., Armstrong R., et al. (2009b). Neonatal enteral feeding tubes as loci for colonisation by members of the Enterobacteriaceae. BMC Infect. Dis. 9:146. 10.1186/1471-2334-9-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irikura V. M., Kihara M., Yamaguchi S., Sockett H., Macnab R. M. (1993). Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J. Bacteriol. 175, 802–810. 10.1128/jb.175.3.802-810.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen C., Lane M., Forsythe S. J. (2004). The growth profile, thermotolerance and biofilm formation of Enterobacter sakazakii grown in infant formula milk. Lett. Appl. Microbiol. 38, 378–382. 10.1111/j.1472-765X.2004.01507.x [DOI] [PubMed] [Google Scholar]
- Iversen C., Mullane N., McCardell B., Tall B. D., Lehner A., Fanning S., et al. (2008). Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int. J. Syst. Evol. Microbiol. 58, 1442–1447. 10.1099/ijs.0.65577-0 [DOI] [PubMed] [Google Scholar]
- Jason J. (2012). Prevention of invasive Cronobacter infections in young infants fed powdered infant formulas. Pediatrics 130, e1076–e1084. 10.1542/peds.2011-3855 [DOI] [PubMed] [Google Scholar]
- Jung J.-H., Choi N.-Y., Lee S.-Y. (2013). Biofilm formation and exopolysaccharide (EPS) production by Cronobacter sakazakii depending on environmental conditions. Food Microbiol. 34, 70–80. 10.1016/j.fm.2012.11.008 [DOI] [PubMed] [Google Scholar]
- Kim H., Ryu J.-H., Beuchat L. R. (2006). Attachment of and biofilm formation by Enterobacter sakazakii on stainless steel and enteral feeding tubes. Appl. Environ. Microbiol. 72, 5846–5856. 10.1128/AEM.00654-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S., Blair D. F. (2001). Conformational change in the stator of the bacterial flagellar motor. Biochemistry (Mosc.) 40, 13041–13050. 10.1021/bi011263o [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. 10.1016/0022-2836(82)90515-0 [DOI] [PubMed] [Google Scholar]
- Lai K. K. (2001). Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and a review of the literature. Medicine (Baltimore) 80, 113–122. 10.1097/00005792-200103000-00004 [DOI] [PubMed] [Google Scholar]
- Lehner A., Riedel K., Eberl L., Breeuwer P., Diep B., Stephan R. (2005). Biofilm formation, extracellular polysaccharide production, and cell-to-cell signaling in various Enterobacter sakazakii strains: aspects promoting environmental persistence. J. Food Prot. 68, 2287–2294. 10.4315/0362-028X-68.11.2287 [DOI] [PubMed] [Google Scholar]
- Lloyd S. A., Tang H., Wang X., Billings S., Blair D. F. (1996). Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J. Bacteriol. 178, 223–231. 10.1128/jb.178.1.223-231.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mehall J. R., Kite C. A., Saltzman D. A., Wallett T., Jackson R. J., Smith S. D. (2002). Prospective study of the incidence and complications of bacterial contamination of enteral feeding in neonates. J. Pediatr. Surg. 37, 1177–1182. 10.1053/jpsu.2002.34467 [DOI] [PubMed] [Google Scholar]
- Nakao R., Ramstedt M., Wai S. N., Uhlin B. E. (2012). Enhanced biofilm formation by Escherichia coli LPS mutants defective in Hep biosynthesis. PLoS ONE 7:e51241. 10.1371/journal.pone.0051241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataro J. P., Deng Y., Maneval D. R., German A. L., Martin W. C., Levine M. M. (1992). Aggregative adherence fimbriae I of enteroaggregative Escherichia coli mediate adherence to HEp-2 cells and hemagglutination of human erythrocytes. Infect. Immun. 60, 2297–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näther D. J., Rachel R., Wanner G., Wirth R. (2006). Flagella of Pyrococcus furiosus: multifunctional organelles, made for swimming, adhesion to various surfaces, and cell-cell contacts. J. Bacteriol. 188, 6915–6923. 10.1128/JB.00527-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaili T. M., Shaker R. R., Al-Haddaq M. S., Al-Nabulsi A. A., Holley R. A. (2009). Heat resistance of Cronobacter species (Enterobacter sakazakii) in milk and special feeding formula. J. Appl. Microbiol. 107, 928–935. 10.1111/j.1365-2672.2009.04271.x [DOI] [PubMed] [Google Scholar]
- Osterman I. A., Dikhtyar Y. Y., Bogdanov A. A., Dontsova O. A., Sergiev P. V. (2015). Regulation of flagellar gene expression in bacteria. Biochem. Biokhimiia 80, 1447–1456. 10.1134/S000629791511005X [DOI] [PubMed] [Google Scholar]
- Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., et al. (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Prigent-Combaret C., Prensier G., Le Thi T. T., Vidal O., Lejeune P., Dorel C. (2000). Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2, 450–464. 10.1046/j.1462-2920.2000.00128.x [DOI] [PubMed] [Google Scholar]
- Proudy I., Bouglé D., Coton E., Coton M., Leclercq R., Vergnaud M. (2008). Genotypic characterization of Enterobacter sakazakii isolates by PFGE, BOX-PCR and sequencing of the fliC gene. J. Appl. Microbiol. 104, 26–34. 10.1111/j.1365-2672.2007.03526.x [DOI] [PubMed] [Google Scholar]
- Ravisankar S., Syed S. S., Garg P., Higginson J. (2014). Is Cronobacter sakazakii infection possible in an exclusively breastfed premature neonate in the neonatal intensive care unit? J. Perinatol. Off. J. Calif. Perinat. Assoc. 34, 408–409. 10.1038/jp.2014.14 [DOI] [PubMed] [Google Scholar]
- Riedel K., Lehner A. (2007). Identification of proteins involved in osmotic stress response in Enterobacter sakazakii by proteomics. Proteomics 7, 1217–1231. 10.1002/pmic.200600536 [DOI] [PubMed] [Google Scholar]
- Rocha S. P. D., Elias W. P., Cianciarullo A. M., Menezes M. A., Nara J. M., Piazza R. M. F., et al. (2007). Aggregative adherence of uropathogenic Proteus mirabilis to cultured epithelial cells. FEMS Immunol. Med. Microbiol. 51, 319–326. 10.1111/j.1574-695X.2007.00308.x [DOI] [PubMed] [Google Scholar]
- Schembri M. A., Christiansen G., Klemm P. (2001). FimH-mediated autoaggregation of Escherichia coli. Mol. Microbiol. 41, 1419–1430. 10.1046/j.1365-2958.2001.02613.x [DOI] [PubMed] [Google Scholar]
- Schluter J., Nadell C. D., Bassler B. L., Foster K. R. (2015). Adhesion as a weapon in microbial competition. ISME J. 9, 139–149. 10.1038/ismej.2014.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlock O., Vejborg R. M., Klemm P. (2005). The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infect. Immun. 73, 1954–1963. 10.1128/IAI.73.4.1954-1963.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons B. P., Gelfand M. S., Haas M., Metts L., Ferguson J. (1989). Enterobacter sakazakii infections in neonates associated with intrinsic contamination of a powdered infant formula. Infect. Control Hosp. Epidemiol. 10, 398–401. 10.2307/30144207 [DOI] [PubMed] [Google Scholar]
- Sjoblad R. D., Doetsch R. N., Emala C. W. (1985). Novel function of eubacterial flagella: role in aggregation of a marine bacterium. Arch. Microbiol. 142, 101–102. 10.1007/BF00409246 [DOI] [Google Scholar]
- Sourjik V., Wingreen N. S. (2012). Responding to chemical gradients: bacterial chemotaxis. Curr. Opin. Cell Biol. 24, 262–268. 10.1016/j.ceb.2011.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington A., Ballantyne M., Pinelli J., Stevens B. (1995). Indwelling versus intermittent feeding tubes in premature neonates. J. Obstet. Gynecol. Neonatal Nurs. 24, 321–326. 10.1111/j.1552-6909.1995.tb02483.x [DOI] [PubMed] [Google Scholar]
- Vadyvaloo V., Viall A. K., Jarrett C. O., Hinz A. K., Sturdevant D. E., Joseph Hinnebusch B. (2015). Role of the PhoP-PhoQ gene regulatory system in adaptation of Yersinia pestis to environmental stress in the flea digestive tract. Microbiol. Read. Engl. 161, 1198–1210. 10.1099/mic.0.000082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Acker J., de Smet F., Muyldermans G., Bougatef A., Naessens A., Lauwers S. (2001). Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J. Clin. Microbiol. 39, 293–297. 10.1128/JCM.39.1.293-297.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hu X., Tao G., Wang X. (2012). Outer membrane defect and stronger biofilm formation caused by inactivation of a gene encoding for heptosyltransferase I in Cronobacter sakazakii ATCC BAA-894. J. Appl. Microbiol. 112, 985–997. 10.1111/j.1365-2672.2012.05263.x [DOI] [PubMed] [Google Scholar]
- Wang L., Qin W., Yang S., Zhai R., Zhou L., Sun C., et al. (2015). The Adh adhesin domain is required for trimeric autotransporter Apa1-mediated Actinobacillus pleuropneumoniae adhesion, autoaggregation, biofilm formation and pathogenicity. Vet. Microbiol. 177, 175–183. 10.1016/j.vetmic.2015.02.026 [DOI] [PubMed] [Google Scholar]
- Yang J., Zhang Y. (2015). I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 43, W174–W181. 10.1093/nar/gkv342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Norris S. J., Liu J. (2014). Molecular architecture of the bacterial flagellar motor in cells. Biochemistry (Mosc.) 53, 4323–4333. 10.1021/bi500059y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.