Abstract
The trace element copper serves as cofactor for many enzymes but is toxic at elevated concentrations. In bacteria, the intracellular copper level is maintained by copper efflux systems including the Cue system controlled by the transcription factor CueR. CueR, a member of the MerR family, forms homodimers, and binds monovalent copper ions with high affinity. It activates transcription of the copper tolerance genes copA and cueO via a conserved DNA-distortion mechanism. The mechanism how CueR-induced transcription is turned off is not fully understood. Here, we report that Escherichia coli CueR is prone to proteolysis by the AAA+ proteases Lon, ClpXP, and ClpAP. Using a set of CueR variants, we show that CueR degradation is not altered by mutations affecting copper binding, dimerization or DNA binding of CueR, but requires an accessible C terminus. Except for a twofold stabilization shortly after a copper pulse, proteolysis of CueR is largely copper-independent. Our results suggest that ATP-dependent proteolysis contributes to copper homeostasis in E. coli by turnover of CueR, probably to allow steady monitoring of changes of the intracellular copper level and shut-off of CueR-dependent transcription.
Keywords: AAA+ proteases, proteolysis, Lon, ClpXP, ClpAP, CueR, copper homoeostasis, MerR family
Introduction
Copper is a trace element required as cofactor for full functionality of several enzymes, such as cytochrome c oxidase of the respiratory chain (van der Oost et al., 1994). The intracellular copper concentration must be strictly maintained since elevated copper levels are toxic for the cell, e.g., by generation of reactive oxygen species (Rensing and Grass, 2003; Grass et al., 2011). In Escherichia coli two copper efflux systems, the Cue and the Cus system, adjust the intracellular copper level to the cellular demand (Rensing and Grass, 2003; Rademacher and Masepohl, 2012). While the Cus system operates under anaerobic conditions, the Cue system is predominantly active under aerobic conditions (Outten et al., 2001). CueR, the key regulator of the Cue system, activates transcription of the copper tolerance genes copA and cueO (Outten et al., 2000; Stoyanov et al., 2001). CopA is a P-type ATPase located in the cytoplasmic membrane and pumps monovalent copper ions (Cu+) into the periplasm (Petersen and Møller, 2000; Rensing et al., 2000). The multi-copper oxidase CueO is located in the periplasm and oxidizes Cu+ to the divalent form, Cu2+, which is not able to pass the inner membrane by simple diffusion (Grass and Rensing, 2001; Rensing and Grass, 2003).
The transcription factor CueR is a member of the MerR family named after the mercury resistance regulator MerR (Brown et al., 2003). Proteins of this family typically form homodimers and are comprised of three characteristic domains: the N-terminal DNA-binding domain, the central dimerization helix, and the C-terminal metal-binding domain (Brown et al., 2003; Changela et al., 2003). CueR contains two copper-binding cysteines in its metal-binding domain (C112, C120), which are essential for covalent binding of monovalent copper ions. An active CueR homodimer, binding two Cu+ ions (holo-CueR), induces the expression of copA and cueO by binding to their promoter regions which induces torsional transformations in the DNA conformation (Changela et al., 2003; Chen et al., 2003; Stoyanov and Brown, 2003; Philips et al., 2015). By kinks and undertwisting, the DNA switches from a B-form into an A-form-like conformation that allows access of the RNA polymerase. The metal-free CueR dimer (apo-CueR) is also able to bind to the promoter region resulting in a tight DNA conformation, which represses copA and cueO expression (Philips et al., 2015).
CueR binds copper with high affinity (Changela et al., 2003). An open question is how CueR-mediated expression of copper detoxification systems is turned off when necessary or how the cellular CueR pool is maintained to allow continuous sensing of the actual intracellular copper level. Several studies have implicated a role of proteolysis in the regulation of metal homeostasis (Lu and Solioz, 2001; Solioz, 2002; Lu et al., 2003; Solioz and Stoyanov, 2003; Liu et al., 2007; Pruteanu et al., 2007; Pruteanu and Baker, 2009). Regulated proteolysis is a universal post-translational strategy adapting the existing protein pool to the cellular demand. In E. coli five different ATP-dependent proteases (AAA+ proteases, ATPases associated with a variety of cellular activities), namely ClpXP, ClpAP, HslUV, Lon, and FtsH, are responsible for quality control of proteins as well as for the regulated turnover of intact proteins (Baker and Sauer, 2006; Sauer and Baker, 2011; Bittner et al., 2016). AAA+ proteases are comprised of two functional domains, the ATPase and protease domain. While the proteases ClpP and HslV associate with separate ATPases to form ClpXP, ClpAP, or HslUV complexes, the two domains of Lon and FtsH are encoded by a single gene. The ATPase domain is needed for ATP-dependent unfolding and translocation of a substrate into the proteolytic chamber of the protease domain, in which the substrate is degraded (Bittner et al., 2016; Sauer and Baker, 2011). AAA+ proteases recognize their substrates via exposed recognition motifs, so-called degrons and also adaptor proteins can be involved in recognition (Sauer et al., 2004; Baker and Sauer, 2006; Gur et al., 2011, 2013; Sauer and Baker, 2011). An example for proteolysis of proteins involved in metal homeostasis is the MerR-like regulator ZntR, which binds zinc (Changela et al., 2003) and activates expression of the zinc exporter ZntA (Brocklehurst et al., 1999; Outten et al., 1999). ZntR is a substrate of the Lon and ClpXP proteases in E. coli (Chivers, 2007; Pruteanu et al., 2007; Pruteanu and Baker, 2009). Moreover, the metallochaperone CopZ from Enterococcus hirae and the Saccharomyces cerevisiae proteins Ctr1p (plasma membrane transporter for high-affinity copper uptake) and Mac1 (copper-sensing transcriptional activator) are degraded upon increased copper levels (Ooi et al., 1996; Zhu et al., 1998; Lu and Solioz, 2001; Solioz, 2002; Lu et al., 2003; Solioz and Stoyanov, 2003; Liu et al., 2007). Here, we report proteolysis of the metalloregulator CueR by Lon and the ClpP machineries in E. coli.
Materials and methods
Bacterial strains and growth conditions
E. coli strains used in this study are listed in Table 1. Cells were grown in liquid LB, 2YT, or M9 minimal medium in a water bath shaker (180 rpm) or on LB agar plates at 30 or 37°C. When required, antibiotics were used as follows: ampicillin (Amp) 100 μg/ml, chloramphenicol (Cm) 25 μg/ml, kanamycin (Kan) 50 μg/ml, or tetracycline (Tet) 10 μg/ml.
Table 1.
E. coli strains used in this study.
| E. coli strain | Relevant characteristics | Source |
|---|---|---|
| DH5α | supE44, ΔlacU169 (Y80lacZDM15), hsdR17, recA1, gyrA96, thi1, relA1 | Sambrook and Russell, 2001 |
| K12 | wild type | Bachmann, 1972 |
| MC4100 (RH166) | MC4100 Δara/Δleu, lac− | Becker and Hengge-Aronis, 2001 |
| Δlon | RH166, lon:Tn10 | Barembruch and Hengge, 2007 |
| ΔclpP | MC4100 ΔclpP::kan | Schmidt et al., 2009 |
| BW25113 | F−, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ−, rph-1, Δ(rhaD-rhaB)568, hsdR514 (CGSC # 7636) | Baba et al., 2006 |
| ΔclpA | BW25113; F−, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ−, ΔclpA783::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514 (JW0866-1; CGSC # 8898) | Baba et al., 2006 |
| ΔclpX | BW25113; F−, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), ΔclpX724::kan, λ−, rph-1, Δ(rhaD-rhaB)568, hsdR514 (JW0428-1; CGSC # 8591) | Baba et al., 2006 |
| ΔhslUV | MC4100, hslUV::kan | Barembruch and Hengge, 2007 |
| W3110 | F−, IN(rrnD-rrnE)1 | Tatsuta et al., 1998 |
| ΔftsH | W3110, zad220::Tn10 sfhC21ΔftsH3::kan | Tatsuta et al., 1998 |
| MG1655 | F−, λ−, rph-1 | Bachmann, 1972 |
| KY2981 | MG1655 Δ(clpPX-lon)1196::cat, ΔhslVU1172::tet, sulA2981 | Kanemori et al., 1999 |
| WOII260A | lacIq, lacZWJ16, ΔcueR, Φ(copA-lacZ) | Outten et al., 2000 |
| WOII248B | BW25113, ΔcueR | Outten et al., 2000 |
| BL21[DE3] | F−, ompT, gal (dcm) (lon), hsdSB (rB−mB−), λ[DE3] | Studier et al., 1990 |
| CH1019 | X90ssrA:cat[DE3] ΔyefM-yoeB::kan | R.T. Sauer |
Construction of plasmids
Plasmids and oligonucleotides used in this study are listed in Tables 2, 3, respectively. Recombinant DNA techniques were performed using standard protocols (Sambrook and Russell, 2001). E. coli DH5α cells served as cloning host. For construction of inducible CueR expression plasmids, genomic E. coli K12 DNA was used as template for PCR amplification of the cueR gene for full-length or C-terminally truncated CueR variants. The PCR product was cloned into pASK-IBA5(+) or pASK-IBA3 via primer-derived restriction sites to create pBO2584, pBO2585, pBO2860, or pBO2862, respectively. CueR variants with amino acid substitutions were generated by QuikChange® PCR using pBO2584 as template and mutagenized primers to create pBO2591, pBO2595, or pBO4800, respectively. For construction of pBO3687, a plasmid encoding constitutively expressed CueR, the cueR gene was amplified from genomic E. coli K12 DNA and cloned into pACYC184 via primer-derived restriction sites. This plasmid was used for QuikChange® PCR to create constitutively expressed CueR_C112S variant (pBO4801). All cloning results were confirmed by sequencing.
Table 2.
Plasmids used in this study.
| Plasmids | Relevant characteristics | Source |
|---|---|---|
| pASK-IBA5(+) | Ampr, P/Otet, tetR, encodes for N-terminal Strep-tag fusions | IBA GmbH |
| pASK-IBA3 | Ampr, P/Otet, tetR, encodes for C-terminal Strep-tag fusions | IBA GmbH |
| pACYC184 | Low copy number cloning vector, Cmr, Tetr | New England Biolabs |
| pBO2584 | pASK-IBA5(+) derivative encoding Strep_CueR (N-term. Strep-tag) | This study |
| pBO2585 | pASK-IBA3 derivative encoding CueR_Strep (C-term. Strep-tag) | This study |
| pBO2591 | pASK-IBA5(+) derivative encoding Strep_CueRR18A (N-term. Strep-tag) | This study |
| pBO2595 | pASK-IBA5(+) derivative encoding Strep_CueRA78C (N-term. Strep-tag) | This study |
| pBO2860 | pASK-IBA5(+) derivative encoding Strep_CueRΔC5 (N-term. Strep-tag) | This study |
| pBO2862 | pASK-IBA5(+) derivative encoding untagged CueR | This study |
| pBO3687 | pACYC184 derivative encoding for constitutive expression of CueR | This study |
| pBO4800 | pASK-IBA5(+) derivative encoding Strep_CueRC112S (N-term. Strep-tag) | This study |
| pBO4801 | pACYC184 derivative encoding for constitutive expression of CueR_C112S | This study |
| pBO1115 | pET19b derivative encoding His6_CspD | Langklotz and Narberhaus, 2011 |
| pET21b-Lon | Ampr; PT7; encodes for Lon with a C-terminal His6-tag fusion | R.T. Sauer |
Table 3.
Oligonucleotides used in this study.
| Name | Variant | Template | Sequence (5′-3′) | Plasmid |
|---|---|---|---|---|
| Strep-CueR-IBA.fw | cueR | gDNA | AAAAGAATTCAAACATCAGCGATGTAGCAAAAATTACC | pBO2584 |
| CueR.rv | TTTTAAGCTTTCACCCTGCCCGATGATGAC | |||
| Cuer-Strep.fw | cueR | gDNA | AAAAGAATTCAACATCAGCGATGTAGCAAAAATTACC | pBO2585 |
| CueR-Strep.rv | TTTTCCATGGGGCCCTGCCCGA | |||
| CueR_R18A.fw | cueR_R18A | pBO2584 | TGACCAGCAAAGCAATTGCCTTCTAT | pBO2591 |
| CueR_R18A.rv | CTTCTCTTCATAGAAGGCAATTGCTTTGC | |||
| CueR_A78C.fw | cueR_A78C | pBO2584 | GGCACAGCTGCGATGTCAAACG | pBO2595 |
| CueR_A78C.rv | GCCGTTTGACATCGCAGCTGTG | |||
| CueR.fw | cueR_ΔC5 | gDNA | AAAATGTACAAACATCAGCGATGTAGCAAAAATTACCG | pBO2860 |
| CueR_dC5.rv | TTTTAAGCTTTCAACAGCAGCCGGAGAGATTTTC | |||
| CueR-untagged_fw | cueR | gDNA | AAAAGCTAGCAACATCAGCGATGTAGCAAAAATTACC | pBO2862 |
| CueR.rv | TTTTAAGCTTTCACCCTGCCCGATGATGAC | |||
| CueR_ACYC.fw | cueR | gDNA | AAAAGATATCTAACAAAGCACAGGAGGCGTTGCG | pBO3687 |
| CueR_ACYC.rv | AAAAGGATCCTCACCCTGCCCGATGATGA | |||
| cueR_QC.fw | cueR_C112S | pBO2584 | GCTAGCCCTGGCGATGACAGCGCCGACAGC | pBO4800 |
| cueR_overlap_new.rv | CGCCAGGGCTAGCATTCGCCAGTGCCAGCAG | |||
| cueR_QC_fwd | cueR_C112S | pBO3687 | GCTAGCCCTGGCGATGACAGCGCCGACAGC | pBO4801 |
| cueR_overlap_new.rv | CGCCAGGGCTAGCATTCGCCAGTGCCAGCAG |
In vivo degradation experiments
To analyze the stability of different CueR variants, cells containing inducible expression plasmids encoding for corresponding CueR proteins were grown overnight in M9 minimal medium containing corresponding antibiotics for selection at 30°C. Fifteen milliliters of M9 minimal medium supplemented with corresponding antibiotics were inoculated with the overnight culture to an optical density (A580) of 0.05. Cells were grown to an A580 of 0.5 and protein expression was induced by adding 15 ng/ml anhydrotetracycline (AHT) for 20 min. Translation was blocked by addition of 200 μg/ml Cm. As an exception, translation of the strain lacking all three proteases (ΔclpXP, Δlon, ΔhslUV) and its parental strain E. coli Wt MG1655 was blocked by addition of 300 μg/ml spectinomycin (Sp) since the triple knockout strain is resistant to Cm. Samples were taken at different time points, frozen into liquid nitrogen and subjected to SDS-PAGE, Western transfer, and immunodetection as described below.
To analyze the stability of Strep_CueR under defined copper concentrations the same in vivo degradation experiments were performed as described above with minor modifications: To avoid copper contamination all steps were performed in plastic ware and all M9 minimal medium components except trace elements were previously incubated overnight with 50 g/l Chelex 100 resin (Bio-Rad) to remove trace metals. Before usage trace metals (without copper component) were added to the medium, mixed and sterile-filtered. Cells were grown to an A580 of 0.5, defined copper concentrations (CuSO4) were supplemented for 1 h and the in vivo degradation experiments were performed as described above.
For analyses of Strep_CueR stability over the entire growth curve cells were grown in LB medium + Amp at 37°C to different growth phases and in vivo degradation experiments were performed in every growth phase as described above. To analyze Strep_CueR stability over the whole growth curve under different copper concentrations, defined copper concentrations (CuSO4) were added to the main cultures at the time of inoculation or a copper pulse was given to the main culture after the second in vivo degradation experiment had been started (~2.5 h after inoculation and 60 min before the third degradation experiment was started).
Preparation of protein extracts and immunodetection
Cell pellets were resuspended in TE buffer depending on their optical density (10 mM Tris/HCl, pH 8; 1 mM EDTA; 50 μl TE buffer per A580 of 1.0) and mixed with protein sample buffer (final concentrations of 2% SDS (w/v), 0.1% (w/v) bromophenol blue, 10% (v/v) glycerol, 1% (v/v) β-mercaptoethanol, 50 mM Tris/HCl, pH 6.8). Samples were incubated for 5 min at 95°C, centrifuged (1 min, 16,000 × g) and subjected to SDS-PAGE and Western transfer using standard protocols (Sambrook and Russell, 2001). Strep-tagged fusion proteins were detected using a Strep-tag-HRP conjugate (IBA GmbH). Endogenous CueR and untagged CueR were detected using a polyclonal anti CueR antibody (Yamamoto and Ishihama, 2005) and a goat-anti-rabbit IgG (H+L) HRP conjugate (BioRad) as second antibody. Protein signals were visualized using Luminata Forte Western HRP substrate (Millipore) and the Chemi Imager Ready (Alpha Innotec). Half-lives of proteins were calculated by pixel counting with AlphaEaseFC software (version 4.0.0, Alpha Innotec).
Protein purification
Strep_CueR (pBO2584), His6_CspD (pBO1115), or Lon_His6 (pET21b-Lon) were transformed in E. coli Δlon, BL21 or CH1019, respectively. Cells were grown to an A580 of 0.5 at 37°C in LB (Strep_CueR) or 2YT (His6_CspD and Lon_His6) medium and gene expression was induced by addition of 150 ng/ml AHT (Strep_CueR) or 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside) (His6_CspD and Lon_His6). Cells were harvested after 3 h of overexpression at 30°C, resuspended in lysis buffer containing 20 mM Tris/HCl, pH 7.5, 200 mM NaCl, 1 mM DTT, 0.35 mg/ml lysozyme, 0.2 mg/ml DNase, and 0.2 mg/ml RNase and were disrupted via French Press. Strep- or His-tagged proteins were purified using streptactin sepharose (IBA GmbH) or Ni-NTA agarose (Qiagen), respectively. Purification of His-tagged proteins was performed as described previously (Langklotz and Narberhaus, 2011). Strep_CueR purification was performed using standard protocols of the purification kit (IBA GmbH). Protein concentrations were determined via Bradford assay (Bradford, 1976).
In vitro degradation experiments
Fifteen micromolars of Strep_CueR or His6_CspD and 600 nM Lon_His6 were incubated for 2 min at 37°C in the degradation buffer described in Bissonnette et al. (2010). In vitro degradation was initialized by addition of 20 mM ATP. Degradation experiments without addition of ATP were performed as controls. Results were visualized by SDS-PAGE and Coomassie staining or Western transfer following standard protocols (Sambrook and Russell, 2001).
In vivo CueR activity assays
Cultures with inducible expression plasmids encoding different CueR variants were grown in plastic ware in copper-free M9 minimal medium treated with 50 g/l Chelex 100 resin (Bio-Rad) to remove trace metals. Before use trace metals (without copper component) and 30 ng/ml AHT were added to the medium, mixed and sterile-filtered. Cells were grown to an A580 of 0.5 and defined copper concentrations (CuSO4) were adjusted in the cultures. After 1 h, 1 ml of the culture was harvested for β-galactosidase activity assay. The assay was performed as described previously (Miller, 1972).
Results and discussion
CueR is a target of ATP-dependent proteolysis in E. coli
Transcriptional regulators differentially control genes in order to adapt the proteome to the ambient conditions. Both, level and activity of transcription regulators can be tuned to the cellular need. For instance, the basal level of the copper efflux regulator CueR always present in the cell is elevated at increasing copper concentrations (Yamamoto and Ishihama, 2005). The activity of transcriptional regulators is often controlled by modification or oligomerization. In case of CueR, only the Cu+-bound dimer (holo-CueR) is capable of activating expression of the copper tolerance genes copA and cueO (Outten et al., 2000). Just as important as activation of transcriptional regulators is their inactivation since the cell would waste valuable resources for expression of pathways not needed under the given condition. Moreover, uncontrolled overexpression of membrane proteins like CopA might compromise membrane integrity. Since CueR covalently binds Cu+ with high affinity in the zeptomolar range, it is unlikely that the transcription factor is inactivated by simple dissociation of copper from its metal-binding pocket (Changela et al., 2003). We postulate that E. coli might shut down the copper-stress response by proteolysis of the metal-loaded transcription factor.
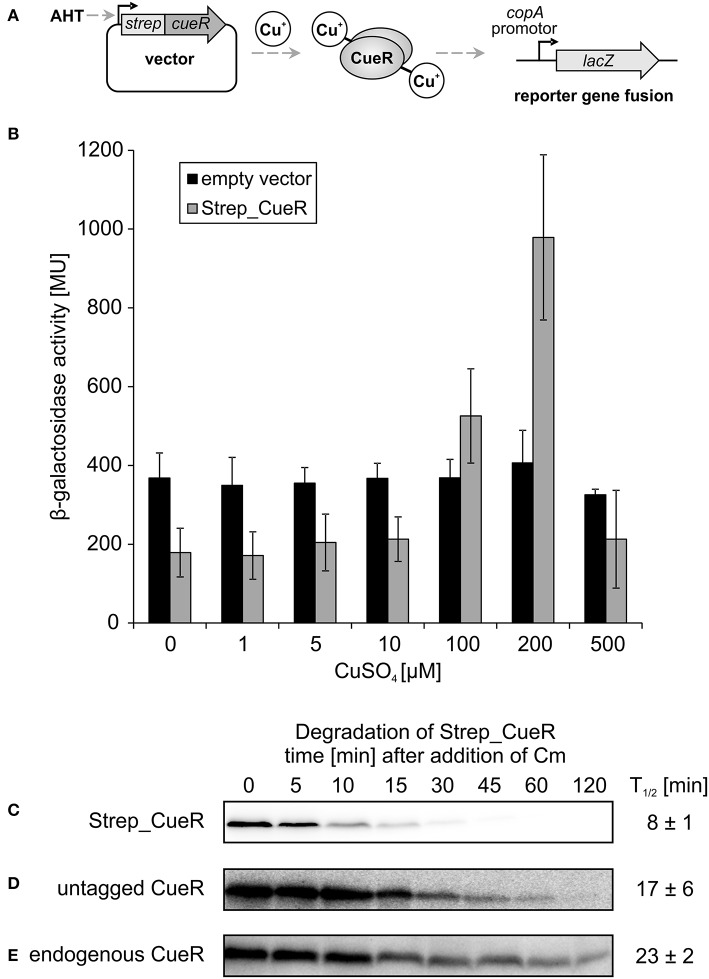
To be able to address whether CueR is a protease substrate in E. coli, we expressed it as N-terminally Strep-tagged variant (Strep_CueR) that facilitates immunodetection of the protein. First, we used an activity assay previously described by Outten et al. to ascertain that the tagged protein is functionally active as transcription factor. The original assay is based on a ΔcueR strain encoding the CueR-dependent copA promoter fused to lacZ on the chromosome, and a plasmid encoding constitutively expressed cueR (Outten et al., 2000). To establish the assay we constitutively expressed untagged CueR and an inactive CueR variant (CueR_C112S) not able to bind Cu+ ions (Chen et al., 2003; Stoyanov and Brown, 2003). As expected, β-galactosidase activity increased with increasing copper concentration in the presence of CueR (Figure S1). The CueR_C112S variant was unable to activate copA expression and produced copper-independent background activity like the empty vector control strain (Figure S1). The assay worked equally well with Strep_CueR produced from an AHT-inducible plasmid (Figure 1A). Copper-controlled copA expression showed that the N-terminal Strep-tag did not interfere with transcriptional activation (Figure 1B). The stability of Strep_CueR was analyzed in an E. coli wildtype strain (MC4100) during exponential growth in M9 minimal medium after translation was blocked by addition of chloramphenicol. The protein was rapidly degraded with a half-life of about 8 min (Figure 1C) indicating that Strep_CueR is a target of proteolysis in E. coli. As control we performed in vivo degradation experiments with Strep_CueR in a strain lacking cueR, which had no effect on stability (Figure S2). Furthermore, both plasmid-encoded untagged and endogenous CueR were prone to proteolysis, yet with higher half-lives compared to the Strep-tagged version (Figures 1D,E). A similar effect on the half-life of tagged proteins was observed for the related transcription factor ZntR (Pruteanu et al., 2007).
Figure 1.
Activity and stability of CueR in E. coli. Schematic presentation of the in vivo CueR activity assay (A). E. coli ΔcueR, Φ(copA-lacZ) cells were transformed with the empty vector pASK-IBA5(+) or the inducible plasmid encoding Strep_CueR and grown to exponential growth phase (M9 minimal medium; with the addition of 30 ng/ml AHT; 30°C). Cells were stressed with increasing CuSO4 concentrations for 1 h and β-galactosidase activity was measured in Miller Units (MU). Standard deviations were calculated from at least two independent experiments (B). Plasmid-encoded Strep_CueR was expressed for 20 min in exponential growth phase (M9 minimal medium; 30°C) in E. coli (MC4100). Translation was blocked by addition of Cm. Samples were taken at indicated time points, subjected to SDS-PAGE, Western transfer, and immunodetection. Half-lives (T1/2) and standard deviations were calculated from 10 independent experiments (C). In vivo degradation experiments with plasmid-encoded untagged CueR were performed as described above. Half-lives (T1/2) and standard deviations were calculated from five independent experiments (D). Stability of endogenous CueR was determined in E. coli MC4100 as described above. Half-lives (T1/2) and standard deviations were calculated from two independent experiments (E).
Strep_CueR is degraded by Lon, ClpXP and ClpAP
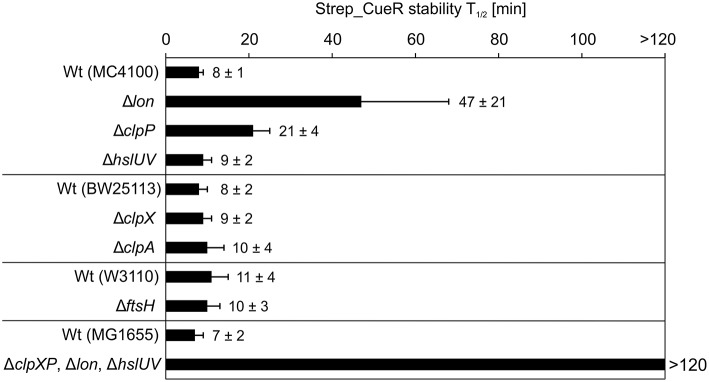
To identify the protease responsible for Strep_CueR degradation, we monitored the stability of the protein in various protease-deficient E. coli strains and their corresponding parental strains. In all parental strains and in strains lacking the membrane-anchored FtsH (ΔftsH) or the cytosolic HslUV (ΔhslUV) protease, the half-life of Strep_CueR was not altered. Hence, FtsH and HslUV are not involved in proteolysis of the transcription factor (Figure 2). In contrast, Strep_CueR was stabilized about sixfold in the Δlon strain. Endogenous CueR also was equally stabilized with a half-life around 2 h in the lon mutant (Figure S3). In a strain lacking the proteolytic ClpP subunit of the ClpXP and ClpAP complexes Strep_CueR was stabilized about two to threefold. On the contrary, in strains lacking only one of the ATPases of the ClpP-containing proteases (either ClpX or ClpA) Strep_CueR was degraded wild-type-like suggesting that both ATPase subunits contribute to CueR proteolysis. As expected, Strep_CueR was completely stable in a strain void of all cytosolic AAA+ proteases (Figure 2). Substrate sharing by different AAA+ proteases has been described previously and contributes to robust post-translational regulation. For instance, the MerR family member ZntR is degraded by Lon and ClpXP but not by ClpAP (Pruteanu et al., 2007). It seems that regulated proteolysis of MerR-like regulators is a commonly used mechanism to control metal homeostasis in E. coli.
Figure 2.
Strep_CueR is degraded by Lon, ClpXP, and ClpAP protease. Plasmid-encoded Strep_CueR was expressed for 20 min in exponential growth phase (M9 minimal medium; 30°C) in different protease-deficient E. coli strains and their corresponding wild-type (Wt) strains. Translation was blocked by addition of Cm or with spectinomycin for the strain lacking all three proteases (ΔclpXP, Δlon, ΔhslUV) and its parental strain (MG1655) since the triple knockout strain is resistant to Cm. Samples were taken at indicated time points, subjected to SDS-PAGE, Western transfer, and immunodetection. Half-lives (T1/2) and standard deviations were calculated from at least two or three independent experiments.
Activity of CueR does not influence its stability
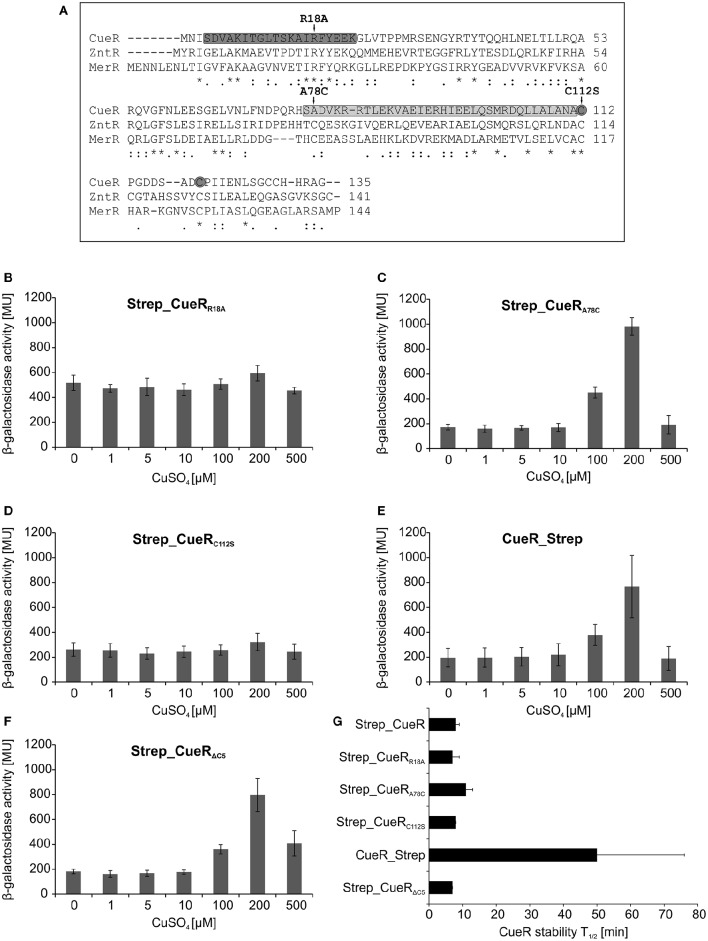
Next, we analyzed whether already known recognition strategies of Lon or ClpP-containing AAA+ proteases apply to CueR. The mechanisms how AAA+ proteases recognize their substrates are highly diverse (Hoskins et al., 2002; Sauer et al., 2004; Baker and Sauer, 2006; Sauer and Baker, 2011). Lon predominantly recognizes proteins with exposed aromatic and hydrophobic residues as it is often the case in unfolded or unassembled proteins (Chung and Goldberg, 1981; Gur and Sauer, 2008). Terminal degrons recognized by Lon have also been identified (Ishii et al., 2000; Ishii and Amano, 2001; Shah and Wolf, 2006). Among them is the SsrA-tag, which is C-terminally added via the tmRNA system to polypeptides stalled during translation. However, SsrA-tagged proteins are predominantly recognized by ClpXP (Keiler et al., 1996; Flynn et al., 2001), a protease that is also known to utilize N-terminal degrons (Flynn et al., 2003). ClpAP recognizes several substrates via the so-called N-end rule pathway, in which the first N-terminal amino acid is critical for degradation (Erbse et al., 2006; Mogk et al., 2007; Dougan et al., 2010; Román-Hernández et al., 2011). Comparison of residues in the N or C terminus of CueR with known degrons of Lon, ClpXP and ClpAP did not reveal similarities to other protease substrates. The same was reported for the zinc-dependent transcriptional regulator ZntR, degraded by Lon and ClpXP in E. coli (Pruteanu et al., 2007) suggesting that yet unknown mechanisms of recognition may apply to these MerR-like proteins. For ZntR it was shown that mutation of the conserved arginine in the helix-turn-helix motif of the DNA-binding region results in faster degradation of the protein (Pruteanu et al., 2007). Therefore, we constructed a corresponding Strep_CueRR18A variant (Figure 3A), which as expected (Philips et al., 2015) failed to induce copA-lacZ transcription since DNA binding is impaired (Figure 3B). Yet, degradation of the inactive CueR variant was not affected (Figure 3G and Figure S4).
Figure 3.
Activity and stability of various CueR variants in E. coli. Comparison of the amino acid sequence of CueR, ZntR, and MerR. DNA-binding domain and dimerization domain of CueR are marked in dark gray and light gray, respectively. Amino acids, which were substituted in different variants used in this study, are highlighted with arrows and the two copper-binding cysteines of CueR are indicated with gray circles. (* = identical amino acid; : = conserved substitution;. = semi conserved substitution; − = lacking amino acid). Alignment was performed by using the align tool of the uniprot database (http://www.uniprot.org/) (A). E. coli ΔcueR, Φ(copA-lacZ) cells harboring inducible plasmids encoding Strep_CueRR18A (B), Strep_CueRA78C (C), Strep_CueRC112S (D), CueR_Strep (E), or Strep_CueRΔC5 (F) were grown in M9 minimal medium with 30 ng/ml AHT at 30°C to log phase. Cells were then treated with increasing CuSO4 concentrations for 1 h. β-galactosidase activity and standard deviations were calculated from at least two independent experiments (B–F). Plasmid-encoded CueR variants were expressed for 20 min in exponential growth phase (M9 minimal medium; 30°C). Translation was blocked by addition of Cm. Samples were taken at indicated time points, subjected to SDS-PAGE, Western transfer, and immunodetection. Half-lives (T1/2) and standard deviations were calculated from at least three independent experiments. For comparison half-life of Strep_CueR is presented (G).
Two additional variants of N-terminally Strep-tagged CueR with amino acid substitutions in functionally relevant regions of the protein were analyzed (Figure 3A). Strep_CueRA78C is a variant with a substitution at the very beginning of the dimerization helix that differs from ZntR and MerR, which have a conserved cysteine at this position. Strep_CueRC112S carries a substitution of a copper-binding cysteine in the metal-binding domain. Strep_CueRA78C is able to activate copA expression in a copper-responsive manner (Figure 3C), while substitution of one of the two copper-binding cysteines (Strep_CueRC112S) inactivated CueR (Figure 3D). Regardless of whether they were active as transcription factor or not, both point-mutated variants were degraded like Strep_CueR (Figure 3G and Figure S4). Therefore, like for ZntR (Pruteanu et al., 2007), mutations in the dimerization and metal binding regions do not influence proteolysis.
Since some degrons are exposed at the termini of a substrate, we placed a Strep-tag at the C terminus to see whether it affects protein stability. Terminal tags have previously been shown to block proteolysis, for example of the Lon substrate SoxS (Griffith et al., 2004). Although activity of CueR_Strep was unaffected (Figure 3E), the protein was stabilized about six-fold (Figure 3G and Figure S4), suggesting a contribution of the C-terminal end to protease targeting. We also constructed a C-terminally truncated, active version of Strep_CueR lacking the last five C-terminal residues (Figure 3F). Strep_CueRΔC5 was degraded like Strep_CueR (Figure 3G and Figure S4) excluding that the last five amino acids of the C terminus are critical for recognition.
Sometimes the recognition process is aided by adaptor proteins (Battesti and Gottesman, 2013). Given that Strep_CueR is primarily degraded by the Lon protease in vivo (Figure 2), we analyzed if it is degraded by Lon in a reconstituted in vitro system. For this purpose, Strep_CueR and Lon_His6 were purified and subjected to in vitro degradation experiments. The replication inhibitor His6_CspD served as a control protein as it is known to be a direct substrate of Lon in vitro (Langklotz and Narberhaus, 2011) (Figure 4A). In contrast to His6_CspD, Strep_CueR remained stable when incubated without (Figure S5) or with Lon_His6, both in the absence and presence of ATP (Figure 4B). This is in contrast to ZntR, which is degraded by Lon but not by ClpXP in vitro (Pruteanu et al., 2007). On the one hand it is possible that Lon needs to be allosterically activated for CueR degradation as it was shown for the replication initiator DnaA from Caulobacter crescentus. DnaA is degraded in vivo but remains stable in in vitro degradation experiments. When Lon is allosterically activated by the addition of unfolded proteins, DnaA is degraded in vitro (Jonas et al., 2013; Joshi and Chien, 2016). On the other hand a factor mediating Strep_CueR degradation might be missing in the purified system. Putative non-proteinaceous regulatory molecules might be guanosine pentaphosphate/tetraphosphate ((p)ppGpp) and inorganic poly phosphates (polyP), which are known to influence proteolysis of several AAA+ protease substrates (Kuroda et al., 2001, 2006; Kuroda, 2006; Schäkermann et al., 2013; Bittner et al., 2015). However, we can exclude an involvement of (p)ppGpp and polyP in CueR proteolysis since the protein was wild-type-like degraded in strains lacking these regulatory molecules (data not shown).
Figure 4.
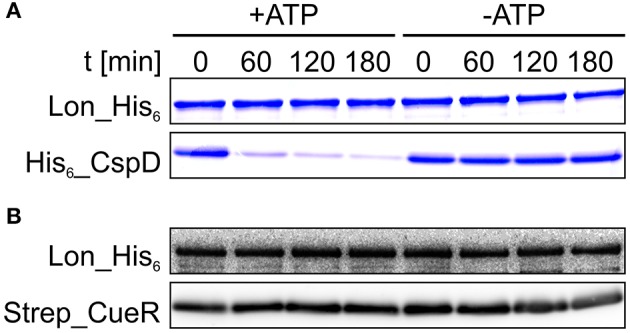
Strep_CueR is not degraded by Lon in vitro. Lon_His6, His6_CspD, and Strep_CueR were purified and used for in vitro degradation experiments (A,B). Degradation experiments were initialized by addition of 20 mM ATP (+ATP). An approach without ATP addition (-ATP) served as control. Samples were taken at indicated time points, subjected to SDS-PAGE and Coomassie staining for His6_CspD (A) or were subjected to Western transfer, and immunodetection for Strep_CueR (B). Data are representative of five independent experiments.
A putative adaptor protein lacking in the in vitro system might sense the cellular copper status. This is reminiscent of the adaptor protein YjbH that is able to coordinate zinc ions and is involved in ClpXP-dependent degradation of the transcriptional regulator Spx in Bacillus subtilis and Staphylococcus aureus (Garg et al., 2009; Engman et al., 2012). To date little is known about adaptors involved in Lon-dependent degradation. Recently, degradation of the master regulator of flagellar biosynthesis SwrA in B. subtilis was reported to be assisted by the swarming motility inhibitor A (SmiA), in vivo and in vitro. Hence, SmiA is the first described adaptor protein for Lon-dependent proteolysis (Mukherjee et al., 2015). Further, studies targeted at identifying the CueR degron and potential adaptor proteins might reveal similarities and differences in the recognition logics of ZntR and CueR.
Is proteolysis of CueR regulated?
As proteolysis of some metalloregulators, like ZntR, Ctr1p, or Mac1 is metal-dependent (Ooi et al., 1996; Zhu et al., 1998; Liu et al., 2007; Pruteanu et al., 2007), we analyzed the effect of defined copper concentrations on the stability of Strep_CueR. E. coli cells harboring an AHT-inducible plasmid encoding Strep_CueR were grown to exponential growth phase under copper-limited conditions. Cells were then supplemented with various copper concentrations for 1 h followed by in vivo degradation experiments. The half-life of Strep_CueR remained similar at CuSO4 concentrations between 0 and 200 μM (Figure 5) indicating that the cellular copper level has little effect on CueR stability.
Figure 5.
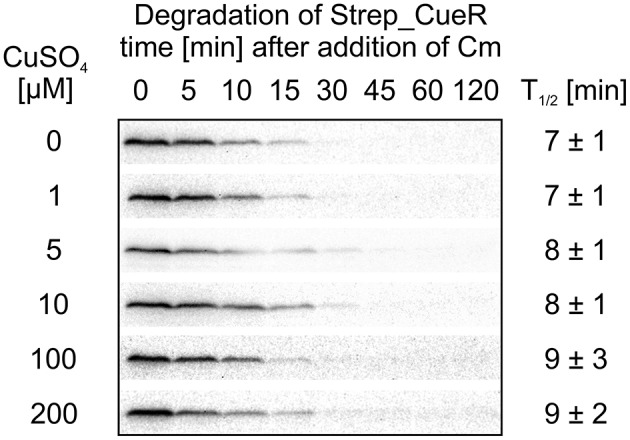
Stability of Strep_CueR in response to increasing CuSO4 concentrations. E. coli MC4100 (Wt) cells harboring a plasmid encoding for Strep_CueR were grown to exponential phase (M9 minimal medium at 30°C). Cultures were supplemented with varying CuSO4 concentrations for 1 h followed by in vivo degradation experiments. Translation was blocked by addition of Cm. Samples were taken at indicated time points, subjected to SDS-PAGE, Western transfer, and immunodetection. Half-lives (T1/2) and standard deviations were calculated from at least two independent experiments.
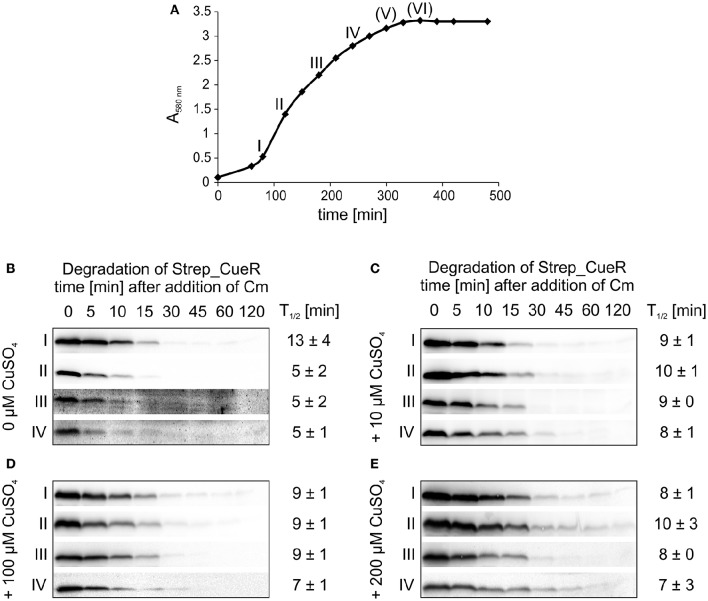
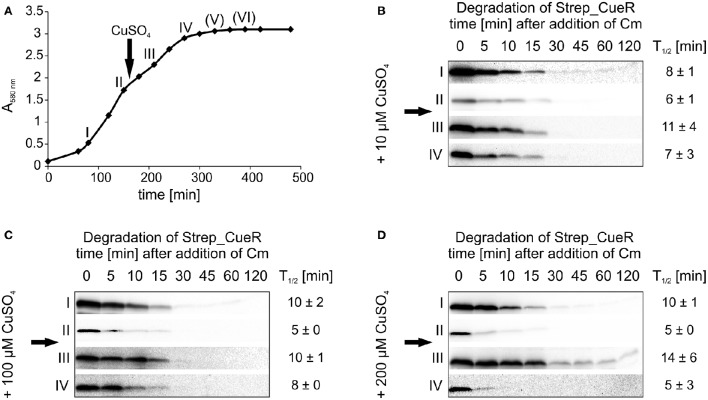
It recently turned out that degradation for several protease substrates is growth phase-dependent (Langklotz and Narberhaus, 2011; Westphal et al., 2012; Bittner et al., 2015). Therefore, we examined whether CueR stability depends on the growth status of E. coli and performed in vivo degradation experiments with Strep_CueR in different growth phases. All experiments described above were performed in M9 minimal medium at 30°C. To allow optimal growth, LB medium, and a temperature of 37°C were chosen for this experiment (Figure 6A). When no additional copper was added to the culture, degradation was accelerated about twofold from early exponential to exponential growth phase but remained the same in late exponential growth phase (Figure 6B). Strep_CueR was not detectable in later growth phases. Addition of external copper to the growth medium right from the beginning of the experiment led to constant half-lives in the range between 8 and 10 min (Figures 6C–E).
Figure 6.
Degradation of Strep_CueR in different growth phases under varying CuSO4 concentrations. Stability of Strep_CueR was determined in LB medium at 37°C in different growth phases (I–VI) (A) in E. coli MC4100 (Wt) under varying CuSO4 concentrations (B-E). Defined CuSO4 concentrations (0-200 μM) were added right from inoculation of the main culture. In vivo degradation experiments were performed after 20 min of Strep_CueR induction in every growth phase. Translation was blocked by addition of Cm. Samples were taken at indicated time points, subjected to SDS-PAGE, Western transfer, and immunodetection. Half-lives (T1/2) and standard deviations were calculated from at least two independent experiments. Strep_CueR was not detectable in in vivo degradation experiments (V) and (VI).
To address whether sudden copper stress affects CueR degradation, we carried out in vivo degradation experiments over the entire growth curve with a copper pulse ~2.5 h after inoculation (Figure 7A). As shown above (Figure 6B), Strep_CueR showed a slightly accelerated degradation upon entry into exponential growth prior to copper treatment (Figures 7B–D; time points I and II). Immediately after a copper pulse of 10, 100, or 200 μM CuSO4, the stability of Strep_CueR increased about twofold before it returned to pre-shock values (Figures 7B–D, time points III and IV) indicating that the cells sensed and slightly reacted to altered copper concentrations. Again, the transcription factor was not detectable in late growth phases. Accelerated degradation of CueR in copper-starved fast-growing cells and transient stabilization of the protein after copper shock are consistent with the physiological demand for this copper export regulator. This is in good agreement with (i) ZntR, which is also only stabilized about two-fold after the addition of zinc (Pruteanu et al., 2007) and (ii) the estimation that newly synthesized CopA proteins reach sufficient efflux power about 2 min after addition of copper to clear excess copper from the cytosol (Tottey et al., 2007).
Figure 7.
Stability of Strep_CueR in different growth phases before and after a CuSO4 pulse. Stability of Strep_CueR was determined in different growth phases in E. coli MC4100 (Wt) before and after a copper pulse (arrow) with different CuSO4 concentrations added to the main culture. Cells were grown to different growth phases (I-VI) in LB medium at 37°C (A). The first two in vivo degradation experiments were performed without CuSO4 treatment (I and II) (B–D). After 20 min of Strep_CueR induction in every growth phase, translation was blocked by addition of Cm. Samples were taken at indicated time points, subjected to SDS-PAGE, Western transfer, and immunodetection. Approx. 2.5 h after inoculation a CuSO4 pulse (10–200 μM CuSO4) was given to the main cultures and in vivo degradation experiments in further growth phases (III–VI) followed like described before. Half-lives (T1/2) and standard deviations were calculated from at least two independent experiments. Strep_CueR was not detectable in in vivo degradation experiments (V) and (VI).
Overall, it seems that E. coli continuously degrades CueR with minor adjustments to the external copper status. We propose that this is due to the fast clearance of excess copper after CueR activation of the Cue system, which might not require a long-term stabilization of CueR. Secondly, proteolysis might preferentially erase the overrepresented copper-loaded form of CueR, thereby contributing to the maintenance of a copper-free CueR pool derived from new synthesis to allow measuring the acute copper level in the cell. Apo-CueR may exchange holo-CueR dimers bound to the corresponding promoters via the recently postulated mechanisms of direct substitution or assisted dissociation (Joshi et al., 2012; Chen et al., 2013). Both pathways are based on the formation of a very short-lived transition state (determined as Protein2-DNA ternary complex), in which two CueR dimers (e.g., apo- and holo-CueR) bind to the extended spacer sequence of the -35 and -10 regions of copA or cueO with one of their DNA-binding domains. Given the instability of this state, one CueR protein, e.g., holo-CueR, loses its grip on the dyad giving the other CueR dimer, apo-CueR, the chance to fully bind to the dyad with both of its DNA-binding domains (direct substitution) or both dimers fall off the DNA (assisted dissociation) (Joshi et al., 2012; Chen et al., 2013, 2015). The constitutive proteolysis of CueR described in this study might contribute to an accurate adjustment of the CueR pool always prepared to react to the current cellular copper level to efficiently maintain copper homeostasis.
Author contributions
LB, SS, AK, and FN designed the study. LB and AK performed the experiments. LB and FN wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Julia Horstmann is acknowledged for plasmid construction and initial steps in this project. The authors gratefully thank Regine Hengge, Eliora Ron, Axel Mogk, Eberhard Klauck, and Thomas V. O'Halloran for kind gift of E. coli strains, Akira Ishihama for the CueR antibody, and Robert T. Sauer for providing the Lon expression system. Jan Arends, Johanna Roßmanith, and Linna Danne are acknowledged for critical reading of the manuscript. We gratefully acknowledge financial support by a grant from the German Research Foundation (DFG; SFB642, GTP-, and ATP-dependent membrane processes) to FN.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmolb.2017.00009/full#supplementary-material
References
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. (1972). Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 36, 525–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T. A., Sauer R. T. (2006). ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem. Sci. 31, 647–653. 10.1016/j.tibs.2006.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barembruch C., Hengge R. (2007). Cellular levels and activity of the flagellar sigma factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis. Mol. Microbiol. 65, 76–89. 10.1111/j.1365-2958.2007.05770.x [DOI] [PubMed] [Google Scholar]
- Battesti A., Gottesman S. (2013). Roles of adaptor proteins in regulation of bacterial proteolysis. Curr. Opin. Microbiol. 16, 140–147. 10.1016/j.mib.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker G., Hengge-Aronis R. (2001). What makes an Escherichia coli promoter σS dependent? role of the -13/-14 nucleotide promoter positions and region 2.5 of σS. Mol. Microbiol. 39, 1153–1165. 10.1111/j.1365-2958.2001.02313.x [DOI] [PubMed] [Google Scholar]
- Bissonnette S. A., Rivera-Rivera I., Sauer R. T., Baker T. A. (2010). The IbpA and IbpB small heat-shock proteins are substrates of the AAA+ Lon protease. Mol. Microbiol. 75, 1539–1549. 10.1111/j.1365-2958.2010.07070.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner L. M., Arends J., Narberhaus F. (2016). Mini review: ATP-dependent proteases in bacteria. Biopolymers 105, 505-517. 10.1002/bip.22831 [DOI] [PubMed] [Google Scholar]
- Bittner L. M., Westphal K., Narberhaus F. (2015). Conditional proteolysis of the membrane protein YfgM by the FtsH protease depends on a novel N-terminal degron. J. Biol. Chem. 290, 19367–19378. 10.1074/jbc.m115.648550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Brocklehurst K. R., Hobman J. L., Lawley B., Blank L., Marshall S. J., Brown N. L., et al. (1999). ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol. Microbiol. 31, 893–902. 10.1046/j.1365-2958.1999.01229.x [DOI] [PubMed] [Google Scholar]
- Brown N. L., Stoyanov J. V., Kidd S. P., Hobman J. L. (2003). The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27, 145–163. 10.1016/S0168-6445(03)00051-2 [DOI] [PubMed] [Google Scholar]
- Changela A., Chen K., Xue Y., Holschen J., Outten C. E., O'Halloran T. V., et al. (2003). Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301, 1383–1387. 10.1126/science.1085950 [DOI] [PubMed] [Google Scholar]
- Chen K., Yuldasheva S., Penner-Hahn J. E., O'Halloran T. V. (2003). An atypical linear Cu(I)-S2 center constitutes the high-affinity metal-sensing site in the CueR metalloregulatory protein. J. Am. Chem. Soc. 125, 12088–12089. 10.1021/ja036070y [DOI] [PubMed] [Google Scholar]
- Chen P., Keller A. M., Joshi C. P., Martell D. J., Andoy N. M., Benítez J. J., et al. (2013). Single-molecule dynamics and mechanisms of metalloregulators and metallochaperones. Biochemistry. 52, 7170–7183. 10.1021/bi400597v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. Y., Santiago A. G., Jung W., Krzeminski L., Yang F., Martell D. J., et al. (2015). Concentration- and chromosome-organization-dependent regulator unbinding from DNA for transcription regulation in living cells. Nat. Commun. 6, 7445. 10.1038/ncomms8445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivers P. T. (2007). A galvanizing story-protein stability and zinc homeostasis. J. Bacteriol. 189, 2953–2954. 10.1128/jb.00173-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C. H., Goldberg A. L. (1981). The product of the lon (capR) gene in Escherichia coli is the ATP-dependent protease, protease La. Proc. Natl. Acad. Sci. U.S.A. 78, 4931–4935. 10.1073/pnas.78.8.4931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan D. A., Truscott K. N., Zeth K. (2010). The bacterial N-end rule pathway: expect the unexpected. Mol. Microbiol. 76, 545–558. 10.1111/j.1365-2958.2010.07120.x [DOI] [PubMed] [Google Scholar]
- Engman J., Rogstam A., Frees D., Ingmer H., von Wachenfeldt C. (2012). The YjbH adaptor protein enhances proteolysis of the transcriptional regulator Spx in Staphylococcus aureus. J. Bacteriol. 194, 1186–1194. 10.1128/jb.06414-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbse A., Schmidt R., Bornemann T., Schneider-Mergener J., Mogk A., Zahn R., et al. (2006). ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature 439, 753–756. 10.1038/nature04412 [DOI] [PubMed] [Google Scholar]
- Flynn J. M., Levchenko I., Seidel M., Wickner S. H., Sauer R. T., Baker T. A. (2001). Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. U.S.A. 98, 10584–10589. 10.1073/pnas.191375298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J. M., Neher S. B., Kim Y. I., Sauer R. T., Baker T. A. (2003). Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11, 671–683. 10.1016/S1097-2765(03)00060-1 [DOI] [PubMed] [Google Scholar]
- Garg S. K., Kommineni S., Henslee L., Zhang Y., Zuber P. (2009). The YjbH protein of Bacillus subtilis enhances ClpXP-catalyzed proteolysis of Spx. J. Bacteriol. 191, 1268–1277. 10.1128/jb.01289-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass G., Rensing C. (2001). CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem. Biophys. Res. Commun. 286, 902–908. 10.1006/bbrc.2001.5474 [DOI] [PubMed] [Google Scholar]
- Grass G., Rensing C., Solioz M. (2011). Metallic copper as an antimicrobial surface. Appl. Environ. Microbiol. 77, 1541–1547. 10.1128/aem.02766-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith K. L., Shah I. M., Wolf R. E., Jr. (2004). Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Mol. Microbiol. 51, 1801–1816. 10.1046/j.1365-2958.2003.03952.x [DOI] [PubMed] [Google Scholar]
- Gur E., Biran D., Ron E. Z. (2011). Regulated proteolysis in Gram-negative bacteria-how and when? Nat. Rev. Microbiol. 9, 839–848. 10.1038/nrmicro2669 [DOI] [PubMed] [Google Scholar]
- Gur E., Ottofueling R., Dougan D. A. (2013). Machines of destruction - AAA+ proteases and the adaptors that control them. Subcell. Biochem. 66, 3–33. 10.1007/978-94-007-5940-4_1 [DOI] [PubMed] [Google Scholar]
- Gur E., Sauer R. T. (2008). Recognition of misfolded proteins by Lon, a AAA+ protease. Genes Dev. 22, 2267–2277. 10.1101/gad.1670908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins J. R., Yanagihara K., Mizuuchi K., Wickner S. (2002). ClpAP and ClpXP degrade proteins with tags located in the interior of the primary sequence. Proc. Natl. Acad. Sci. U.S.A. 99, 11037–11042. 10.1073/pnas.172378899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y., Amano F. (2001). Regulation of SulA cleavage by Lon protease by the C-terminal amino acid of SulA, histidine. Biochem. J. 358, 473–480. 10.1042/bj3580473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y., Sonezaki S., Iwasaki Y., Miyata Y., Akita K., Kato Y., et al. (2000). Regulatory role of C-terminal residues of SulA in its degradation by Lon protease in Escherichia coli. J. Biochem. 127, 837–844. 10.1093/oxfordjournals.jbchem.a022677 [DOI] [PubMed] [Google Scholar]
- Jonas K., Liu J., Chien P., Laub M. T. (2013). Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell 154, 623–636. 10.1016/j.cell.2013.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi C. P., Panda D., Martell D. J., Andoy N. M., Chen T. Y., Gaballa A., et al. (2012). Direct substitution and assisted dissociation pathways for turning off transcription by a MerR-family metalloregulator. Proc. Natl. Acad. Sci. U.S.A. 109, 15121–15126. 10.1073/pnas.1208508109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi K. K., Chien P. (2016). Regulated Proteolysis in Bacteria: Caulobacter. Annu. Rev. Genet. 50, 423–445. 10.1146/annurev-genet-120215-035235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemori M., Yanagi H., Yura T. (1999). The ATP-dependent HslVU/CplQY protease participates in turnover of cell division inhibitor SulA in Escherichia coli. J. Bacteriol. 181, 3674–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler K. C., Waller P. R., Sauer R. T. (1996). Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–993. 10.1126/science.271.5251.990 [DOI] [PubMed] [Google Scholar]
- Kuroda A. (2006). A polyphosphate-Lon protease complex in the adaptation of Escherichia coli to amino acid starvation. Biosci. Biotechnol. Biochem. 70, 325–331. 10.1271/bbb.70.325 [DOI] [PubMed] [Google Scholar]
- Kuroda A., Nomura K., Ohtomo R., Kato J., Ikeda T., Takiguchi N., et al. (2001). Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293, 705–708. 10.1126/science.1061315 [DOI] [PubMed] [Google Scholar]
- Kuroda A., Nomura K., Takiguchi N., Kato J., Ohtake H. (2006). Inorganic polyphosphate stimulates Lon-mediated proteolysis of nucleoid proteins in Escherichia coli. Cell. Mol. Biol. 52, 23–29. [PubMed] [Google Scholar]
- Langklotz S., Narberhaus F. (2011). The Escherichia coli replication inhibitor CspD is subject to growth-regulated degradation by the Lon protease. Mol. Microbiol. 80, 1313–1325. 10.1111/j.1365-2958.2011.07646.x [DOI] [PubMed] [Google Scholar]
- Liu J., Sitaram A., Burd C. G. (2007). Regulation of copper-dependent endocytosis and vacuolar degradation of the yeast copper transporter, Ctr1p, by the Rsp5 ubiquitin ligase. Traffic 8, 1375–1384. 10.1111/j.1600-0854.2007.00616.x [DOI] [PubMed] [Google Scholar]
- Lu Z. H., Dameron C. T., Solioz M. (2003). The Enterococcus hirae paradigm of copper homeostasis: copper chaperone turnover, interactions, and transactions. Biometals 16, 137–143. 10.1023/A:1020709307589 [DOI] [PubMed] [Google Scholar]
- Lu Z. H., Solioz M. (2001). Copper-induced proteolysis of the CopZ copper chaperone of Enterococcus hirae. J. Biol. Chem. 276, 47822–47827. 10.1074/jbc.M106218200 [DOI] [PubMed] [Google Scholar]
- Miller J. H. (1972). Experiments in Molecular Genetics. Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Mogk A., Schmidt R., Bukau B. (2007). The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol. 17, 165–172. 10.1016/j.tcb.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Bree A. C., Liu J., Patrick J. E., Chien P., Kearns D. B. (2015). Adaptor-mediated Lon proteolysis restricts Bacillus subtilis hyperflagellation. Proc. Natl. Acad. Sci. U.S A. 112, 250–255. 10.1073/pnas.1417419112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi C. E., Rabinovich E., Dancis A., Bonifacino J. S., Klausner R. D. (1996). Copper-dependent degradation of the Saccharomyces cerevisiae plasma membrane copper transporter Ctr1p in the apparent absence of endocytosis. EMBO J. 15, 3515–3523. [PMC free article] [PubMed] [Google Scholar]
- Outten C. E., Outten F. W., O'Halloran T. V. (1999). DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli. J. Biol. Chem. 274, 37517–37524. 10.1074/jbc.274.53.37517 [DOI] [PubMed] [Google Scholar]
- Outten F. W., Huffman D. L., Hale J. A., O'Halloran T. V. (2001). The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276, 30670–30677. 10.1074/jbc.m104122200 [DOI] [PubMed] [Google Scholar]
- Outten F. W., Outten C. E., Hale J., O'Halloran T. V. (2000). Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, cueR. J. Biol. Chem. 275, 31024–31029. 10.1074/jbc.M006508200 [DOI] [PubMed] [Google Scholar]
- Petersen C., Møller L. B. (2000). Control of copper homeostasis in Escherichia coli by a P-type ATPase, CopA, and a MerR-like transcriptional activator, CopR. Gene 261, 289–298. 10.1016/S0378-1119(00)00509-6 [DOI] [PubMed] [Google Scholar]
- Philips S. J., Canalizo-Hernandez M., Yildirim I., Schatz G. C., Mondragón A., O'Halloran T. V. (2015). Allosteric transcriptional regulation via changes in the overall topology of the core promoter. Science 349, 877–881. 10.1126/science.aaa9809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruteanu M., Baker T. A. (2009). Proteolysis in the SOS response and metal homeostasis in Escherichia coli. Res. Microbiol. 160, 677–683. 10.1016/j.resmic.2009.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruteanu M., Neher S. B., Baker T. A. (2007). Ligand-controlled proteolysis of the Escherichia coli transcriptional regulator ZntR. J. Bacteriol. 189, 3017–3025. 10.1128/jb.01531-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher C., Masepohl B. (2012). Copper-responsive gene regulation in bacteria. Microbiology 158, 2451–2464. 10.1099/mic.0.058487-0 [DOI] [PubMed] [Google Scholar]
- Rensing C., Fan B., Sharma R., Mitra B., Rosen B. P. (2000). CopA: An Escherichia coli Cu(I)-translocating P-type ATPase. Proc. Natl. Acad. Sci. U.S.A. 97, 652–656. 10.1073/pnas.97.2.652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing C., Grass G. (2003). Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27, 197–213. 10.1016/S0168-6445(03)00049-4 [DOI] [PubMed] [Google Scholar]
- Román-Hernández G., Hou J. Y., Grant R. A., Sauer R. T., Baker T. A. (2011). The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease. Mol. Cell 43, 217–228. 10.1016/j.molcel.2011.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn. Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sauer R. T., Baker T. A. (2011). AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80, 587–612. 10.1146/annurev-biochem-060408-172623 [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Bolon D. N., Burton B. M., Burton R. E., Flynn J. M., Grant R. A., et al. (2004). Sculpting the proteome with AAA+ proteases and disassembly machines. Cell 119, 9–18. 10.1016/j.cell.2004.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäkermann M., Langklotz S., Narberhaus F. (2013). FtsH-mediated coordination of lipopolysaccharide biosynthesis in Escherichia coli correlates with the growth rate and the alarmone (p)ppGpp. J. Bacteriol. 195, 1912–1919. 10.1128/jb.02134-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R., Zahn R., Bukau B., Mogk A. (2009). ClpS is the recognition component for Escherichia coli substrates of the N-end rule degradation pathway. Mol. Microbiol. 72, 506–517. 10.1111/j.1365-2958.2009.06666.x [DOI] [PubMed] [Google Scholar]
- Shah I. M., Wolf R. E., Jr. (2006). Sequence requirements for Lon-dependent degradation of the Escherichia coli transcription activator SoxS: identification of the SoxS residues critical to proteolysis and specific inhibition of in vitro degradation by a peptide comprised of the N-terminal 21 amino acid residues. J. Mol. Biol. 357, 718–731. 10.1016/j.jmb.2005.12.088 [DOI] [PubMed] [Google Scholar]
- Solioz M. (2002). Role of proteolysis in copper homoeostasis. Biochem. Soc. Trans. 30, 688–691. 10.1042/bst0300688 [DOI] [PubMed] [Google Scholar]
- Solioz M., Stoyanov J. V. (2003). Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 27, 183–195. 10.1016/S0168-6445(03)00053-6 [DOI] [PubMed] [Google Scholar]
- Stoyanov J. V., Brown N. L. (2003). The Escherichia coli copper-responsive copA promoter is activated by gold. J. Biol. Chem. 278, 1407–1410. 10.1074/jbc.C200580200 [DOI] [PubMed] [Google Scholar]
- Stoyanov J. V., Hobman J. L., Brown N. L. (2001). CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol. Microbiol. 39, 502–511. 10.1046/j.1365-2958.2001.02264.x [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. (1990). Use of T7 RNA-polymerase to direct expression of cloned genes. Methods Enzymol. 185, 60–89. 10.1016/0076-6879(90)85008-C [DOI] [PubMed] [Google Scholar]
- Tatsuta T., Tomoyasu T., Bukau B., Kitagawa M., Mori H., Karata K., et al. (1998). Heat shock regulation in the ftsH null mutant of Escherichia coli: dissection of stability and activity control mechanisms of σ32 in vivo. Mol. Microbiol. 30, 583–593. 10.1046/j.1365-2958.1998.01091.x [DOI] [PubMed] [Google Scholar]
- Tottey S., Harvie D. R., Robinson N. J. (2007). Understanding how cells allocate metals. Berlin; Heidelberg: Springer-Verlag. [DOI] [PubMed] [Google Scholar]
- van der Oost J., de Boer A. P., de Gier J. W., Zumft W. G., Stouthamer A. H., van Spanning R. J. (1994). The heme-copper oxidase family consists of three distinct types of terminal oxidases and is related to nitric oxide reductase. FEMS Microbiol. Lett. 121, 1–9. 10.1111/j.1574-6968.1994.tb07067.x [DOI] [PubMed] [Google Scholar]
- Westphal K., Langklotz S., Thomanek N., Narberhaus F. (2012). A trapping approach reveals novel substrates and physiological functions of the essential protease FtsH in Escherichia coli. J. Biol. Chem. 287, 42962–42971. 10.1074/jbc.m112.388470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Ishihama A. (2005). Transcriptional response of Escherichia coli to external copper. Mol. Microbiol. 56, 215–227. 10.1111/j.1365-2958.2005.04532.x [DOI] [PubMed] [Google Scholar]
- Zhu Z., Labbé S., Peña M. M., Thiele D. J. (1998). Copper differentially regulates the activity and degradation of yeast Mac1 transcription factor. J. Biol. Chem. 273, 1277–1280. 10.1074/jbc.273.3.1277 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.