Abstract
The phytohormone abscisic acid (ABA) is essential in plant responding to biotic and abiotic stresses. Although ABA signaling model is well established in Arabidopsis, ABA receptor PYL family and clade A PP2C subfamily are not yet characterized in monocot model plant Brachypodium distachyon. In this study, we identified 12 PYLs and 8 clade A PP2Cs from B. distachyon genome and successfully cloned 12 PYLs and 7 clade A PP2Cs. Bioinformatic and expression analyses showed that most of the identified genes respond to several signal molecules and abiotic stresses. Protein–protein interaction analysis revealed that many BdPYLs and BdPP2CAs participate in the classic ABA-PYL-PP2C-SnRK2 signaling pathway. A clade A PP2C, designated BdPP2CA6, interacted with BdPYL11 in the absence of ABA and localized in nucleus. Most clade A PP2C members from Arabidopsis showed negatively regulation in ABA signaling pathway, whereas BdPP2CA6-overexpression transgenic Arabidopsis showed ABA hypersensitive phenotype, resulting in enhanced stomatal closure and salinity tolerance. Our results indicate that BdPP2CA6 positively regulates ABA and stress signal pathway in transgenic Arabidopsis plant seedlings.
Keywords: Brachypodium distachyon, abiotic stress, salinity, ABA, PP2C
Introduction
Plants are constantly challenged by diverse biotic and abiotic stresses throughout their lifetime. Complex stress-related signal transduction pathways have been evolved to cope with those biotic and abiotic stresses (Bohnert et al., 2006). Abscisic acid (ABA) is essential in plants responding to biotic and abiotic stresses and ABA signaling model is well established in model plant species Arabidopsis (Lee and Luan, 2012). Type 2C phosphatase (PP2C) is a kind of protein phosphatase that dephosphorylates Ser/Thr residues (Cohen, 1989). Clade A PP2C is a subgroup of PP2C family that inhibits ABA signaling in Arabidopsis (Lee and Luan, 2012). Core ABA signaling module is composed of three main components: PYR/PYL/RCAR (pyrabactin resistance/PYR-like protein/regulatory component of ABA receptor), clade A PP2C, and SnRK2 (SNF1-related protein kinase 2) (Fujii et al., 2009; Ma et al., 2009; Park et al., 2009). In this model, PP2Cs physically interact with SnRK2s and inhibit SnRK2 kinase activities in the absence of ABA. In the presence of ABA, an ABA molecule binds to ABA receptor PYR/PYL/RCAR to form a ternary complex PYR/PYL/RCAR-ABA-PP2C, thus causing the release of SnRK2s from PP2Cs. Subsequently, SnRK2s phosphorylate and activate downstream targets to respond to external stresses (Klingler et al., 2010; Lee and Luan, 2012; Osakabe et al., 2014).
To date, plenty of studies have revealed involvement of PP2Cs and PYR/PYL/RCARs in stress signaling pathways in plants. By inhibiting PP2C activities, PYR/PYL/RCARs function positively in major abiotic stress signal pathways. Overexpressing of PYL4A194T, a mutant that can form stable complex with PP2CA without exogenous ABA, reduces stomatal conductance and enhances water use efficiency in transgenic seedlings in comparison with wild type (WT) or PYL4-overexpressing seedlings, thus enhancing drought and dehydration tolerance (Pizzio et al., 2013). PYL5 is a positive regulator of ABA signaling and enhances drought resistance through inhibition of HAB1 (Santiago et al., 2009). PYL9 inhibits ABI1, HAI1, HAI2, HAI3, HAB1, HAB2, and PP2CA, and promotes leaf senescence and resistance to extreme drought stress in Arabidopsis (Zhao et al., 2016). Studies also demonstrate that clade A PP2Cs in Arabidopsis, such as ABI1, ABI2, PP2CA, and HAB1, negatively regulate ABA signaling, while their loss-of-function mutants display improved tolerance to abiotic stress such as salinity and drought (Merlot et al., 2001; Saez et al., 2004, 2006; Zhang et al., 2013). However, recently a clade A PP2C gene isolated from rice, namely OsPP108, is found to positively regulate abiotic stress signaling through ABA independent pathway, indicating that clade A PP2Cs have multiple functions in stress signal pathways (Singh et al., 2015).
By now, putative ABA signaling genes in monocot such as rice, barley, and maize have been identified. The rice PYR/PYL/RCAR family has 12 members and its PP2C superfamily consists of 78 members, 10 of which belong to clade A subfamily (Xue et al., 2008; He et al., 2014). In barley, nine HvPYR/PYLs and six clade A HvPP2Cs have been identified (Seiler et al., 2014). While in maize, 11 ZmPYLs and 5 clade A ZmPP2Cs have been identified (Fan et al., 2016). However, PYL family and clade A PP2C subfamily have not yet been characterized in monocot grass species Brachypodium distachyon, which is an excellent model species for functional genomics research in temperate grasses. In this study, we identified 12 PYL and 8 clade A PP2C genes from B. distachyon, and analyzed their chromosomal distributions and expression patterns. Additionally, interaction analyses were performed between PP2Cs and PYR/PYL/RCARs or PP2Cs and SnRK2s to determine their interaction network in response to abiotic stresses. Furthermore, functional analyses of BdPP2CA6 revealed its positive regulatory role in response to salinity through ABA dependent pathway. These analyses are helpful for understanding core ABA signaling in B. distachyon and may shed light on functional characterization of PP2Cs in monocot crop plants, such as wheat.
Materials and Methods
Plant Materials
The B. distachyon Bd21 was cultured and used for gene cloning and expression analysis. The Arabidopsis thaliana ecotype Colombia (Col-0) was used as WT control. The pSN1301-BdPP2CA6 plasmid was transformed into Arabidopsis using Agrobacterium tumefaciens strain EHA105 as described previously (Clough and Bent, 1998). Arabidopsis transformed with pSN1301 empty vector was used as vacant vector control (VC). Seeds of transformed Arabidopsis were selected using MS medium supplemented with 20 mg/L hygromycin. Homozygous lines of T3 and T4 generations were used for further analysis.
Plant Growth Conditions and Treatments
All plant materials were grown under a 14 h light/10 h dark cycle and 70% relative humidity at 22°C. For organ-specific expression assay, different organs including root, stem, leaf, and caryopsis were taken, respectively, from 3-week-old B. distachyon plants. For abiotic stress treatments, 3-week-old B. distachyon were treated with 100 μM ABA, 200 mM NaCl, or 20% (w/v) PEG-6000, respectively, and then leaves were sampled at 0, 1, 3, 6, 12, and 24 h after treatments.
Identification and Analysis of BdPYLs and Clade A BdPP2Cs
To identify PYLs and clade A PP2Cs in B. distachyon, PYL and clade A PP2C gene sequences of Arabidopsis were downloaded from TAIR database1, which were then used to do BLAST searches against Ensembl Plants database2. All resulting proteins were confirmed to contain certain domains using Pfam database3. Identified sequences of B. distachyon were aligned using ClustalX2.0 with default parameters (Larkin et al., 2007). Phylogenetic trees were constructed using bootstrap neighbor-joining (NJ) method and bootstrap analysis (1,000 replicates) by MEGA6.0 (Tamura et al., 2013). Promoter sequences and exon-intron data were obtained from Plaza database4. Abiotic stress-responsive cis-elements in promoters were analyzed using PlantCare database5. Expression heatmaps of BdPP2CAs and BdPYLs were constructed as described previously (Chen L. et al., 2016).
Plasmid Construction
A total of 7 BdPP2CAs and 12 BdPYLs were amplified by RT-PCR. For yeast two-hybrid assay, ORFs of BdPP2CAs and BdPYLs were amplified from B. distachyon cDNA and cloned into vectors pGBKT7 and pGADT7, respectively. The pGADT7-BdSnRK2.2, pGADT7-BdSnRK2.3, and pGADT7-BdSnRK2.6 vectors were obtained from our laboratory (Wang et al., 2015). The ORFs of the AtSnRK2.2, AtSnRK2.3, and AtSnRK2.6 were amplified from Arabidopsis Col-0 WT cDNA and were cloned into vector pGADT7. For BiFC assay, ORFs of BdPP2CA6 were cloned into vector pUC-SPYNE, while BdPYL11, and AtSnRK2.3 were cloned into vector pUC-SPYCE. For GFP fusion protein expression, 35S::BdPYL11::GFP, 35S::BdPP2CA6::GFP, and 35S::AtSnRK2.3::GFP were constructed. For plant transformation, ORF of BdPP2CA6 was cloned into vector pSN1301 driven by CaMV 35S promoter.
All primers used are listed in Supplementary Table S2 and all amplified fragments were confirmed by sequencing. All accession numbers of the genes mentioned in manuscript are listed in Supplementary Table S3.
Yeast Two-Hybrid Assay
Each pair of constructed plasmids were co-transformed into yeast strain AH109 following the manufacturer’s protocol (Clontech Inc., USA) and subsequently plated onto SD minimal medium without leucine and tryptophan (SD/-Leu-Trp). Then yeast cells were transferred to specific selection media for yeast growth assessments.
Subcellular Localization and BiFC Assay
The constructs of BdPYL11-GFP and BdPP2CA6-GFP were transformed, and constructs of BdPP2CA6-YNE and BdPYL11-YCE or AtSnRK2.3-YCE were co-transformed into onion epidermal cells by using biolistic method (Biolistic PDS-1000/He Particle Delivery System, Bio-Rad, USA), respectively (Hanson and Kohler, 2001). Fluorescence was observed by fluorescence microscope (IX71, Olympus, Japan) 1 day after transformation. DAPI (4′,6-diamidino-2-phenylindole) staining was performed as described previously (Sun et al., 2015).
RNA Extraction and qRT-PCR Analysis
Total RNAs were isolated using Plant RNA Extraction Kit (ZOMANBIO, China). The cDNAs were obtained by using FastQuant RT Kit (TIANGEN, China). The qRT-PCR analysis was performed using SYBR Green Kit (TIANGEN, China) on a real-time PCR machine (CFX96TM Real-time Detection System, Bio-Rad, USA). Gene-specific primer sets were listed in Supplementary Table S2 and some of them were designed according to previous studies (Fujii et al., 2007; Feng et al., 2014; Chen Z.H. et al., 2016). All amplified fragments were confirmed by sequencing. The qRT-PCR analysis was performed as described previously (Hu et al., 2013).
Stomatal Aperture Analysis
For stomatal aperture measurement, leaves of 4-week-old WT and transgenic plants were collected and incubated in solution containing 30 mM KCl, 10 mM MES-Tris, and 50 μM CaCl2 (pH 6.15) with light for 2 h. Then 50 μM ABA or 150 mM NaCl was added to the solution. After 2 h treatments, leaf samples were examined using microscope (IX71, Olympus, Japan). Widths and lengths of stomata were measured to figure out stomatal apertures.
Stress Tolerance Assay
For ABA treatment, 7-day-old seedlings germinated on 1/2 MS medium were transferred to MS medium or MS media supplemented with 1, 5, or 10 μM ABA for 10 days. For salinity stress tolerance assay at early development stage, 7-day-old seedlings were transferred onto MS medium or MS medium supplemented with 150 mM NaCl for 10 days. For physiological index measurement and qRT-PCR analysis, 4-week-old seedlings cultured in soil were watered with or without 150 mM NaCl for 10 days and then leaf samples were collected. Survival rates of the treated Arabidopsis seedlings were measured 35 days post NaCl treatment and pictures of the seedlings were taken 30 days post NaCl treatment.
Ion Content Measurement
For ion content determination, treatment of plants and sampling of leaves were performed as described previously with slight modification (Deng et al., 2013). One-week-old seedlings culturing on MS medium were transferred onto MS medium supplemented with 150 mM NaCl for 1 week and then leaves were sampled. Measurement was carried out by using Atomic Absorption Spectrometry (ContrAA 700, Analytik Jena, Germany).
Physiological Index Measurement
Measurements of ion leakage (IL), H2O2, proline, chlorophyll, and malondialdehyde (MDA) content were performed as described previously (Huang et al., 2015).
Statistical Analysis
Statistical analyses were performed using Excel (Microsoft, USA) and SPSS (IBM, USA). Figures were plotted by using OriginPro (OriginLab, USA).
Results
Identification of BdPYL Family and Clade A BdPP2C Subfamily in B. distachyon
To identify BdPYL and clade A BdPP2C proteins from B. distachyon, BLAST searches were performed. Twelve putative BdPYL proteins and eight putative clade A BdPP2C proteins were identified from the B. distachyon genome. Phylogenetic analyses were performed to construct phylogenetic trees by using full length amino acid sequences of BdPYL family or clade A BdPP2C subfamily, and the results showed that key domains of BdPYL proteins or clade A BdPP2C proteins are evolutionary conserved (Figure 1).
FIGURE 1.
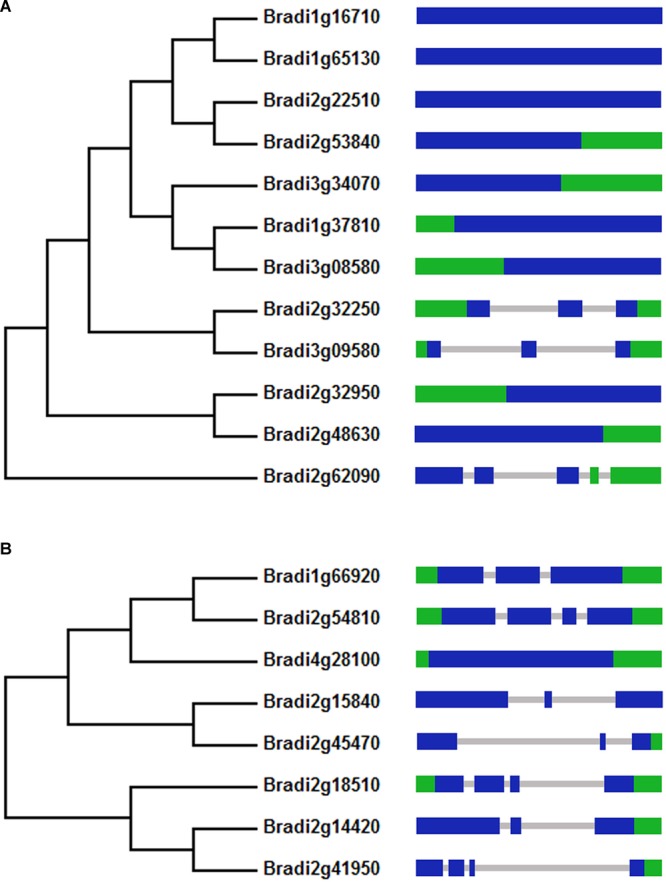
Phylogenetic and exon-intron analyses of BdPYLs (A) and BdPP2CAs (B). The green area represents UTR exon, the blue area represents exon, and the gray area represents intron.
Chromosomal Locations of BdPYLs and Clade A BdPP2Cs
To analyze the genomic distribution of BdPYLs and clade A BdPP2Cs, their approximate positions on each chromosome were marked. The results showed that six BdPYLs distributed on chromosome 2 and the rest members were localized on chromosomes 1 and 3, while six clade A BdPP2Cs distributed on chromosome 2 and the rest members were mapped on chromosomes 1 and 4 (Supplementary Figure S1). All BdPYLs and clade A BdPP2Cs were designated according to their distribution on chromosomes as listed in Supplementary Table S1.
Analyses of Gene Structures and Cis-elements in Promoters
The exon–intron structures of BdPYLs and BdPP2CAs were also analyzed (Figure 1). For BdPYL family, only three BdPYLs contains two or more introns and the rests contain no intron at all. For BdPP2CA subfamily, all members except BdPP2CA8 have two or three introns. These conserved intron and extron numbers in both BdPYL family and BdPP2CA subfamily support their close evolutionary relationships.
To identify abiotic stress-responsive cis-elements in promoters, 1,500 bp sequences upstream of BdPYL and BdPP2CA CDSs were obtained, and several abiotic stress-responsive cis-elements (ABRE, CE3, CGTCA-motif, GARE-motif, HSE, TGACG-motif, MBS, TCA-element) were identified. The results demonstrate that 50% of the BdPYLs contain ABRE elements in their promoter regions and only BdPP2CA5 has no ABRE element in its promoter region, implying that many BdPYLs and BdPP2CAs maybe ABA responsive. Results also show that these genes have one or more additional stress-responsive cis-elements in their promoter regions, suggesting their potential involvements in various stress responses (Supplementary Table S1).
Expressions of BdPP2CA6 and BdPP2CA8 Are Responsive to Salt or Drought Treatments
To analyze the potential functions of BdPYLs and BdPP2CAs under environmental stresses, public microarray data were obtained to analyze their expressions under cold, heat, drought, and high-salinity conditions (Priest et al., 2014). The results showed that expressions of most BdPYLs were not obviously changed under stress treatments, while BdPP2CA6 was significantly up-regulated under high-salinity stress and BdPP2CA8 was significantly up-regulated under drought stress (Supplementary Figure S2).
BdPYLs and BdPP2CAs Cloning and Yeast Two-Hybrid Assay
Although we tried hard to clone BdPP2CA3 from the B. distachyon cDNA, we failed to clone the full length of it. The rest 7 BdPP2CAs and all identified BdPYLs were successfully cloned. In Arabidopsis, ABA-bound PYLs interact with clade A PP2Cs and inhibit their activities (Melcher et al., 2009; Miyazono et al., 2009; Yin et al., 2009). To investigate their interaction networks in B. distachyon, we examined the interactions between BdPP2CAs and BdPYLs in yeast two-hybrid assay. Data of those BdPP2CAs/BdPYLs which could not interact with any BdPYLs/BdPP2CAs were not shown. The result showed that BdPP2CA4, BdPP2CA5, BdPP2CA6, BdPP2CA8 interact with 7 or 8 or 9 BdPYLs when supplied with exogenous ABA (Figure 2A). In the absence of exogenous ABA, fewer BdPYLs interact with these four BdPP2CAs (BdPP2CA4, BdPP2CA5, BdPP2CA6, and BdPP2CA8) and only BdPYL11 can interact with all of them (Figure 2B).
FIGURE 2.
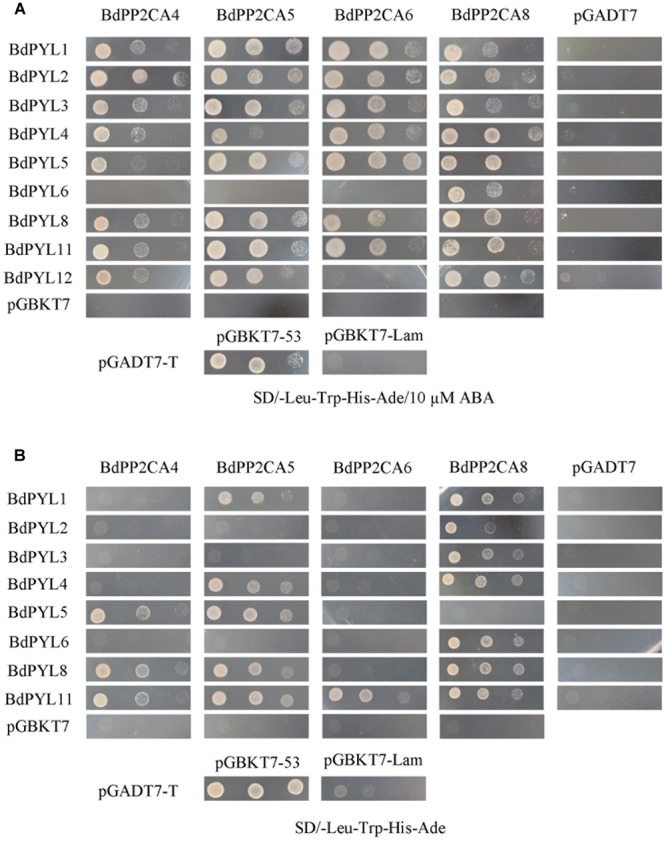
Interaction analyses of BdPYLs-BdPP2CAs. The yeast strains AH109 transformed with different combinations of plasmids were cultivated on selective medium (A) SD/-Trp-Leu-His-Ade/10 μM ABA or (B) SD/-Trp-Leu-His-Ade. Interactions between SV40-T and p53 or SV40-T and human lamin C were set as positive or negative control, respectively. Yeast strains were assessed by different dilution rates (1, 1/10, and 1/100). Data of those BdPP2CAs/BdPYLs which could not interact with any BdPYLs/BdPP2CAs were not shown.
Expression Patterns of BdPYL11 and BdPP2CAs
As interactions between BdPYL11 and BdPP2CA4, BdPP2CA5, BdPP2CA6, or BdPP2CA8 are strong and ABA independent, qRT-PCR analyses were performed to study their potential functions in response to abiotic stresses. Organ-specific expression analysis was also carried out, and the results demonstrated that BdPP2CA8 expressed only in roots while BdPYL11, BdPP2CA4, BdPP2CA5, and BdPP2CA6 were detected in roots, as well as in leaves, stems, and caryopses (Figure 3A).
FIGURE 3.
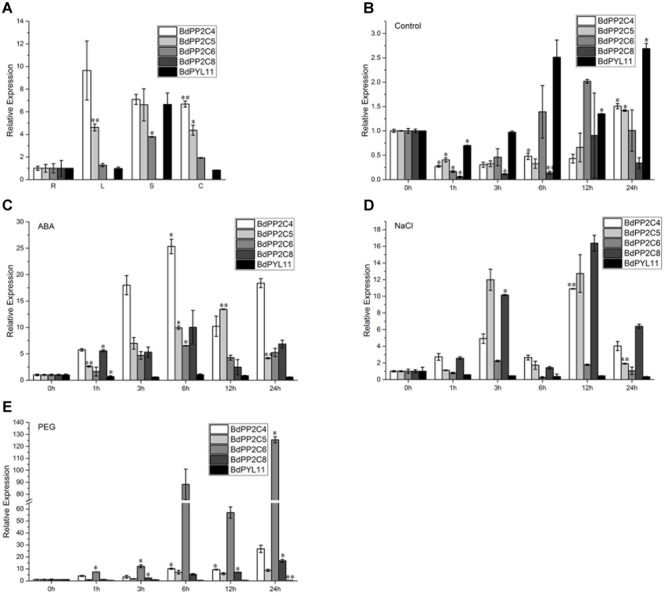
Organ-specific expression patterns of BdPYL11, BdPP2CA4, BdPP2CA5, BdPP2CA6, and BdPP2CA8 (A), and expression patterns of these genes under normal (B), 100 μM ABA (C), 200 mM NaCl (D), and 20% (w/v) PEG-6000 (E) treatments. The organs (root, stem, leaf, and caryopsis) were represented by R, S, L, and C, respectively. Data are means ± SE of three replicates. Significances of differences are indicated as ∗P < 0.05; ∗∗P < 0.01.
In Arabidopsis, PYLs were strongly down-regulated while clade A PP2Cs were strongly up-regulated by ABA treatment (Merlot et al., 2001; Saez et al., 2004; Santiago et al., 2009). The qRT-PCR results reveal that expression patterns of the investigated genes in B. distachyon are consistent with their homologs in Arabidopsis. BdPYL11 was down-regulated under ABA, NaCl and PEG6000 treatments while BdPP2CA4, BdPP2CA5, BdPP2CA6, and BdPP2CA8 were up-regulated (Figures 3B–E). These results suggest that these genes respond to various abiotic stresses.
Interaction between BdPP2CA6 and BdPYL11 Confirmed by BiFC Assay
Without exogenous ABA, BdPP2CA6 only interacts with BdPYL11 in yeast two-hybrid assay. So we were more interested in interaction between these two proteins. GFP fusion vectors were constructed and transferred into onion epidermal cells to investigate their subcellular localizations. BdPYL11-GFP fusion protein was observed in the nucleus, plasma membrane, and cytoplasm, while BdPP2CA6-GFP fusion protein was observed only in nucleus (Figure 4A). To examine whether BdPP2CA6 interacts with BdPYL11 in vivo, we further carried out BiFC assay. YFP fluorescence was observed in nucleus, suggesting that BdPP2CA6 interact with BdPYL11 in vivo (Figure 4B).
FIGURE 4.
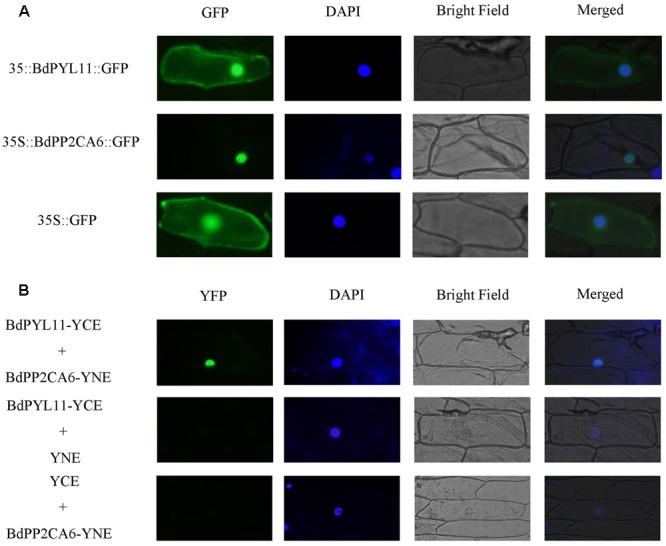
Subcellular localization of BdPYL11-GFP and BdPP2CA6-GFP (A), and BiFC visualization of BdPYL11 interaction with BdPP2CA6 (B). BdPYL11-GFP, BdPP2CA6-GFP, BdPYL11-YCE, and BdPP2CA6-YNE fusion proteins transiently expressed in onion epidermal cells were observed. DAPI staining for the nucleus was performed after GFP/YFP and bright field visualization. Subcellular localization of GFP was set as control. BdPYL11-YCE and YNE, BdPP2CA6-YNE and YCE were set as negative controls.
Overexpression of BdPP2CA6 in Arabidopsis Affects ABA Sensitivity
BdPP2CA6, a clade A PP2C gene, was greatly induced by ABA, PEG6000 and NaCl treatments (Figure 3). BdPP2CA6 exhibited a strong interaction with BdPYL11 in the absence of ABA (Figures 2A and 4A). Transient expression of BdPP2CA6-GFP fusion protein in onion epidermal cells demonstrates that BdPP2CA6 localizes only at the nucleus (Figure 4B). Therefore we focused on investigating BdPP2CA6 and obtained BdPP2CA6 overexpression Arabidopsis plants for further analyses.
To investigate whether overexpression of BdPP2CA6 could affect ABA response, 7-day-old WT, VC and BdPP2CA6-overexpression Arabidopsis seedlings were placed on MS medium and MS media supplemented with different concentrations of ABA, respectively, for 10 days. ABA-mediated inhibition of root growth was affected by overexpression of BdPP2CA6 (Figures 5A–D). Root elongations of BdPP2CA6-overexpression plants were inhibited with higher degrees than that of WT and VC by all concentrations of ABA. Fresh weights and root lengths were measured and were significantly higher for WT and VC plants than for transgenic plants at all ABA concentrations (Figures 5G,H). The results demonstrated that overexpression of BdPP2CA6 enhanced ABA sensitivity at early growing stage of plants.
FIGURE 5.
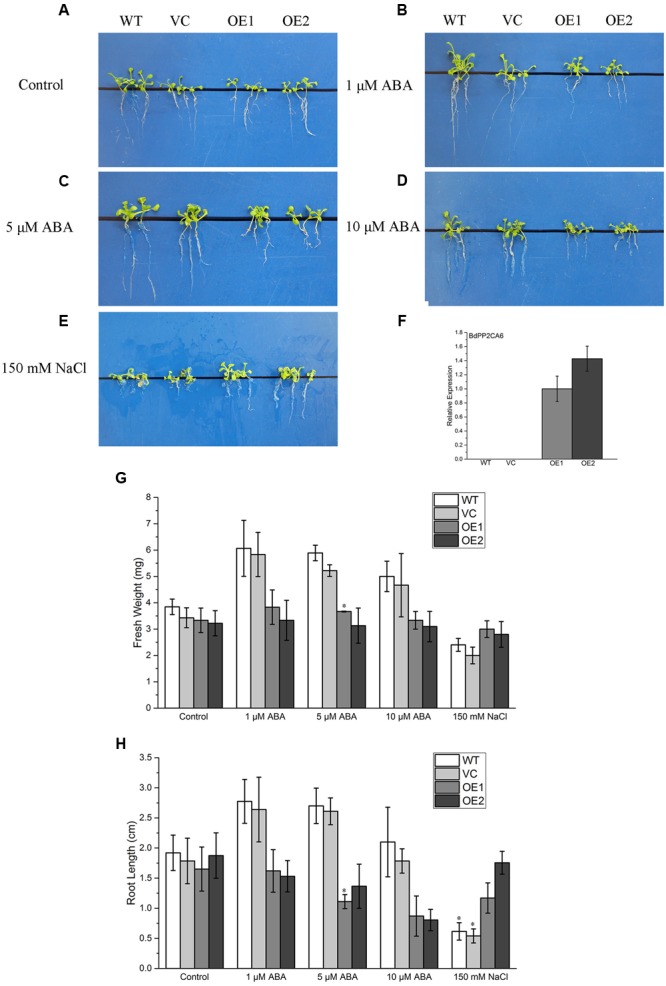
Root elongation analyses of BdPP2CA6-overexpression Arabidopsis plants under ABA and NaCl treatment. (A–E) Root growths of WT, VC and transgenic lines on MS medium, or MS medium supplemented with different concentrations of ABA or NaCl. (F) BdPP2CA6 expression levels in WT, VC and transgenic lines. (G) Fresh weights of WT, VC and transgenic lines. (H) Root lengths of WT, VC and transgenic lines. Data are means ± SE of three replicates. Significances of differences are indicated as ∗P < 0.05.
BdPP2CA6 Regulates ABA-Mediated Stomatal Closure
Whether BdPP2CA6 could regulate ABA-mediated stomatal closure was investigated. Under normal condition, stomatal apertures of transgenic seedlings were a little bit smaller than that of WT. After ABA treatment, stomatal apertures of all seedlings decreased, with a much more significant decrease in transgenic plants compared with WT (Figure 6). These results suggest that overexpression of BdPP2CA6 promotes ABA-mediated stomatal closure.
FIGURE 6.
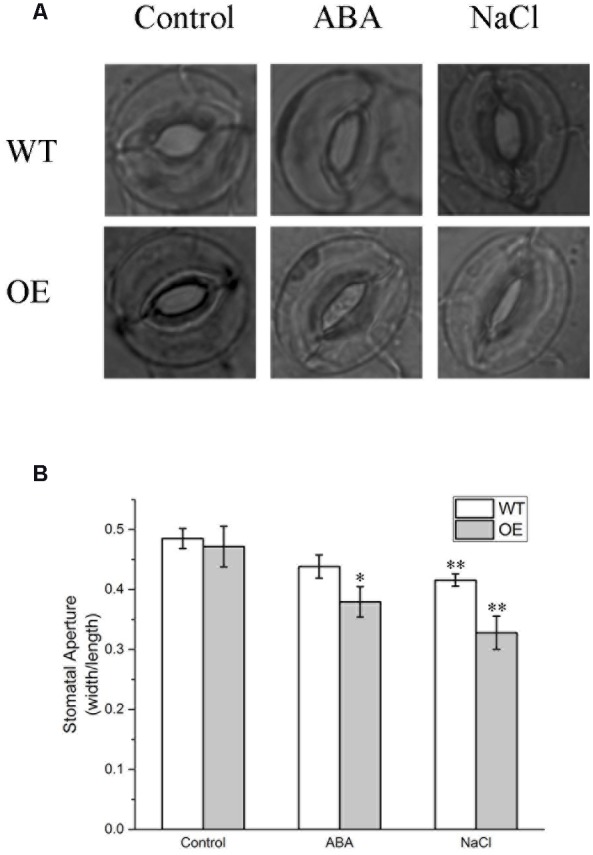
Stomatal aperture analyses of WT and BdPP2CA6 overexpression Arabidopsis plants under ABA and NaCl treatment. (A) Representative images; (B) Ratio of width to length (n = 20–30). Data are means ± SE and significances of differences are indicated as ∗P < 0.05, ∗∗P < 0.01.
Overexpression of BdPP2CA6 Increases Salinity Tolerance in Arabidopsis Plants
To evaluate the effects of salinity stress on growth of BdPP2CA6-overexpression plants, 7-day-old WT, VC and BdPP2CA6-overexpression plants were placed on MS medium and MS medium supplemented with 150 mM NaCl, respectively. After 10 days, no obvious difference was discovered at post-germination developmental stage on MS media. The addition of 150 mM NaCl significantly inhibited root elongation of WT but not of BdPP2CA6-overexpression plants (Figures 5A,E). Fresh weights and root lengths were evaluated and the average fresh weight and root length of BdPP2CA6-overexpression plants were higher than that of WT and VC at 150 mM NaCl concentration (Figures 5G,H). In addition, growth of 2-week-old seedlings under salinity treatment was measured. Two-week-old WT and BdPP2CA6-overexpression plants growing on MS medium were transferred to Hoagland solution supplemented with 0 or 150 mM NaCl. Most WT plants wilted after 1 day, while most BdPP2CA6-overexpression transgenic Arabidopsis remained green, indicating that BdPP2CA6 is involved in responses to salinity stress at the early growing stage (Figure 7B).
FIGURE 7.
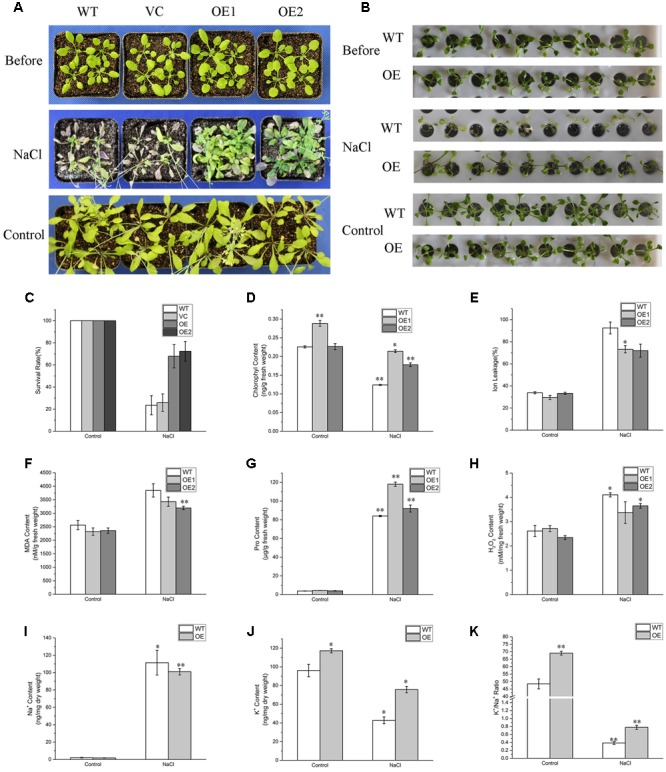
Analyses of enhanced salinity tolerance of BdPP2CA6-overexpressing Arabidopsis seedlings. (A) Phenotype of 3-week-old WT, VC and transgenic seedlings before NaCl treatment, 30 days post treatment, and 30 days later without treatment (control). (B) Phenotype of 2-week-old WT and transgenic seedlings culturing in Hoagland solution before NaCl treatment, 1 days after treatment, and 1 day later without treatment (control). (C) Survival rates were measured 35 days post NaCl treatment and 35 days later without treatment (control). (D) Chlorophyll content. (E) Ion leakage content. (F) MDA content. (G) Proline content. (H) H2O2 content. (I) Na+ content. (J) K+ content. (K) K+/Na+ ratio. Data are means ± SE of three replicates. Significances of differences are indicated as ∗P < 0.05, ∗∗P < 0.01.
For NaCl treatment, 3-week-old WT, VC and BdPP2CA6-overexpression plants growing in soil were watered with 150 mM NaCl for 30 days. Most WT plants turned purple or wilted whereas most transgenic plants remained green (Figure 7A). Survival rates of transgenic seedlings were measured 35 days post NaCl treatment and were significantly higher than that of WT and VC plants (Figure 7C). As we found that overexpression of BdPP2CA6 could promote ABA-mediated stomatal closure, we assumed that BdPP2CA6-overexpression plants might be able to withhold more water to cope with external salinity through stomatal closure. To confirm this possibility, we measured stomatal apertures of leaves from WT and BdPP2CA6-overexpression plants under NaCl treatment. Under normal condition, the differences between WT and BdPP2CA6-overexpression plants were not significant, the stomatal aperture indices of transgenic plants after salinity treatment decreased more rapidly than that of WT, indicating that transgenic plants could withhold more water to cope with salinity stress (Figure 6).
Overexpression of BdPP2CA6 Increases K+/Na+ Ratio under Salinity Stress
A higher K+/Na+ ratio is important for plants to cope with salinity stress (Zhu, 2003). To investigate whether overexpression of BdPP2CA6 could affect Na+ and K+ accumulations in plants, the Na+ and K+ contents were measured. No significant difference in Na+ contents was observed between WT and transgenic plants under normal condition or NaCl treatment. Whereas K+ contents of transgenic plants were significantly higher than that of WT under both normal condition and NaCl treatment (Figures 7I–K). The results demonstrated that overexpression of BdPP2CA6 increased K+ accumulation, thus increasing the ratio of K+/Na+ under salinity stress condition.
Overexpression of BdPP2CA6 Increases Chlorophyll, Proline Content, and Decreased H2O2, MDA, and IL Level under Salinity Stress
Several physiological indices including IL, H2O2, proline, chlorophyll, and MDA contents were measured to investigate the physiological mechanisms in BdPP2CA6-overexpression plants in response to salinity stress. BdPP2CA6-overexpression plants exhibited lower contents of IL, MDA, and H2O2, and higher levels of chlorophyll and proline compared with WT under NaCl treatment (Figures 7D–H). The results suggested that transgenic seedlings suffered less membrane damage and intracellular oxidative stress, therefore were more tolerant to external salinity stress.
BdPP2CA6 Interacts with Both AtSnRK2 and BdSnRK2
To further understand the molecular mechanisms of BdPP2CA6 function in abiotic stress tolerance, interactions between BdPP2CA6 and AtSnRK2/BdSnRK2 were investigated. SnRK2.2/2.3/2.6 were selected as potential targets because SnRK2.2/2.3/2.6 are essential in ABA signaling and interact with several clade A PP2Cs in Arabidopsis (Fujita et al., 2009; Nakashima et al., 2009). We chose SD/-Trp-Leu-His/0.1 μM 3-AT to assess growth status of yeast cells co-transformed with BdPP2CA6 and BdSnRKs because they were not able to grow on SD/-Trp-Leu-His-Ade. The results showed that BdPP2CA6 had a mild interaction with BdSnRK2.2 but had a strong interaction with AtSnRK2.3 (Figures 8A,B). This strong interaction between BdPP2CA6 and AtSnRK2.3 was further confirmed by BiFC assay (Figure 8C).
FIGURE 8.
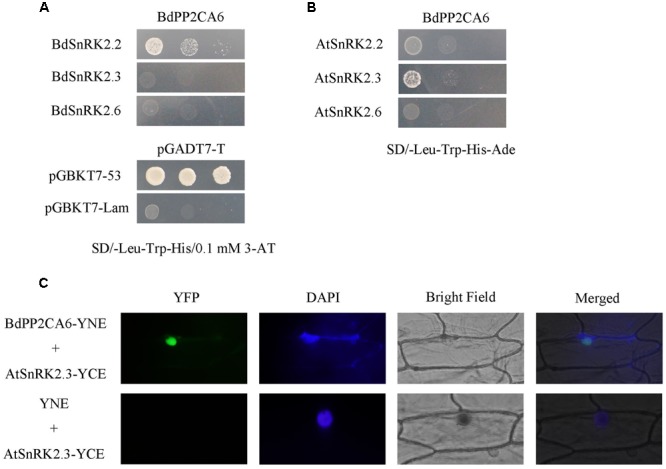
Interaction analyses of BdPP2CA6-BdSnRKs and BdPP2CA6-AtSnRKs. (A) pGBKT7-BdPP2CA6 and pGADT7-BdSnRKs were co-transformed and transformed yeast cells were cultivated on selective medium SD/-Trp-Leu-His/0.1 μM 3-AT, respectively. Interactions between SV40-T and p53 or SV40-T and human lamin C were set as positive or negative control, respectively. Yeast strains were assessed by different dilution rates (1, 1/10, and 1/100). (B) pGBKT7-BdPP2CA6 and pGADT7-BdSnRKs were co-transformed and transformed yeast cells were cultivated on nutritional selective medium SD/-Trp-Leu-His-Ade. (C) BiFC visualization of BdPP2CA6 interaction with AtSnRK2.3 transiently expressed in onion epidermal cells. DAPI staining for the nucleus was performed after YFP and bright field visualization. AtSnRK2.3-YCE and YNE was set as negative control.
Altered Expressions of Related Stress-Responsive Genes in BdPP2CA6-Overexpression Plants
To identify molecular events involved in BdPP2CA6-mediated signal pathway, expressions of several ABA- and stress-responsive genes were investigated by qRT-PCR technique. The results showed that certain genes (ABF2, ABF3, DREB2A, MYB15, RD26, GORK, and SLAH3) were significantly induced by salinity stress and were induced to higher expression levels in transgenic plants than WT while expression levels of MYB44, RD29B, and ABF4 were not significantly different between transgenic plants and WT under salinity stress (Figure 9). These results imply that BdPP2CA6 positively regulate the response to salinity stress by modulating expression of diverse stress-related genes in Arabidopsis plants.
FIGURE 9.
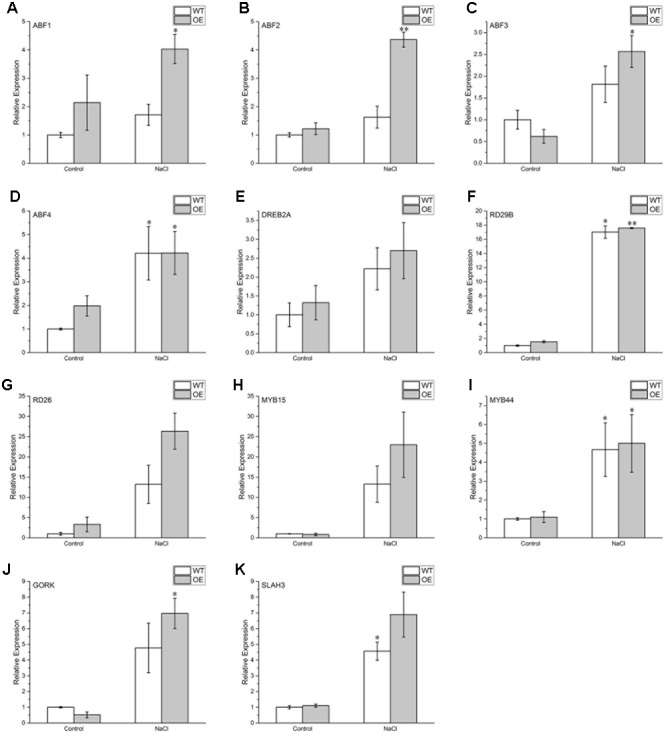
Expression pattern of relevant genes in WT and BdPP2CA6-overexpression seedlings. (A) ABF1; (B) ABF2; (C) ABF3; (D) ABF4; (E) DREB2A; (F) RD29B; (G) RD26; (H) MYB15; (I) MYB44; (J) GORK; (K) SLAH3. Seedlings of WT and OE were treated with 150 mM NaCl for 10 days and total RNAs were extracted from leaves. Data are means ± SE of three replicates. Significances of differences are indicated as ∗P < 0.05, ∗∗P < 0.01.
Discussion
Abscisic acid is a key phytohormone in abiotic stress response, and understanding regulatory mechanism of ABA signaling is therefore very important. The ABA-PYL-PP2C-SnRK2 signal pathway has been demonstrated in Arabidopsis (Fujii et al., 2009). Here, we identified PYLs and PP2CAs in B. distachyon to investigate their functions in ABA signaling as gene function analysis of monocot model plant species B. distachyon can help the similar researches being conducted in other important cereal plant species such as wheat. In our study, we found that many interactions between BdPYLs and BdPP2CAs were dependent on the presence of ABA in yeast two-hybrid assay. Strong interaction between BdPYL11 and BdPP2CA6 was confirmed in vivo by BiFC assay, therefore we focused on functional analysis of BdPP2CA6 (Figures 2 and 4A). Moreover, BdPP2CA6 interacts with BdSnRK2.2 and AtSnRK2.3, suggesting the participation of BdPP2CA6 in core ABA signaling (Figure 8).
The subcellular localization of BdPP2CA6 is different from those of clade A PP2Cs in Arabidopsis, but consistent with several clade A PP2Cs in rice (Kim et al., 2012; Singh et al., 2015). Our studies showed that BdPP2CA6 and complexes consisting of BdPP2CA6 and its interacting partners, BdPYL11 or AtSnRK2.3 were localized to the nucleus (Figures 4 and 8C). This indicates that BdPP2CA6 recruits BdPYL11 to the nucleus and BdPYL11 might function as a nuclear ABA receptor.
We found that overexpression of BdPP2CA6 in Arabidopsis enhances ABA sensitivity (Figure 5). Moreover, overexpression of BdPP2CA6 in Arabidopsis promotes stomatal closure under both ABA and NaCl treatments (Figure 6). Accumulation of ABA under stress conditions leads to stomatal closure, thus conserving water to survive abiotic stress such as drought (Schroeder et al., 2001; Wasilewska et al., 2008; Kim et al., 2010; Lee and Luan, 2012). In our study, BdPP2CA6 overexpression plants showed enhanced tolerance to salinity stress. We suppose that ABA-dependent enhanced stomatal closure of BdPP2CA6 overexpression plants attributes to better tolerance under salinity treatment (Figure 6).
A higher K+/Na+ ratio is important for plants to maintain cellular metabolism and to survive under salinity stress (Zhu, 2003). Higher K+/Na+ ratio in BdPP2CA6 overexpressing plants enhances salinity stress tolerance to plants. Regulation of K+ channel activity by protein kinases and phosphatases could control K+ transport during stresses (Cherel, 2002; Lan et al., 2011). BdPP2CA6 might enhance salinity stress tolerance by regulating K+ channels activity.
To gain a further insight into how BdPP2CA6 regulates gene expression under salinity stress, expressions of some stress-related genes were detected. Previous studies have revealed many positive regulators of ABA and stress signaling pathways. ABF1, ABF2, ABF3, and ABF4 are predominant TFs downstream of SnRK2.2/2.3/2.6 in ABA signal pathway, responding to osmotic stresses at vegetative growth stage (Fujita et al., 2005, 2013; Abdeen et al., 2010; Yoshida et al., 2010, 2015). Beside AREB/ABF TFs, many other TFs have been reported to be involved in ABA-mediated gene expression. For instance, DREB2A, MYB15, MYB44, and RD26 are positive regulators of ABA signaling and are strongly induced by osmotic stress (Nakashima et al., 2000; Fujita et al., 2004; Jung et al., 2008; Ding et al., 2009). Higher expression levels of some stress-related genes (ABF2, ABF3, ABF4, DREB2A, MYB15, and RD26) in BdPP2CA6 overexpression plants than in WT under salinity treatment are contributive to enhanced salt tolerance.
Previous studies show that several clade A PP2Cs in Arabidopsis negatively regulate ABA and stress signaling pathways (Merlot et al., 2001; Saez et al., 2004, 2006; Zhang et al., 2013). In our study, BdPP2CA6 was found to be a positive regulator in both ABA and stress signaling pathways. To uncover how BdPP2CA6 functions is an interesting task. By now, few studies have demonstrated that the clade A PP2Cs could contribute to stress tolerance. Tobacco plants overexpressing ZmPP2C2 are highly tolerant to cold stress (Hu et al., 2010). Overexpression of OsPP108 in Arabidopsis enhanced high salt and mannitol stress tolerance at stages of seed germination, early development, and adult (Singh et al., 2015). Our finding, that BdPP2CA6 confers salinity tolerance to transgenic Arabidopsis plant seedlings, is consistent with the reported results that PP2C genes from monocot plants confer stress tolerance to transgenic plants. However, the mechanism underlying how BdPP2CA6 regulates ABA signaling and stress signaling in Arabidopsis is still is unclear, and we cannot rule out that the native function in Brachypodium could be different. Previous studies reveal that abi1-1, a mutant of ABI1, lacks PYR/PYL/RCAR-binding ability and constitutively inactivates SnRK2s even in the presence of PYR1 and ABA (Ma et al., 2009; Park et al., 2009; Umezawa et al., 2009). In our study, BdPP2CA6 might share some similarity with the phenotype of abi1-1 but in a converse way. BdPP2CA6 can interact with BdPYL11 in the absence of ABA (Figure 4B). Though interaction between BdPP2CA6 and AtSnRK2.3 is confirmed in the BiFC assay, BdPP2CA6 might not inhibit activity of AtSnRK2.3 properly like its homologs do in Arabidopsis. Moreover, we tried to confirm the interaction between BdPP2CA6 and BdSnRK2.2 by using BiFC but failed to observe fluorescence signal, suggesting that BdPP2CA6 interacts with BdSnRK2.2 weakly. We were curious if there is any other SnRK2 that could interact with BdPP2CA6 apart from these three commonly known ABA related SnRK2s (SnRK2.2/2.3/2.6), so we cloned the rest SnRK2s from both Arabidopsis and Brachypodium to carry out yeast-two-hybrid assay and we failed to detect any interaction signal between BdPP2CA6 and SnRK2s (data not shown). Based on above-mentioned observation, we assume that SnRK2s are not the key regulators at downstream of BdPP2CA6. Previous studies reveal that PP2CA specifically interacts with CIPK6 and directly interacts with the CIPK6 kinase domain to inhibit AKT1 activation induced by CIPK6 (Lan et al., 2011). Therefore, the function diversity of clade A PP2Cs and the fact that clade A PP2Cs regulate both ABA-PYL-PP2C-SnRK2 signal pathway and CBL-CIPK-PP2CA network give us new insight into the function mechanism of BdPP2CA6. We also assume that BdPP2CA6 might have undiscovered substrates that function in ABA and stress signaling pathway to confer enhanced stomatal closure, with increased expressions of some stress-responsive genes, and higher K+/Na+ ratio to the BdPP2CA6-overexpressing Arabidopsis plant seedlings. On one hand, BdPP2CA6 participates in the classic ABA-PYL-PP2C-SnRK2 core signal pathway but with less inhibition to SnRK2s. On the other hand, BdPP2CA6 positively regulate ABA and stress signaling pathways through certain uncovered substrates. The latter is predominate, therefore overexpression of BdPP2CA6 in Arabidopsis exhibits hypersensitivity to ABA and tolerance to salinity stress.
Conclusion
BdPP2CA6 is a clade A PP2C from B. distachyon and unlike most other clade A PP2Cs in Arabidopsis, it positively regulates ABA signaling and salinity stress signaling. BdPP2CA6 overexpression in Arabidopsis leads to hypersensitivity to ABA at early growing stage and enhanced salinity tolerance at both early developing stage and adult stage through ABA-dependent pathway. These findings give us insight into cross talk between ABA and abiotic stress signaling.
Author Contributions
GH, GY, JC, FZ, and QW designed the experiments. FZ and QW participated in all experiments, analyzed results and wrote the manuscript. JS performed the physiological experiments. XJ constructed the vectors. YH, QL, and YZ performed the qRT-PCR assay. GH, GY, JC, FZ, and YW revised the manuscript. All authors read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Analytical and Testing Center of Wuhan University for providing atomic absorption spectroscopy assay.
Funding. This work was supported by National Genetically Modified New Varieties of Major Projects of China (2016ZX08010004-004), and the National Natural Science Foundation of China (No. 31570261).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00264/full#supplementary-material
References
- Abdeen A., Schnell J., Miki B. (2010). Transcriptome analysis reveals absence of unintended effects in drought-tolerant transgenic plants overexpressing the transcription factor ABF3. BMC Genomics 11:69 10.1186/1471-2164-11-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert H. J., Gong Q., Li P., Ma S. (2006). Unraveling abiotic stress tolerance mechanisms–getting genomics going. Curr. Opin. Plant Biol. 9 180–188. 10.1016/j.pbi.2006.01.003 [DOI] [PubMed] [Google Scholar]
- Chen L., Han J., Deng X., Tan S., Li L., Li L., et al. (2016). Expansion and stress responses of AP2/EREBP superfamily in Brachypodium distachyon. Sci. Rep. 6:21623 10.1038/srep21623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. H., Wang Y., Wang J. W., Babla M., Zhao C., Garcia-Mata C., et al. (2016). Nitrate reductase mutation alters potassium nutrition as well as nitric oxide-mediated control of guard cell ion channels in Arabidopsis. New Phytol. 209 1456–1469. 10.1111/nph.13714 [DOI] [PubMed] [Google Scholar]
- Cherel I. (2002). Physical and functional interaction of the Arabidopsis K+ channel AKT2 and phosphatase AtPP2CA. Plant Cell 14 1133–1146. 10.1105/tpc.000943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cohen P. (1989). The structure and regulation of protein phosphatases. Annu. Rev. Biochem. 58 453–508. 10.1146/annurev.bi.58.070189.002321 [DOI] [PubMed] [Google Scholar]
- Deng X., Zhou S., Hu W., Feng J., Zhang F., Chen L., et al. (2013). Ectopic expression of wheat TaCIPK14, encoding a calcineurin B-like protein-interacting protein kinase, confers salinity and cold tolerance in tobacco. Physiol. Plant 149 367–377. 10.1111/ppl.12046 [DOI] [PubMed] [Google Scholar]
- Ding Z., Li S., An X., Liu X., Qin H., Wang D. (2009). Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genomics 36 17–29. 10.1016/s1673-8527(09)60003-5 [DOI] [PubMed] [Google Scholar]
- Fan W., Zhao M., Li S., Bai X., Li J., Meng H., et al. (2016). Contrasting transcriptional responses of PYR1/PYL/RCAR ABA receptors to ABA or dehydration stress between maize seedling leaves and roots. BMC Plant Biol. 16:99 10.1186/s12870-016-0764-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C. Z., Chen Y., Wang C., Kong Y. H., Wu W. H., Chen Y. F. (2014). Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 80 654–668. 10.1111/tpj.12670 [DOI] [PubMed] [Google Scholar]
- Fujii H., Chinnusamy V., Rodrigues A., Rubio S., Antoni R., Park S. Y., et al. (2009). In vitro reconstitution of an abscisic acid signalling pathway. Nature 462 660–664. 10.1038/nature08599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Verslues P. E., Zhu J. K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19 485–494. 10.1105/tpc.106.048538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Maruyama K., Seki M., Hiratsu K., Ohme-Takagi M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39 863–876. 10.1111/j.1365-313X.2004.02171.x [DOI] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Satoh R., Maruyama K., Parvez M. M., Seki M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17 3470–3488. 10.1105/tpc.105.035659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Nakashima K., Yoshida T., Katagiri T., Kidokoro S., Kanamori N., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50 2123–2132. 10.1093/pcp/pcp147 [DOI] [PubMed] [Google Scholar]
- Fujita Y., Yoshida T., Yamaguchi-Shinozaki K. (2013). Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant 147 15–27. 10.1111/j.1399-3054.2012.01635.x [DOI] [PubMed] [Google Scholar]
- Hanson M. R., Kohler R. H. (2001). GFP imaging: methodology and application to investigate cellular compartmentation in plants. J. Exp. Bot. 52 529–539. 10.1093/jexbot/52.356.529 [DOI] [PubMed] [Google Scholar]
- He Y., Hao Q., Li W., Yan C., Yan N., Yin P. (2014). Identification and characterization of ABA receptors in Oryza sativa. PLoS ONE 9:e95246 10.1371/journal.pone.0095246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Huang C., Deng X., Zhou S., Chen L., Li Y., et al. (2013). TaASR1, a transcription factor gene in wheat, confers drought stress tolerance in transgenic tobacco. Plant Cell Environ. 36 1449–1464. 10.1111/pce.12074 [DOI] [PubMed] [Google Scholar]
- Hu X., Liu L., Xiao B., Li D., Xing X., Kong X., et al. (2010). Enhanced tolerance to low temperature in tobacco by over-expression of a new maize protein phosphatase 2C, ZmPP2C2. J. Plant Physiol. 167 1307–1315. 10.1016/j.jplph.2010.04.014 [DOI] [PubMed] [Google Scholar]
- Huang Q., Wang Y., Li B., Chang J., Chen M., Li K., et al. (2015). TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 15:268 10.1186/s12870-015-0644-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C., Seo J. S., Han S. W., Koo Y. J., Kim C. H., Song S. I., et al. (2008). Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 146 623–635. 10.1104/pp.107.110981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Hwang H., Hong J. W., Lee Y. N., Ahn I. P., Yoon I. S., et al. (2012). A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 63 1013–1024. 10.1093/jxb/err338 [DOI] [PubMed] [Google Scholar]
- Kim T. H., Bohmer M., Hu H., Nishimura N., Schroeder J. I. (2010). Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61 561–591. 10.1146/annurev-arplant-042809-112226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingler J. P., Batelli G., Zhu J. K. (2010). ABA receptors: the START of a new paradigm in phytohormone signalling. J. Exp. Bot. 61 3199–3210. 10.1093/jxb/erq151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W. Z., Lee S. C., Che Y. F., Jiang Y. Q., Luan S. (2011). Mechanistic analysis of AKT1 regulation by the CBL-CIPK-PP2CA interactions. Mol. Plant 4 527–536. 10.1093/mp/ssr031 [DOI] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lee S. C., Luan S. (2012). ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 35 53–60. 10.1111/j.1365-3040.2011.02426.x [DOI] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324 1064–1068. 10.1126/science.1172408 [DOI] [PubMed] [Google Scholar]
- Melcher K., Ng L. M., Zhou X. E., Soon F. F., Xu Y., Suino-Powell K. M., et al. (2009). A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462 602–608. 10.1038/nature08613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S., Gosti F., Guerrier D., Vavasseur A., Giraudat J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25 295–303. 10.1046/j.1365-313x.2001.00965 [DOI] [PubMed] [Google Scholar]
- Miyazono K., Miyakawa T., Sawano Y., Kubota K., Kang H. J., Asano A., et al. (2009). Structural basis of abscisic acid signalling. Nature 462 609–614. 10.1038/nature08583 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., et al. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50 1345–1363. 10.1093/pcp/pcp083 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Shinwari Z. K., Sakuma Y., Seki M., Miura S., Shinozaki K., et al. (2000). Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration–and high-salinity-responsive gene expression. Plant Mol. Biol. 42 657–665. 10.1023/A:1006321900483 [DOI] [PubMed] [Google Scholar]
- Osakabe Y., Yamaguchi-Shinozaki K., Shinozaki K., Tran L. S. (2014). ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 202 35–49. 10.1111/nph.12613 [DOI] [PubMed] [Google Scholar]
- Park S. Y., Fung P., Nishimura N., Jensen D. R., Fujii H., Zhao Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324 1068–1071. 10.1126/science.1173041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzio G. A., Rodriguez L., Antoni R., Gonzalez-Guzman M., Yunta C., Merilo E., et al. (2013). The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance. Plant Physiol. 163 441–455. 10.1104/pp.113.224162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest H. D., Fox S. E., Rowley E. R., Murray J. R., Michael T. P., Mockler T. C. (2014). Analysis of global gene expression in Brachypodium distachyon reveals extensive network plasticity in response to abiotic stress. PLoS ONE 9:e87499 10.1371/journal.pone.0087499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A., Apostolova N., Gonzalez-Guzman M., Gonzalez-Garcia M. P., Nicolas C., Lorenzo O., et al. (2004). Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 37 354–369. 10.1046/j.1365-313X.2003.01966.x [DOI] [PubMed] [Google Scholar]
- Saez A., Robert N., Maktabi M. H., Schroeder J. I., Serrano R., Rodriguez P. L. (2006). Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol. 141 1389–1399. 10.1104/pp.106.081018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J., Rodrigues A., Saez A., Rubio S., Antoni R., Dupeux F., et al. (2009). Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 60 575–588. 10.1111/j.1365-313X.2009.03981.x [DOI] [PubMed] [Google Scholar]
- Schroeder J. I., Kwak J. M., Allen G. J. (2001). Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410 327–330. 10.1038/35066500 [DOI] [PubMed] [Google Scholar]
- Seiler C., Harshavardhan V. T., Reddy P. S., Hensel G., Kumlehn J., Eschen-Lippold L., et al. (2014). Abscisic acid flux alterations result in differential abscisic acid signaling responses and impact assimilation efficiency in barley under terminal drought stress. Plant Physiol. 164 1677–1696. 10.1104/pp.113.229062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Jha S. K., Bagri J., Pandey G. K. (2015). ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS ONE 10:e0125168 10.1371/journal.pone.0125168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Hu W., Zhou R., Wang L., Wang X., Wang Q., et al. (2015). The Brachypodium distachyon BdWRKY36 gene confers tolerance to drought stress in transgenic tobacco plants. Plant Cell Rep. 34 23–35. 10.1007/s00299-014-1684-6 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106 17588–17593. 10.1073/pnas.0907095106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hu W., Sun J., Liang X., Yang X., Wei S., et al. (2015). Genome-wide analysis of SnRK gene family in Brachypodium distachyon and functional characterization of BdSnRK2.9. Plant Sci. 237 33–45. 10.1016/j.plantsci.2015.05.008 [DOI] [PubMed] [Google Scholar]
- Wasilewska A., Vlad F., Sirichandra C., Redko Y., Jammes F., Valon C., et al. (2008). An update on abscisic acid signaling in plants and more. Mol. Plant 1 198–217. 10.1093/mp/ssm022 [DOI] [PubMed] [Google Scholar]
- Xue T., Wang D., Zhang S., Ehlting J., Ni F., Jakab S., et al. (2008). Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics 9:550 10.1186/1471-2164-9-550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P., Fan H., Hao Q., Yuan X., Wu D., Pang Y., et al. (2009). Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat. Struct. Mol. Biol. 16 1230–1236. 10.1038/nsmb.1730 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Maruyama K., Mogami J., Todaka D., Shinozaki K., et al. (2015). Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38 35–49. 10.1111/pce.12351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61 672–685. 10.1111/j.1365-313X.2009.04092.x [DOI] [PubMed] [Google Scholar]
- Zhang J., Li X., He Z., Zhao X., Wang Q., Zhou B., et al. (2013). Molecular character of a phosphatase 2C (PP2C) gene relation to stress tolerance in Arabidopsis thaliana. Mol. Biol. Rep. 40 2633–2644. 10.1007/s11033-012-2350-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Chan Z., Gao J., Xing L., Cao M., Yu C., et al. (2016). ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. U.S.A. 113 1949–1954. 10.1073/pnas.1522840113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.-K. (2003). Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6 441–445. 10.1016/s1369-5266(03)00085-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


