Abstract
Background
Scorpion venoms contain toxins that modulate ionic channels, among which are the calcins, a small group of short, basic peptides with an Inhibitor Cystine Knot (ICK) motif that target calcium release channels/ryanodine receptors (RyRs) with high affinity and selectivity. Here we describe the heterologous expression of Intrepicalcin, identified by transcriptomic analysis of venomous glands from Vaejovis intrepidus.
Methods
Recombinant Intrepicalcin was obtained in Escherichia coli BL21-DE3 (periplasm) by fusing the intrepicalcin gene to sequences coding for signal-peptide, thioredoxin, His-tag and enterokinase cleavage site.
Results
[3H]Ryanodine binding, used as a functional index of RyR activity, revealed that recombinant Intrepicalcin activates skeletal RyR (RyR1) dose-dependently with Kd = 17.4 ± 4.0 nM. Intrepicalcin significantly augments the bell-shaped [Ca2+]-[3H]ryanodine binding curve at all [Ca2+] ranges, as is characteristic of the calcins. In single channel recordings, Intrepicalcin induces the appearance of a subconductance state in RyR1 with a fractional value ~55% of the full conductance state, very close to that of Vejocalcin. Furthermore, Intrepicalcin stimulates Ca2+ release at an initial dose = 45.3 ± 2.5 nM, and depletes ~50% of Ca2+ load from skeletal sarcoplasmic reticulum vesicles.
Conclusions
We conclude that active recombinant Intrepicalcin was successfully obtained without the need of manual oxidation, enabling it to target RyR1s with high affinity.
General Significance
This is the first calcin heterologously expressed in the periplasma of Escherichia coli BL21-DE3, shown to be pharmacologically effective, thus paving the way for the generation of Intrepicalcin variants that are required for structure-function relationship studies of calcins and RyRs.
Keywords: Scorpion, Vaejovis intrepidus, Intrepicalcin, ryanodine receptor, sarcoplasmic reticulum, calcin
1. Background
Scorpions are fascinating animals existing on Earth for over 400 million years, allowing them to develop more than 2 200 (Santibáñez-López et al., 2015) different species with sophisticated venoms that have been playing a pivotal role for their survival and successful evolution, fundamentally, to protect themselves against predators, as well as to provide them with effective tools for capturing their preys (Dunlop and Selden, 2009; Jeyaprakash and Hoy, 2009). The venoms contain a molecular arsenal of different toxins that can be considered as a true combinatorial library of peptides with diversified biological activites such as anti-cancer (Dardevet et al., 2015), antimicrobial (Torres-Larios et al., 2000; Luna-Ramirez et al., 2013; Harrison et al., 2014), antiviral (Yan et al., 2011; Chen et al., 2012), antimalarial (Conde et al., 2000; Carballar-Lejarazu et al., 2008), immune-modulatory (Gurrola et al., 2012; Remijsen et al., 2010; Varga et al., 2012), anti-epilepsy (Wang et al., 2001) and bradykinin potentiating components (Almaaytah and Albalas, 2014). In fact, these toxins/peptides have been extensively exploited as a source of specific probes and potential therapeutic molecules for their targeting proteins, e.g. Na+, K+, Cl− and Ca2+ channels (Possani et al., 1999; Quintero-Hernandez et al., 2013).
The scorpion peptides that target sarcoplasmic reticulum (SR) Ca2+ release channel/ryanodine receptors (RyRs) are referred to as calcins (Xiao et al., 2016). They are structurally characterized by the presence of an Inhibitor Cystine Knot (ICK) motif, a folding that is commonly found in toxins affecting voltage-gated Ca2+ channels from spiders and snails (Narasimhan et al., 1994; Pallaghy et al., 1994), and thus setting calcins structurally apart from the Na+, K+ and Cl− channel scorpion toxins (Darbon, 1999). Calcins are also able to penetrate cell membranes (Estève et al., 2005; Schwartz et al., 2009). The founding member of the calcin family, Imperacalcin (previously called Imperatoxin A), was isolated from the venom of the African scorpion Pandinus imperator. Imperacalcin is a 33-amino acid peptide with ICK motif that binds with high affinity (~8 nM) to RyRs and increases their activity by inducing the appearance of a long-lived subconductance state. Other calcins including Maurocalcin from Scorpio maurus palmatus (Fajloun et al., 2000), Hemicalcin from Hemiscorpus lepturus (Shahbazzadeh et al., 2007), Opicalcin1 and Opicalcin2 from Opistophthalmus carinatus (Zhu et al., 2003), Hadrucalcin from Hadrurus gertschi (Schwartz et al., 2009), Urocalcin from Urodacus yaschenkoi (Luna-Ramirez et al., 2013; Luna-Ramirez et al., 2015), and Vejocalcin from Vaejovis mexicanus (Xiao et al., 2016), have been chemically synthesized and functionally characterized (Xiao et al., 2016). Intrepicalcin (ViCaTx1) from Vaejovis intrepidus (Quintero-Hernandez et al., 2015) was subsequently identified as member of the calcin family due to its high sequence similarity (>70%) and predicted ICK motif, but it has not been synthesized nor functionally characterized.
The identification of calcins like Imperacalcin, Hemicalcin and Hadrucalcin was based on classical biochemical methods (chromatographic separation combined with Edman degradation sequencing and mass spectrometry) and guided by pharmacological tests specific for RyRs ([3H]ryanodine binding and single RyR channel recording in lipid bilayers) (Valdivia et al., 1992; Zamudio et al., 1997). Other calcins including Opicalcin1, Opicalcin2, Urocalcin, Vejocalcin, and Intrepicalcin were identified by molecular cloning with either mRNA RACE or massive mRNA sequencing-transcriptomic analysis. Normally, the chemical synthesis of these calcins produces active peptides using the stardard synthesis and folding protocol (Xiao et al., 2016; Zamudio et al., 1997). Recombinant expression is an alternative way to generate competent calcins with correctly oxidized and uniformly folded ICK core (Seo et al., 2006) for immediate use after extraction from lysates of E. coli. In this study we report the results of heterologous expression of active Intrepicalcin without manual oxidation in the periplasma of Escherichia coli BL21 DE3, which was previously identified by transcriptomic analysis from the scorpion Vejovis intrepidus (Quintero-Hernandez et al., 2015). Recombinant Intrepicalcin was obtained by fusing the gene sequence coding for thioredoxin plus a short segment recognizable by enterokinase and a tag sequence of histidine. In addition to sequence alignment, bioinformatic analysis and three-dimensional modeling, the activity of recombinant Intrepicalcin to RyR1 was tested using [3H]ryanodine binding, single RyR recording in lipid bilayer, as well as Ca2+ release from heavy SR.
2. Materials and Methods
2.1 Construction of plasmid pET-Trx-Intrepicalcin
The gene sequence encoding Intrepicalcin was previously reported (Quintero-Hernandez et al., 2015). Four oligonucleotides, including preferential codons optimized for the codon usage of E. coli were designed to cover the entire gene and restriction sites that allow subsequent cloning into the expression vector: Up1-BHEK of 47 nt (5' GA GGA TCC CAC CAC CAC CAC CAC CAC GGT ACC GAT GAC GAT GAC AAG 3') with the BamHI restriction site (underlined), 6 histidines (italics) and the cleavage site of enterokinase enzyme (bold); Icalcin1-Rev of 54 nt (5' GTT TTT TTT GCA CAG TTT CAG GTG CGC CAG GCA ATC CGC CTT GTC ATC GTC ATC 3'), Icalcin2-Dir 53 nt (5' CTG TGC AAA AAA AAC AAA GAT TGC TGC TCT AAA AAA TGC TCT CGT CGT GGT AC 3') and finally the oligonucleotide Icalcin3-Rev of 49 nt (5' CA CTC GAG TTA ACG GCA ACG CTG TTC CGG GTT GGT ACC ACG ACG AGA GC 3') including the restriction site XhoI (underlined).
The PCR assembly of Intrepicalcin was performed using Vent DNA Polymerase (New England Biolabs, USA). The two external oligonucleotides Up1-BHEK and Icalcin3-Rev were used at a final concentration of 0.2 pmol/μL while the internal oligonucleotides Icalcin1-Rev and Icalcin2-Dir at a concentration of 0.02 pmol/μL. PCR conditions were performed as follows: 5 minutes at 94 °C, followed by 8 cycles at low annealing temperature (94 °C 30 sec, 50 °C 1 min, 72 °C 1 min) and then 25 short cycles with high annealing temperature (94 °C 30 sec, 60 °C 30 sec, 72 °C 30 sec), finalizing with 5 min incubation at 72 °C.
The PCR products, which were separated on agarose gel and purified with the kit High Pure PCR Product Purification Kit (Roche), were further cloned with blunt ends into the plasmid pJET (CloneJET™ PCR Cloning Kit, Fermentas) using the T4 DNA ligase. In the following, the ligation product was electrotransformed into electrocompetent E. coli DH5α that were subsequently plated in Petri dishes with the medium 2XYT. For the selection of positive clones, colony PCR was performed with the external oligonucleotides (Up1-BHEK and Icalcin3-Rev). Plasmid DNA of positive colonies (whose PCR product had the expected size) was extracted (High Pure Plasmid Isolation Kit, Roche) and further confirmed by automated sequencing.
The expression vector pET-Trx-Intrepicalcin (pET-Trx-Intrep) was constructed by cloning the thioredoxin gene (obtained from pThioHisC vector, Invitrogen) into the the plasmid pET-22b(+) (Novagen) between the restriction sites Nco I and BamHI, resulting the vector pET-22b-Thio, then the Intrepicalcin gene was cloning into pET-22b-Thio between the restriction sites BamHI and XhoI as follows.
The pJET plasmid carrying Intrepicalcin segment and the expression plasmid pET-22b-Thio were simultaneously digested with the restriction enzymes BamH I and Xho I. The digested expression vector pET-22b-Thio and the fragment encoding the Intrepicalcin were purified using agarose gel electrophoresis, which were further purified and ligated with the the High Pure PCR Product Purification Kit (Roche) and the enzyme T4DNA ligase (Fermentas) respectively. Colony PCR and automated sequencing were performed to finally confirm the built pET-Trx-Intrep.
2.2 Recombinant Intrepicalcin expression and purification
The pET-Trx-Intrep was then transformed by electroporation into E. coli BL21 (DE3) to express the fusion protein Trx-Intrep as follows. A preinoculum of E. coli BL21 (DE3) containing the pET-Trx-Intrep was grown in LB medium plus 200 μg/mL ampicillin, after 1 L of LB medium plus 200 μg/mL ampicillin was inoculated with the preinoculum and cultured at 37 °C with an agitation of 150 rpm (Orbital Shaker Incubator from Accesolab), and when the culture reached a value of optical density between 0.7-1 at 600 nm it was induced with 1 mM IPTG (Isopropyl β-D-1-thiogalactopyranoside) to express the fusion protein Trx-Intrep for a period of 12-16 hours at 30 °C. The pellet of E. coli was harvested by centrifugation at 8 000 rpm (Beckman Coulter Avanti J-30I centrifuge) for 10 min at 4 °C. The cellular pellet was then suspended in 25 mL of buffer PPB (200 mg/mL sucrose, 1 mM EDTA, 30 mM Tris-HCl, pH = 8) and incubated at 4 °C for 30 min before centrifugation for 20 min at 8 000 rpm. The pellet was re-suspended in 25 mL of a 5 mM solution of MgS04, incubated for 20 min at 4 °C, and centrifuged again at 8 000 rpm for 20 min at 4 °C, recovering the supernatant. Both supernatants (periplasmic extracts) were placed on nitrocellulose membranes and dialyzed against 1X PBS (1 L of PBS: 1.5 g of Na2HPO4, 0.2 g of KH2PO4, 8 g of NaCl and 0.2 g of KCl) buffer for 2 hours.
The expressed fusion protein Trx-Intrepicalcin was purified by affinity chromatography using the columns with Ni-NTA agarose resin (QIAGEN). Briefly, after the equilibration with the buffer 1X PBS plus 20 mM imidazole, pH = 8, the total volume of the periplasmic extract was loaded into the column and equilibrated with 1X PBS plus 35 mM imidazole. The fusion protein attached in the column was subsequently eluted with 1X PBS plus 250 mM imidazole. A HPLC purification was further performed using an analytical C18 (25 mm × 4.1 mm) reverse-phase column (Hysperia, CA), which was equilibrated with a buffer A (0.12% TFA in H20), and then eluted with a linear gradient from 0-35% of buffer B (0.1% TFA in acetonitrile) in 35 minutes with a flow rate of 0.8 mL/min under the monitoring at λ=230 nm.
The purified fusion protein was digested with the enzyme enterokinase (New England Biolabs) in 200 mM Tris HCl pH 8.0, 500 mM NaCl, 20 mM CaCl2 for a period of 16 h at 25 °C. The effectiveness of hydrolysis was analyzed by gel electrophoresis using Tricine SDS-PAGE with 12% acrylamide. Pure Intrepicalcin was finally isolated by HPLC as described above, which was running for 60 min instead of 35 min.
2.3 Determination by molecular mass and N-terminal sequencing
The fractions obtained after HPLC (both, the fusion protein and Intrepicalcin alone) were loaded into an ESI-MS, ESI LCQ FLEET spectrometer Thermo Scientific (San José California) with electrospray ionization system (ESI). The mobile phase consisted of 60 % acetonitrile, plus 1% acetic acid; it was injected at a flow rate of 10 μL/min. The spray voltage was 2.1 kV and the ion detection was made on positive mode. Spectrometric data were acquired manually using Thermo Xcalibur for data deconvolution. Molecular masses were determined as average mass considering the contribution of heavy isotopes.
Automatic sequencing determination by Edman degradation was performed using a PPSQ-31A Protein Sequencer from Shimadzu Scientific Biotech, Inc. (Columbia, Maryland, USA). The fraction (approximately 250 pmol) was adsorbed on Glass Fiber Disk TFA treated, distributed by Shimadzu.
2.4 Animals
Animal handling and care conformed to the Guide for Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH publication no. 85-23, revised 1996), and experimental protocols were approved by the local institutional ethical committee. The New Zealand White rabbits used in the experiments of this work were maintained in environmentally controlled rooms with cycles of 12 h light/dark and adequate water and food, where it was supervised daily for well-being by trained veterinary personnel. Back and leg skeletal muscle was obtained after i.p. injection of pentobarbital (100 mg/kg). Adequacy of anesthesia was determinated by pedal reflex and evaluation of jaw and muscle tone before proceeding with surgery (Schwartz et al., 2009; Xiao et al., 2016).
2.5 Rabbit skeletal heavy SR preparation
Heavy SR vesicles were obtained and purified from rabbit white back and leg skeletal muscle, as described previously (Meissner, 1984; Dyck et al., 1987). An estimate of the purity of “heavy” SR vesicles, was assessed by [3H]ryanodine binding assays (~10-15 pmoles/mg protein), which is in line with that reported by Meissner, the original author of this method (Meissner, 1984). The vesicles release equivalent amount of Ca2+ in response to caffeine, 4-cmc and A23187. Protein concentration was determined by the Bradford method.
2.6 [3H]ryanodine binding assay
[3H]Ryanodine binding to rabbit skeletal SR was carried out as described previously (Schwartz et al., 2009; Xiao et al., 2016). Briefly, Intrepicalcin was diluted directly into the incubation medium (10 μM CaCl2, 0.2 M KCl, 10 mM Na-HEPES, pH 7.2) to obtain a final concentration of 1 pM to 20 μM. To evaluate the effect of Intrepicalcin on Ca2+ sensitivity of [3H]ryanodine binding activity, a fixed concentration of Intrepicalcin at 100 nM was added to the standard incubation medium containing 0.2 M KCl, 1 mM Na2EGTA, 10 mM Na-PIPES (pH 7.2) and CaCl2 to set free [Ca2+] in the range of 10 nM to 10 mM. Ca2+/EGTA ratio was determinated with the computer program MaxChelator (http://www.stanford.edu/~cpatton/maxc.html). [3H]Ryanodine (95 Ci/mmol, PerkinElmer, USA) was directly diluted in the incubation medium to a final concentration of 5 nM. Protein concentration of heavy SR was 0.3 mg/mL and was calculated by Bradford method. The incubation lasted for a period of 120 min at 36 °C. Samples (100 μL) were always run by duplicate, filtered on Whatman GF/C glass filters (Whatman, Clifton, NJ, USA) and washed twice with 5 mL of distilled water using a Brandel M24-R cell harvester (Gaithersburg, MD, USA). Non-specific binding was determined in the presence of 20 μM unlabelled ryanodine, and has been subtracted from each sample.
2.7 Single channel recordings
Single-channel recordings of rabbit skeletal RyR (RyR1) incorporated into planar lipid bilayers were carried out as described previously (Schwartz et al., 2009; Xiao et al., 2016). Planar lipid bilayers, constituted by phosphatidylethanolamine and phosphatidylserine (Avanti Polar Lipids, Inc.) (1: 1), were painted with a glass rod across an aperture of 150 μm diameter in a Delrin cup. The cis chamber represented the cytosolic side, which was held at virtual ground and contained the reference electrode. The trans chamber corresponded to the luminal side and contained the voltage command electrode. Electrodes were connected to the head stage of a 200A Axopatch amplifier. Both the cis (0.7 mL) and trans (0.7 mL) chambers were filled with 300 mM caesium methanesulphonate and 20 mM MOPS, pH 7.2. Heavy SR vesicles (20-50 μg) were added to the cis chamber. Channel recordings were collected at a holding potential (+40 mV) before and after addition of Intrepicalcin. Single channel recordings were filtered with an 8-pole low-pass Bessel filter set at 2 kHz and digitized at a rate of 4 kHz by using a Digidata 1440A AD/DA interface. Data acquisition and analysis were performed with Axon Instruments (Burlingame, CA, USA) hardware and software (pClamp 10) and graphed by using Origin 9.0 (Microcal Inc., Northampton, MA, USA).
2.8 SR Ca2+ release measurements
Ca2+ release from heavy SR was determinated using the Ca2+-sensitive dye Arsenazo III (Sigma-Aldrich), modified as previously described (Gurrola et al., 1999; Xiao et al., 2016). Briefly, the absorbance was monitored at 650 nm by a Compact UV-visible spectrophotometer (Eppendorf, Biophotometer Plus). Heavy SR vesicles (20 μg) were actively loaded with Ca2+ at room temperature in a 1 mL basal buffer containing 7.5 mM sodium pyrophosphate, 100 mM KCl, 20 mM MOPS, pH 7.0, supplemented with 5 mM creatine phosphate disodium salt tetrahydrate (ICN Biomedicals Inc), 25 μM Arsenazo III, 1 mM ATP/MgCl2 (Sigma-Aldrich), and 12 μg/mL creatine phosphokinase (porcine heart, Calbiochem). Ca2+ loading was started by consecutive additions of 50 nmol twice and 20 nmol thrice of CaCl2 before the addition of Intrepicalcin. The total [Ca2+] loaded into the SR was quantified by the addition of the Ca2+ ionophore A23187 (5 μM) and the absorbance was transformed to nmoles [Ca2+] by a standard curve generated by consecutive additions of CaCl2 (10-20 μM).
2.9 Bioinformatics
Six calcin sequences (mature peptide and/or precursors) called, Imperacalcin (formerly Imperatoxin A) (P59868.1) (Zamudio et al., 1997), Maurocalcin (P60254.1) (Fajloun et al., 2000), Opicalcin1 (P60252.1) (Zhu et al., 2003), Opicalcin2 (P60253.1) (Zhu et al., 2003), Hadrucalcin (B8QG00.1) (Schwartz et al., 2009) and Urocalcin (AGA82762.1) (Luna-Ramirez et al., 2013) were identified using the Intrepicalcin sequence with BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The correct mature sequences were further confirmed from their original papers. The sequence of Hemicalcin, Intrepicalcin and Vejocalcin were obtained from their corresponding papers (Shahbazzadeh et al., 2007; Quintero-Hernandez et al., 2015; Xiao et al., 2016). Net charge vs. pH plots and hydrophobicity plots were determinated by the online peptide calculator (http://www.chinapeptides.com/tool.php?isCalu=1). Using Imperacalcin as the template (Lee et al., 2004), three-dimensional simulation was carried out by Swiss-pdbViewer 4.1.0 with the mutagenesis function following the minimum energy principle. The Discovery Studio 4.5 program was used to display the modeling molecules. The dipole moment (DM) was analyzed and determinated by the online Protein Dipole Moments Server (http://dipole.weizmann.ac.il/).
2.10 Statistics
Data are presented as the mean ± SEM. Statistical analyses were performed using t-test. Differences of P < 0.05 were considered significant.
3. Results
3.1 Design and generation of Intrepicalcin gene, expression and purification
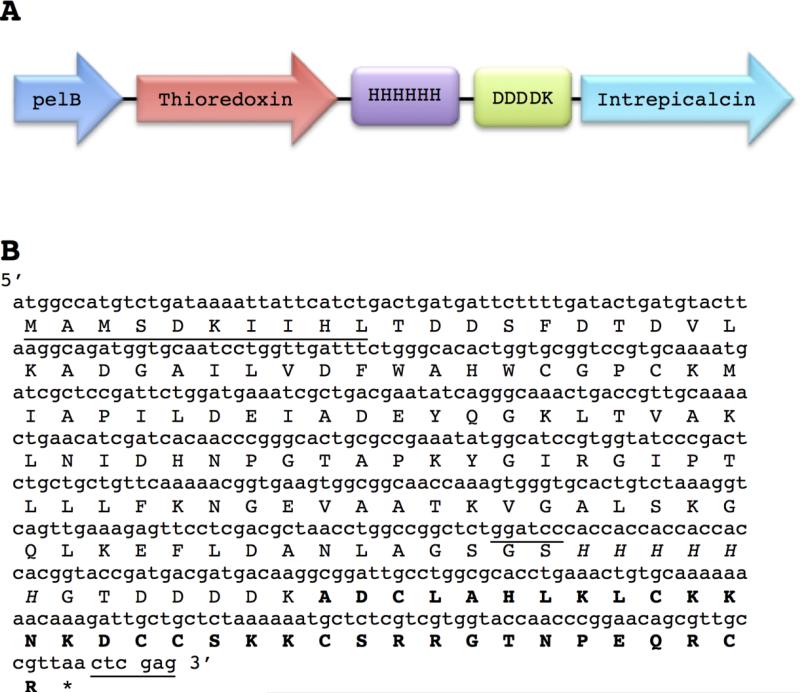
The sequence that encodes Intrepicalcin was previously found in a transcriptomic analysis of scorpion venom gland from Vaejovis intrepidus (Quintero-Hernandez et al., 2015). Based on this sequence, four oligonucleotides were designed and optimized with the preferential codon usage of E. coli (see Materials and Methods), which allowed the cloning of Intrepicalcin fused to thioredoxin (Trx) plus 6 histidines and a cleavage site for enterokinase (Figure 1). In addition, the sequence encoding the fusion protein Trx-Intrepicalcin was cloned at the 5′ end of the signal peptide sequence, pelB, which is contained in the pET-22b (+) plasmid (Fig. 1A). PelB allows the export of the fusion protein to the periplasm of the bacteria. The recombinant expression of Intrepicalcin fusion protein was induced with IPTG and collected from the periplasmic extract as described in Materials and Methods.
Figure 1.
Design of the fusion protein and nucleotide sequences (A) The fusion protein starts with a signal peptide sequence (PelB) that allows exporting the fusion protein to the periplasm of E. coli BL21 DE3, followed by the gene that codes for the thioredoxin protein, plus a histidine tag, a cleavage site for enterokinase enzyme and finally the gene encoding Intrepicalcin. (B) Nucleotide and amino acids sequences corresponding to the elements described in (A) are shown. The first ten amino acids of thioredoxin protein are underlined followed by the remaining sequence of thioredoxin, the histidine tag (in italics) and the five amino acids (DDDDK), of the cleavage site of enterokinase, which is just before the Intrepicalcin sequence (in bold). The nucleotides for BamH I and Xho I sites are also underlined.
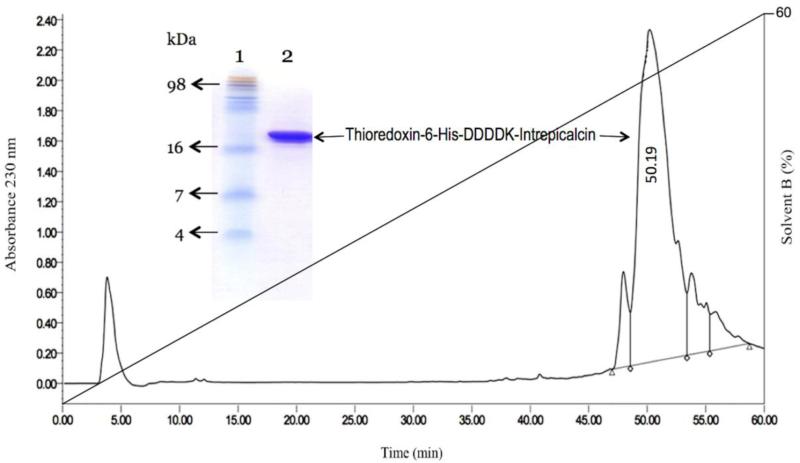
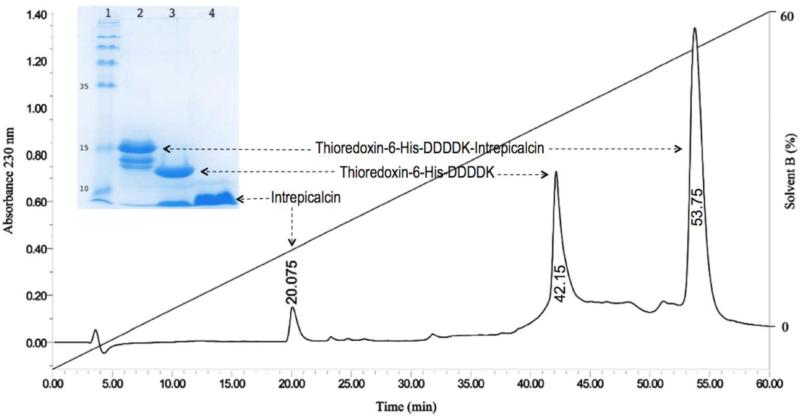
Since the fusion protein contains 6 histidines (Fig. 1), it allows the purification by affinity chromatography. Figure 2 displays the SDS-PAGE electrophoresis results, where a band with the expected molecular weight of the fusion protein is clearly displayed. The molecular weight of the protein as determined by mass spectrometry was 17660.1 Da, as expected. Further confirmation was obtained by N-terminal sequencing, which tested the first 10 amino acid residues corresponding exactly to the expected N-terminal sequence (Fig. 1B). This fusion protein was digested with enterokinase followed by purification with HPLC (Fig. 3). The peptide eluting at 20.07 min corresponds to pure Intrepicalcin. Its molecular weight determined by mass spectrometry (3789.5 Da) is practically the same (within experimental error) as the theoretical molecular weight expected for folded Intrepicalcin (3788.4 Da). The yield of pure fusion protein was 7.2 mg per liter of culture and the amount of pure intrepicalcin was 0.35 mg (after digestion of the fusion protein). The production process was not optimized and a higher yield can be obtained by modifying the conditions of expression and purification of intrepicalcin.
Figure 2.
Expression and purification of Intrepicalcin fusion protein. After His affinity chromatography, the Intrepicalcin fusion protein (Thioredoxin-6-His-DDDDK-Intrepicalcin) was further purified by HPLC. In this figure the HPLC profile of fusion protein is showed (Retention time = 50.19 min). SDS-PAGE analysis of the fusion protein purified by HPLC is indicated as follow: Lane 1: molecular mass marker; lane 2: Purified fusion protein after HPLC.
Figure 3.
Release and purification of Intrepicalcin from fusion protein. After the hydrolysis by enterokinase, Intrepicalcin was released from Thio-His-Intrepicalcin fusion protein and purified by HPLC. The fraction elution time at 20.07 min, 42.15 min and 53.75 min correspond to the released Intrepicalcin, Thiorredoxin-6 His-DDDK and undigested fusion proteins, respectively. The collected fractions were further identified by SDS-PAGE. Lane 1: molecular mass marker; lane 2: The complete fusion protein purified by HPLC; lane 3: fusion protein cleaved by Enterokinase; lane 4: Intrepicalcin purified by HPLC after cleavage step.
This result confirmed that the expression system used and the design of the fusion protein was successful for production of recombinant Intrepicalcin. This procedure allows a determined folding of the recombinant peptide resulting an active calcin. Using this system avoids the production of several isoforms as discussed before (Estrada et al., 2007; Saucedo et al., 2012; Cao et al., 2015). The use of thioredoxin as fusion protein certainly favored the folding of the disulfide bonds because it displays chaperone activity, which provides high stability to the protein with which is fused (LaVallie et al., 1993; Kern et al., 2003). This thioredoxin activity coupled with redox conditions of the E. coli periplasm, where the complex is exported from the cytoplasm, is important for expression of the folded and active Intrepicalcin (Fig. 3).
3.2 Intrepicalcin sequence alignment and bioinformatics
Figure 4A shows the amino acid sequence of all peptide members of the calcin family known to date, aligned with Intrepicalcin as template. All calcins contain 33 amino acids except Hadrucalcin with additional Ser and Glu in its N-terminal for a total of 35 residues. Six cysteines are highly conserved to form three disulfide bonds (Cys3-Cys17, Cys10-Cys21 and Cys16-Cys32) as experimentally determined in Maurocalcin (Mosbah et al., 2000) and Imperacalcin (Lee et al., 2004), contributing significantly to the ICK motif that characterizes this family of peptides. Accordingly, Intrepicalcin and other calcins contain three β strands, 8XXC10, 20XCX22 and 30XXCX33, which include three cysteines (Cys10, Cys21 and Cys32, respectively). As expected from the closeness of the scorpion genealogy, Intrepicalcin from Vejovis intrepidus and Vejocalcin from Vejovis mexicanus differ by only one amino acid (Lys and Asn, respectively, at position 14), displaying 97% similarity overall. More divergent from Intrepicalcin in amino acid sequence, Maurocalcin/Hemicalcin, Hadrucalcin/Opicalcin1, Opicalcin2 and Urocalcin show five, six and seven different residues, respectively, with similarities 81.8%, 78.8% and 75.8%, respectively. Interestingly, the first calcin member, Imperacalcin, displays the lowest (69.7%) sequence identity with Intrepicalcin, with ten different residues, indicating that these two calcins share the farthest evolutionary relationship among all calcins.
Figure 4.
Intrepicalcin sequence alignment and bioinformatics (A) Sequence alignment of the Intrepicalcin (Genbank accession: JZ818387), with the sequences of other eight calcins: Vejocalcin (Xiao et al., 2016), Maurocalcin (UniProt: P60254), Hemicalcin (Shahbazzadeh et al., 2007), Hadrucalcin (UniProt: B8QG00), Opicalcin1 (UniProt: P60252), Opicalcin2 (UniProt: P60253), Urocalcin (UniProt: AGA82762) and Imperacalcin (UniProt: P59868). The identity values to Intrepicalcin are shown in the right side. Positively-charged residues Lysine (K) and Arginine (R), negatively charged residues Aspartic acid (D) and Glutamic acid (E), and the disulfide bond-forming Cysteine (C) are highlighted by the colors blue, red and grey, respectively. Columns with identical residue are marked by asterisk while columns with only one difference are marked by colon in the bottom. Three pairs of highly conserved disulfide bridges (Cys3-Cys17, Cys10-Cys21 and Cys16-Cys32) and the residues forming part of β-strands appear connected to form an ICK motif. (B) Net charge vs. pH plot and Hydrophobicity plot of Intrepicalcin. (C) Three dimensional modeling of Intrepicalcin simulated by Swiss-PdbViewer 4.1.0 and viewed by Discovery Studio 4.0 according to the three-dimensional structure of Imperacalcin resolved by 1H-NMR (Lee et al., 2004). The solid ribbon with disulfide bridge is located in the left and the charged CPK model with “frontal” side (middle) and “dorsal” side (right) are also displayed. Positively charged residues (Lys and Arg), negatively charged residues (Asp and Glu), and neutral residues are colored by blue, red and grey, respectively.
Intrepicalcin conforms to the common rule of amino acid composition and charge distribution that characterizes the calcins (Fig. 4A). The number of positively-charged residues Lys and Arg in Intrepicalcin is 10 (30%), which is among the smallest number in this group of peptides, only higher than that of Vejocalcin (9 basic residues, 27%). Moreover, they share the smallest number (3, 9%) and the same sparse distribution of negatively-charged residues Asp and Glu. Similar to other previously-characterized calcins, the high positive/negative amino acid ratio gives Intrepicalcin high isoelectric point (pI = 9.7) and net positive charge = 6.8 at pH 7.0. Due to the abundance of charged residues overall, the solubility of Intrepicalcin in water is excellent (hydrophilicity = 55%) (Fig. 4B).
We also modeled the three-dimensional structure of Intrepicalcin based on the atomic coordinates of Imperacalcin, whose structure was previously solved by 1H-NMR (Lee et al., 2004). As observed in Fig. 4C, Intrepicalcin is predicted to be a coiled, compact globular structure that embeds an ICK motif featured by the presence of three disulfide bridges interspaced by sections of β strand-forming polypeptides. The “frontal” side of Intrepicalcin presents the majority of the positively-charged residues that form part of clusters 8KLCKK12, 19KKCSRR24 and 31RCR33, however, at variance with the rest of calcins, its “dorsal” side presents two extra basic residues (K12 and K14) and therefore exhibs the smallest charge segregation in this family of peptides, with dielectric moment (DM) ≈ 30 D, even smaller than that of Vejocalcin (DM = 40 D).
3.3 Intrepicalcin stimulates [3H]ryanodine binding
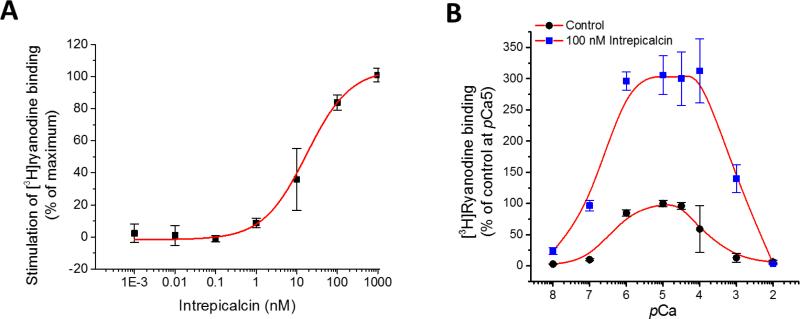
[3H]Ryanodine binds exclusively to the open conformational state of the RyR and therefore, it may be used as an index of the activity of the channel. In a recent study, Xiao et al. (2016) chemically synthesized all eight calcins known and compared side-by-side their properties to stimulate [3H]ryanodine binding. All calcins stimulated [3H]ryanodine binding to RyR1, but also as expected from their unique amino acid sequence, they exhibited different affinity (apparent Kd) and potency (maximal stimulation as percent of control). The apparent Kd surprisingly spanned ≈3 orders of magnitude with the ranking order (in nM): Opicalcin1 0.3±0.04 > Opicalcin2 3.2±0.4 > Vejocalcin 3.7±0.4 > Hemicalcin 6.9±0.7 > Imperacalcin 8.7±0.9 > Hadrucalcin 14.8±1.9 > Maurocalcin 26.4±3.9 >> Urocalcin 376±45 (Xiao et al., 2016). We tested the capacity of Intrepicalcin to stimulate [3H]ryanodine binding at pCa5. Figure 5A shows that Intrepicalcin (0.1 nM to 1 μM) is indeed capable of enhancing [3H]ryanodine binding to skeletal SR vesicles with an apparent Kd = 17.4 ± 4.0 nM. This affinity places Intrepicalcin roughly at the middle of the ranking order for calcins, very similar to Hadrucalcin and Maurocalcin. It is worth noting that Intrepicalcin, which differs by only one amino acid with Vejocalcin (N14 is replaced by K14), displays a ≈ 4.7-fold difference in apparent Kd, much lower than those mutations taken place in other basic residue clusters 19KKCSRR24 and 31RCR33.
Figure 5.
Stimulation of [3H]ryanodine binding to RyR1 by Intrepicalcin (A) Dose-dependent activation of Intrepicalcin on RyR1 between 1 pM and 1 μM (n=3). Heavy SR from rabbit skeletal muscle were incubated with 5 nM [3H]ryanodine in the absence (control) and the presence of indicated concentrations of Intrepicalcin. Binding conditions were specified in Methods. The Kd was determined with the formula B = (Bmax)/[1 + (Kd/[Intrepicalcin])nH), where B is specific binding of [3H]ryanodine, Bmax is the maximum binding stimulated by Intrepicalcin, and nH is the Hill coefficient. (B) Ca2+ sensitivity of [3H]ryanodine binding affected by Intrepicalcin at 100 nM (n=3). The specific [3H]ryanodine binding was standardized with the value of the control at pCa5 as 100%.
We also tested the effect of Intrepicalcin over a wide range of [Ca2+] to determine how it affects the Ca2+-dependent activation and inactivation of RyR1 (Fig. 5B). Specific binding in the absence of Intrepicalcin had a threshold for detection at pCa 7 and was maximal at pCa 6-4.5. Higher [Ca2+] decreased binding. This dual effect of Ca2+ gave rise to the well-known bell-shaped curve that is similar to the Ca2+ dependence of open probability for skeletal RyRs. In the presence of 100 nM Intrepicalcin, the binding curve was also bell-shaped but was dramatically augmented in absolute values, consistent with previous studies on other calcins. At pCa8, [3H]ryanodine binding was already stimulated by Intrepicalcin from 2.9 ± 0.2% (control) to 23.4 ± 5.6% (Intrepicalcin). At pCa 7 and 6, the Intrepicalcin-stimulated binding values were elevated from 10.1 ± 0.2% and 84.6 ± 5.1% to 96.6 ± 8.3% and 296.4 ± 14.6%, respectively. The Intrepicalcin-stimulated binding at pCa6 to 4 exhibit a maximal value close to ~300% compared to that of control at pCa5 that is normalized at 100%. This is characteristic of most calcins, which exhibit a maximal binding stimulation between 300-500% under the conditions of this assay. Moreover, the stimulation rapidly decreases when pCa is higher than 4, and reaches control value (no Intrepicalcin) at pCa2.
3.4 Effect of Intrepicalcin on single RyR1 channels
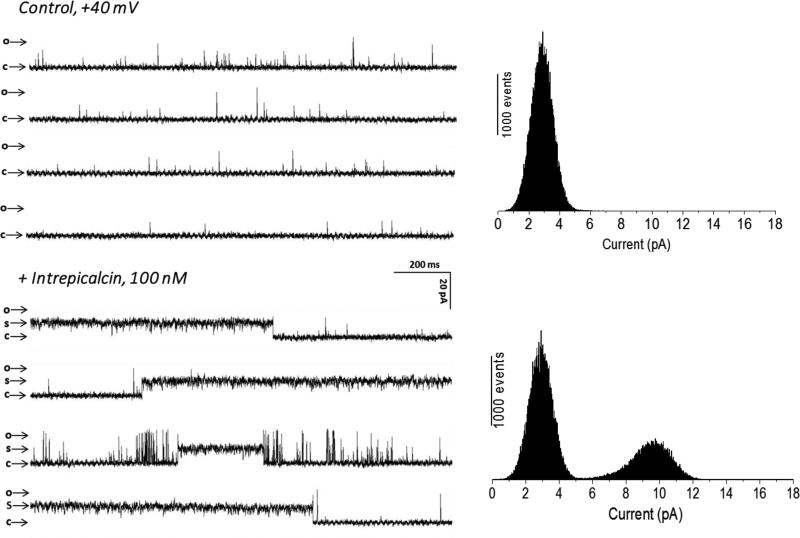
To further investigate the functional properties of Intrepicalcin, we reconstituted RyR1 channels in planar lipid bilayers and determined the effect of Intrepicalcin on single channel gating. Because the probability of calcin binding to RyR1 is Po- and voltage-dependent (Tripathy et al., 1998), we conducted experiments at “Po-clamped” conditions (by fixing [Ca2+] at quasi-optimal levels) and positive holding potential (+40 mV), which favors the natural (lumen to cytosol) flow of ions through RyRs. Our main goal here was to check if Intrepicalcin is able to induce the characteristic long-lasting subconductance of RyR1, and further compare the amplitude of the subconductance state induced by other calcins under identical experimental conditions previously decribed by ourselves (Xiao et al., 2016). Figure 6 displays recordings of RyR1 in the absence and presence of Intrepicalcin. Under these conditions, control (no Intrepicalcin) RyR1 channel recordings showed the characteristic fast-flickering (mean open time 1-3 ms), low Po (0.03 - 0.15) and high-conductance (≈600 pS) gating, with occasional sojourns to native subconducting states that are barely noticeable in the current amplitude histogram (Fig. 6A). Addition of 100 nM Intrepicalcin induced a small increase in the duration of open events, but the most conspicuous effect was the appearance of the ‘signature’ event, a long-lived subconducting state, which corresponded to ~55% of the full-conductance openings. This subconductance state is similar to that induced by Vejocalcin, Urocalcin and Maurocalcin (Fajloun et al., 2000), while higher than that induced by Imperacalcin (Tripathy et al., 1998), Hadrucalcin (Schwartz et al., 2009), Opicalcin1 and Opicalcin2 (Xiao et al., 2016).
Figure 6.
Functional effects of Intrepicalcin on single RyR1 acitvity. RyR1 single channel was incorporated by inducing fusion of skeletal muscle heavy SR vesicles into lipid bilayers. The channel activities were recorded and analyzed as described in Methods. RyR1 single channel in the absence of Intrepicalcin was used as control. Intrepicalcin was added into the cis chamber. The arrow marking ‘c’ shows the zero current level when the channel is in the fully closed state, and the arrow marking ‘s’ shows the Intrepicalcin-stabilized subconductance state of the channel. Current histograms from 10,000–30,000-ms segments of channel activity are shown at the right side of the traces for control or 100 nM Intrepicalcin. Experiments were tested at least three times.
3.5 Ca2+ release from heavy SR
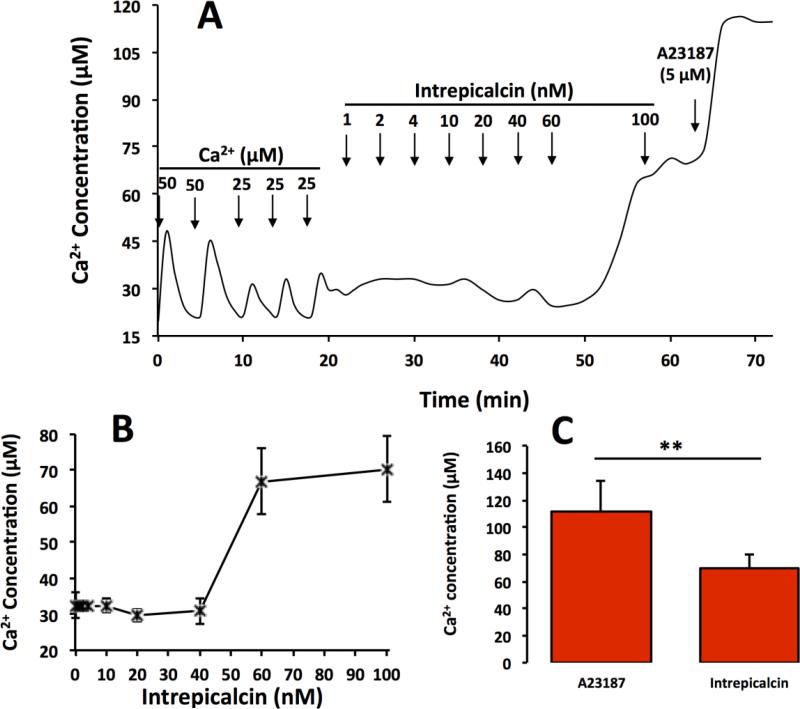
We also tested the capacity of Intrepicalcin to induce Ca2+ release from rabbit skeletal heavy SR vesicles (Fig. 7), another test that is well suited to determine the effect of calcins on RyRs. SR vesicles were loaded with two consecutive additions of 50 μM Ca2+ followed by 25 μM Ca2+ thrice. Each bolus addition of Ca2+ was actively taken up by the SR vesicles and by the 5th bolus extra-vesicular [Ca2+] increased to ≈30 μM, indicating that the vesicles in the system were loaded to full capacity. We then added Intrepicalcin and tested its ability to elicit Ca2+ release. Figure 7A shows a typical trace in which cumulative addition of Intrepicalcin elicits gradual Ca2+ release from SR vesicles. Addition of 1 nM to 40 nM Intrepicalcin barely modifies extravesicular [Ca2+], but the bolus of Intrepicalcin that increases its concentration to 60 nM elicits sharp Ca2+ release, and further Intrepicalcin additions (~100 nM) yield little additional release (Fig. 7A). Addition of the Ca2+ ionophore A23187 (5 μM) to the reaction mixture indicated that Intrepicalcin released a fraction, only, of the total amount of Ca2+ trapped into the SR vesicles. Figure 7B shows the capacity of Intrepicalcin to release Ca2+ in a sharp, stepwise manner once a critical concentration has been reached. The threshold for sharp Ca2+ release by Intrepicalcin is 45.3 ± 2.5 nM. Figure 7C shows that the total Ca2+ released by 100 nM Intrepicalcin is 48.2±4.1%. Thus, Intrepicalcin induces Ca2+ release in a manner that is common of other calcins (Xiao et al., 2016).
Figure 7.
Intrepicalcin-induced Ca2+ release from skeletal heavy SR vesicles (A) Typical trace of Ca2+ release by Intrepicalcin. 60 nM Intrepicalcin increased the Ca2+ release from rabbit skeletal heavy SR. (B) Stastistics on the increase of Ca2+ release by Intrepicalcin 100 nM. (C) At 100 nM concentration, Intrepicalcin caused Ca2+ release from rabbit skeletal heavy SR, as high as approximate 70% (n=3).
4. Discussion
In this study, we generated a biologically active, pharmacologically-competent recombinant calcin (Intrepicalcin), which was obtained by fusing the intrepicalcin gene to the sequence coding for thioredoxin plus a His tag and a cleavage site recognized by the enterokinase enzyme. Recombinant Intrepicalcin enhances the binding of [3H]ryanodine to RyR1s with high affinity (17.4 nM, Figure 5A), and activates RyR1s across a broad range of [Ca2+], as suggested by the [3H]ryanodine binding experiments (Figure 5B). Intrepicalcin induces the appearance of a subconductance state in RyR1s that increases ion flow per unit time (Figure 6) and induces Ca2+ release from isolated SR vesicles (Figure 7). All these properties strongly suggest that recombinant Intrepicalcin is correctly folded and that its three disulfide bonds form a conformationally integral ICK core, thereby targeting RyR1 channels with high affinity and exquisite selectivity.
4.1 Expression of Recombinant Intrepicalcin
Scorpion venoms are rich sources of peptide toxins that target various voltage- and ligand-gated channels including RyRs, although the use of such toxins to study the structure and function of ion channels has been hampered by the lack of accessible, steady and abundant sources of these natural toxins. Chemical synthesis, the most common method to produce calcins in vitro, produces correctly folded peptides that display biological activity indistinguishable from native calcin (Zamudio et al., 1997; Xiao et al., 2016). Imperacalcin was reported to be produced as a fusion protein with the coding sequence fused to the C-terminal portion of the T7 gene9 protein in E. coli (Seo et al., 2006). Activity tests confirmed that recombinant Imperacalcin induced voltage- and concentration-dependent subconductance states in skeletal RyRs, indicating that recombinant Imperacalcin was identical to synthetic as well as native Imperacalcin. Even then, raw extract of recombinant Imperacalcin had to be manually oxidized after purification from the lysate of E. coli to form the canonical ICK core (Seo et al., 2006). To solve this disadvantage, we designed here a heterologous expression system for Intrepicalcin. pET-22b-Thio was used as the expression plasmid that enabled to traffic the recombinant Intrepicalcin to the periplasmic space with the facilitation of the pelB signal sequence in this plasmid. A short segment with His tag and enterokinase cleavage site was also included to favor the purification and release of Intrepicalcin from the fusion protein without extra residues. All the recombinant Intrepicalcin was directed to the periplasmic space under the help of pelB signal peptide and thioredoxin. In the periplasm, the intrepicalcin is folded by the action of thioredoxin, which is a chaperone of intramolecular folding, and due to the oxidizing environment of the periplasmic space which favors the formation of disulfide bridges in the intrepicalcin recombinant (which was proved by the molecular mass determination). It is likely that under these conditions only mature and properly folded Intrepicalcin is obtained, which would endow this method with considerable advantages since the recombinant Imperacalcin of Seo et al. (2006) expressed in the cytosol likely assumed different conformational states and had to undergo disulfide oxidation before obtaining pure and correctly folded Imperacalcin (Seo et al., 2006). In addition to eliminating the step of disulfide oxidation, the methodology used here for recombinant Intrepicalcin may also be readily applied for generation of the calcin variants required for comprehensive structural-functional studies of this family of peptides.
4.2 Structural features of Intrepicalcin
Primary sequence alignment and evolutionary analysis previously revealed a very close relationship among calcins, yet their natural variability yields peptides with binding affinity spanning ≈3 orders of magnitude, highlighting the importance of discrete domains in their interaction with RyR1 channels. In the linear sequence, calcins can be grossly split into the variable N-terminal segment 1G-N14 with the basic cluster1 8KRCR11, whereas the C-terminal segment 15D-R33, comprising the highly positively-charged motifs 19KKCKRR24 (cluster 2) and 30KRCR33 (cluster 3), is relatively conserved among calcins (Ramos-Franco and Fill, 2016; Xiao et al., 2016). In addition to the signature ICK motif folding promoted by the three highly-conserved disulfide bonds, a marked anisotropy of electrostatic charge distribution, where most of the positively-charged residues (cluster 1, 2 and 3) are segregated in one end of the molecule, presents a large functional surface area to interact with the RyRs (Mosbah et al., 2000; Lee et al., 2004) and generates a discrete DM. Intrepicalcin is a novel calcin identified by transcriptome analysis from the scorpion Vaejovis intrepidus, which is closely related to Vaejovis mexicanus that generates Vejocalcin. Intrepicalcin and Vejocalcin differ by only one amino acid (K14 in Intrepicalcin, the most common fourthteen residue in the calcin family, is substituted by N14 in Vejocalcin). It seems that calcins with N14 display larger DMs including Imperacalcin (152 D), Opicalcin1 (172 D) and Opicalcin2 (175 D), whereas calcins with K14 have smaller DMs like Maurocalcin (126 D), Urocalcin (102 D), Hemicalcin (88 D) and Hadrucalcin (65 D). This tendency is perhaps due to the spatial distribution of the fourteenth residue in the ‘dorsal’ side of calcins, which is opposed to the functional surface of basic residues in the ‘frontal’ side and thus decreases the value of DMs. One exception is Vejocalcin that has the amino acid N14 but displays the second smallest DM value (40 D), because the two neutral mutations K22S and K30Q attenuate considerably the positively-charged functional surface of the ‘frontal’ site. The only variation between Intrepicalcin and Vejocalcin (N14K) substantially decreases DM from 40 to ~30, convincingly arguing that K14 is at least one of the natural determinants of the DM of calcins in the ‘dorsal’ or noninteracting surface of this group of peptides.
4.3 DMs and [3H]ryanodine binding assay
In principle, calcins with larger DMs may imply larger surface area to interact with RyRs, therefore DM could be roughly related to stronger binding affinity. In agreement with this assumption, Opicalcin1, Opicalcin2 and Imperacalcin that have DMs 172 D, 175 D and 152 D, respectively, display strong binding affinities (0.3 nM, 3.2 nM and 8.7 nM, respectively) whereas Maurocalcin, Hadrucalcin and Urocalcin that have relatively smaller DMs (126 D, 65 D and 102 D, respectively) display weaker binding affinities (26.4 nM, 14.8 nM, and 376 nM, respectively) (Xiao et al., 2016). However, notable exceptions to this rule undermine the DM-binding affinity relationship. Vejocalcin display the second smallest DM (40 D) because of the two neutral mutants K22S and K30Q in the functional surface but also displays a strong binding affinity (3.7 nM), similar to that of Opicalcin2 (3.2 nM). On the other hand, Hemicalcin has a small DM (88 D) but shows strong binding affinity (6.9 nM), even stronger than that of Imperacalcin (8.7 nM) which displays a larger DM (152 D) (Xiao et al., 2016). When compared to Vejocalcin, the basic mutation N14K from Intrepicalcin structurally distributed in the non-interactive ‘dorsal’ side not only decreases the DM from 40 D (Vejocalcin) to ≈30 D (Intrepicalcin), but also decreases binding affinity 4.7-fold, from 3.7 nM (Vejocalcin) to 17.4 nM (Intrepicalcin). The decrease in binding affinity is somewhat inconsistent with previous reports, specifically, alanine scanning of Imperacalcin reported that the mutation N14A showed almost no influence on binding affinity (Lee et al., 2004), further supporting the notion that the fourteenth residue of calcins is not critical for functional calcin-RyR1 interactions. However, our data here show that substitution of the basic residue (K14 to neutral residue N14) is able to affect the calcin-RyR1 interactions, albeit weakly. This could force a modification to the notion that the “dorsal” side of calcins is not critical for calcin-RyR1 interactions, but more nuances have to be considered, for example, the natural substitution described here is from a positively-charged to a neutral residue (K → N), instead of the neutral-to-neutral residue (N → A) substitution studied in the alanine scanning report (Lee et al., 2004).
4.4 Single channel effect and mechanism
The reversible, concentration- and voltage-dependent, long-lasting subconductance state constitutes the “signature” effect of calcins on RyRs, and this substate has been measured up to now for eight calcins (Tripathy et al., 1998; Gurrola et al., 1999; Fajloun et al., 2000; Chen et al., 2003; Shahbazzadeh et al., 2007; Schwartz et al., 2009), with fractional values of the full conductance state corresponding from 0.2 to 0.6, respectively. Here we report for the first time the fractional conductance of the newly identified Intrepicalcin.
In our experiments, Intrepicalcin induced a subconducting state most similar to that of Vejocalcin (~0.55 of the full conductance value), perhaps not fortuitously, since these two calcins differ by only one amino acid (N14K). But besides the subtly different subconducting value, all calcins known to date induce in RyR1 the same kinetic modifications (fast, reversible, long-lasting subconducting state) that indicate that they all share a common binding site. In this regard, the structural (Samsó et al., 1999) and the functional (Tripathy et al., 1998) data yield apparently conflicting results.
Using cryo-electron microscopy and 3-dimensional single-particle image analysis, Samsó et al. (1999) found that an Imperacalcin-biotin-streptavidin complex bound to a cytoplasmic crevice in each RyR1 monomer (four binding sites per channel tetramer), which is located near the calmodulin-binding site and far (<11 nm) from the center of the transmembrane region of the channel (Samsó et al., 1999). In contrast, using single RyR1 channel analysis, Tripathy et al (1998) concluded that Imperacalcin enters the channel vestibule and binds to a single site located within the ion conduction pathway and close to the voltage drop of the channel. While it is possible that both conclusions are correct because Imperacalcin, like Peptide A, display multiple effects on RyRs that may reflect more than one binding site (Dulhunty et al., 2004), the subconducting state described here is in line with the single binding site proposed by Tripathy et al (1998), and we are conducting cryo-EM experiments at diferent concentrations of Imperacalcin to resolve this issue.
4.5 Ca2+ release from heavy SR
Previous studies showed that calcins induce Ca2+ release at concentrations that are crudely correlated with their [3H]ryanodine binding affinity, with Opicalcin1 and Urocalcin displaying the strongest and weakest capacity, respectively, to stimulate both [3H]ryanodine binding and Ca2+ release. But the correlation was not sensitive enough to discriminate flawlessly for calcins of medium affinity, for example, Vejocalcin, Opicalcin2 and Hemicalcin show strong binding affinity but weaker ability to stimulate Ca2+ release (Xiao et al., 2016). Similar to the tendency of Vejocalcin in this study, Intrepicalcin displays binding affinity = 17.4 nM, very close to that of Hadrucalcin (14.8 nM), but has weaker capacity to trigger Ca2+ release from SR (45.3 nM for Intrepicalcin vs. 11.8 nM for Hadrucalcin). Moreover, the Intrepicalcin-induced Ca2+ release is consistently a fraction (48.2%) of the total SR Ca2+ load, and a residual amount of Ca2+ constantly remains in the SR vesicles even at saturating [Intrepicalcin] (Fig. 7).
This phenomenon of unreleased Ca2+ seems to be common to all calcins, and the mechanism may be related to both the RyR1-unttached SR and the partial induction of Ca2+ release by calcins. The former is not sensitive to calcins but can be released by A23187 while the latter can be deduced from the different values of Ca2+ release from calcins, which should be in balance between Ca2+ release with partial opening of RyR1 by calcins and Ca2+ reuptake by Ca2+-pump.
Caffeine and 4-cmc, two agonists of RyRs, release roughly equivalent levels of Ca2+ in these vesicles as that emptied by A23187 (not shown), indicating that RyR-insensitive stores do not contribute significantly in this assay. Thus, it is likely that the partial release of Ca2+ by Intrepicalcin (and other calcins, see Xiao et al, 2016) is a characteristic borne out of the intrinsic property of these peptides to induce a subconducting state. In support of this hypothesis, the level of Ca2+ release induced by the calcins in the same vesicles varies with the level of calcin-induced subconductance state (Xiao et al 2016). Although the relationship is not completely linear because the dwell time of the calcins in the RyR1 also influences the amount of Ca2+ release, these data strongly suggest that the partial emptying of Ca2+ is a property common to all calcins.
5. Conclusions
In summary, we have heterologously expressed and purified recombinant intrepicalcin in the periplasm of E. coli by fusing the Intrepicalcin gene to the sequence coding for thioredoxin plus a His tag and the cleavage site of enterokinase. Recombinant Intrepicalcin is able to stimulate [3H]ryanodine binding, induce a long-lasting subconductance state confirmed by using single channel recordings in lipid bilayers and trigger Ca2+ release from skeletal SR, in general indicating that biologically-active and folded recombinant Intrepicalcin without extra manual oxidation was successfully obtained. The expression system of recombinant Intrepicalcin described in this study may also be applied to generate peptide variants required for structural and functional investigations between calcins and RyRs.
Highlights.
Active recombinant Intrepicalcin was obtained in the periplasma of E. coli BL21 DE3
Recombinant Intrepicalcin activates skeletal RyR (RyR1) with Kd = 17.4 ± 4.0 nM.
Intrepicalcin induces the appearance of a subconductance state in RyR1
Intrepicalcin stimulates Ca2+ release at an initial dose = 45.3 ± 2.5 nM
Acknowledgments
The authors acknowledge the assistance of Dr. Fernando Zamudio for determination of molecular mass by spectrometry and amino acid sequence by Edman degradation and technical assistance on computer programming performed by M. Sc. Juan Manuel Hurtado Ramírez. Leonel Vargas Jaimes received a scholarship (number 287191) from the National Council of Science and Technology (CONACyT). This work was partially supported by grants from DGAPA-UNAM (IN203416) to LDP and grants from the National Institute of Health, USA (R01-HL055438 and R01-HL120108) to HHV.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Leonel Vargas Jaimes was a master thesis student at the School of Sciences, Universidad Autónoma del Estado de Morelos, Cuernavaca, Morelos 62209, México and Veronica Quintero-Hernández is presently a CONACYT Research Fellow.
References
- Almaaytah A, Albalas Q. Scorpion venom peptides with no disulfide bridges: a review. Peptides. 2014;51:35–45. doi: 10.1016/j.peptides.2013.10.021. [DOI] [PubMed] [Google Scholar]
- Cao Q, Lu W, Cai X, Hu C, Wang C, Ye J, Yan H, Yang Y, Wang Z, Huo J, Liu Y, Yu Y, Ling C, Cao P. In vitro refolding and functional analysis of polyhistidine-tagged Buthus martensii Karsch antitumor-analgesic peptide produced in Escherichia coli. Biotechnology letters. 2015;37:2461–2466. doi: 10.1007/s10529-015-1936-8. [DOI] [PubMed] [Google Scholar]
- Carballar-Lejarazu R, Rodriguez MH, de la Cruz Hernandez-Hernandez F, Ramos-Castaneda J, Possani LD, Zurita-Ortega M, Reynaud-Garza E, Hernandez-Rivas R, Loukeris T, Lycett G, Lanz-Mendoza H. Recombinant scorpine: a multifunctional antimicrobial peptide with activity against different pathogens. Cellular and molecular life sciences : CMLS. 2008;65:3081–3092. doi: 10.1007/s00018-008-8250-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Esteve E, Sabatier JM, Ronjat M, De Waard M, Allen PD, Pessah IN. Maurocalcine and peptide A stabilize distinct subconductance states of ryanodine receptor type 1, revealing a proportional gating mechanism. The Journal of biological chemistry. 2003;278:16095–16106. doi: 10.1074/jbc.M209501200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Cao L, Zhong M, Zhang Y, Han C, Li Q, Yang J, Zhou D, Shi W, He B, Liu F, Yu J, Sun Y, Cao Y, Li Y, Li W, Guo D, Cao Z, Yan H. Anti-HIV-1 activity of a new scorpion venom peptide derivative Kn2-7. PloS one. 2012;7:e34947. doi: 10.1371/journal.pone.0034947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde R, Zamudio FZ, Rodriguez MH, Possani LD. Scorpine, an anti-malaria and anti-bacterial agent purified from scorpion venom. FEBS letters. 2000;471:165–168. doi: 10.1016/s0014-5793(00)01384-3. [DOI] [PubMed] [Google Scholar]
- Darbon H. [Animal toxins and ion channels]. Journal de la Societe de biologie. 1999;193:445–450. [PubMed] [Google Scholar]
- Dardevet L, Rani D, Aziz TA, Bazin I, Sabatier JM, Fadl M, Brambilla E, De Waard M. Chlorotoxin: a helpful natural scorpion peptide to diagnose glioma and fight tumor invasion. Toxins. 2015;7:1079–1101. doi: 10.3390/toxins7041079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulhunty AF, Curtis SM, Watson S, Cengia L, Casarotto MG. Multiple Actions of Imperatoxin A on Ryanodine Receptors. The Journal of biological chemistry. 2004;279(12):11853–62. doi: 10.1074/jbc.M310466200. [DOI] [PubMed] [Google Scholar]
- Dunlop JA, Selden PA. Calibrating the chelicerate clock: a paleontological reply to Jeyaprakash and Hoy. Experimental & applied acarology. 2009;48:183–197. doi: 10.1007/s10493-009-9247-1. [DOI] [PubMed] [Google Scholar]
- Dyck JD, David TE, Burke B, Webb GD, Henderson MA, Fowler RS. Management of coronary artery disease in Hutchinson-Gilford syndrome. J Pediatr. 1987;111:407–410. doi: 10.1016/s0022-3476(87)80466-3. [DOI] [PubMed] [Google Scholar]
- Estève E, Mabrouk K, Dupuis A, Smida-Rezgui S, Altafaj X, Grunwald D, Platel JC, Andreotti N, Marty I, Sabatier JM, Ronjat M, De Waard M. Transduction of the scorpion toxin maurocalcine into cells: Evidence that the toxin crosses the plasma membrane. J. Biol. Chem. 2005;280:12833–12839. doi: 10.1074/jbc.M412521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada G, Garcia BI, Schiavon E, Ortiz E, Cestele S, Wanke E, Possani LD, Corzo G. Four disulfide-bridged scorpion beta neurotoxin CssII: heterologous expression and proper folding in vitro. Biochimica et biophysica acta. 2007;1770:1161–1168. doi: 10.1016/j.bbagen.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Estrada G, Garcia BI, Schiavon E, Ortiz E, Cestele S, Wanke E, Possani LD, Corzo G. Four disulfide-bridged scorpion beta neurotoxin CssII: heterologous expression and proper folding in vitro. Biochimica et biophysica acta. 2007;1770:1161–1168. doi: 10.1016/j.bbagen.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Fajloun Z, Kharrat R, Chen L, Lecomte C, Di Luccio E, Bichet D, El Ayeb M, Rochat H, Allen PD, Pessah IN, De Waard M, Sabatier JM. Chemical synthesis and characterization of maurocalcine, a scorpion toxin that activates Ca(2+) release channel/ryanodine receptors. FEBS letters. 2000;469:179–185. doi: 10.1016/s0014-5793(00)01239-4. [DOI] [PubMed] [Google Scholar]
- Gurrola GB, Arevalo C, Sreekumar R, Lokuta AJ, Walker JW, Valdivia HH. Activation of ryanodine receptors by imperatoxin A and a peptide segment of the II-III loop of the dihydropyridine receptor. The Journal of biological chemistry. 1999;274:7879–7886. doi: 10.1074/jbc.274.12.7879. [DOI] [PubMed] [Google Scholar]
- Gurrola GB, Hernandez-Lopez RA, Rodriguez De La Vega RC, Varga Z, Batista CVF, Salas-Castillo SP, Panyi G, Del Rio-Portilla F, Possani LD. Structure, function, and chemical synthesis of Vaejovis mexicanus peptide 24: A novel potent blocker of Kv1.3 potassium channels of human T lymphocytes. Biochemistry. 2012;51:4049–4061. doi: 10.1021/bi300060n. doi:10.1021/bi300060n. [DOI] [PubMed] [Google Scholar]
- Harrison PL, Abdel-Rahman MA, Miller K, Strong PN. Antimicrobial peptides from scorpion venoms. Toxicon. 2014;88:115–137. doi: 10.1016/j.toxicon.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaprakash A, Hoy MA. First divergence time estimate of spiders, scorpions, mites and ticks (subphylum: Chelicerata) inferred from mitochondrial phylogeny. Experimental & applied acarology. 2009;47:1–18. doi: 10.1007/s10493-008-9203-5. [DOI] [PubMed] [Google Scholar]
- Kern R, Malki A, Holmgren A, Richarme G. Chaperone properties of Escherichia coli thioredoxin and thioredoxin reductase. The Biochemical journal. 2003;371:965–972. doi: 10.1042/BJ20030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVallie ER, DiBlasio EA, Kovacic S, Grant KL, Schendel PF, McCoy JM. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Bio/technology. 1993;11:187–193. doi: 10.1038/nbt0293-187. [DOI] [PubMed] [Google Scholar]
- Lee CW, Lee EH, Takeuchi K, Takahashi H, Shimada I, Sato K, Shin SY, Kim DH, Kim JI. Molecular basis of the high-affinity activation of type 1 ryanodine receptors by imperatoxin A. The Biochemical journal. 2004;377:385–394. doi: 10.1042/BJ20031192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Ramirez K, Quintero-Hernandez V, Juarez-Gonzalez VR, Possani LD. Whole Transcriptome of the Venom Gland from Urodacus yaschenkoi Scorpion. PloS one. 2015;10:e0127883. doi: 10.1371/journal.pone.0127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Ramirez K, Quintero-Hernandez V, Vargas-Jaimes L, Batista CV, Winkel KD, Possani LD. Characterization of the venom from the Australian scorpion Urodacus yaschenkoi: Molecular mass analysis of components, cDNA sequences and peptides with antimicrobial activity. Toxicon : official journal of the International Society on Toxinology. 2013;63:44–54. doi: 10.1016/j.toxicon.2012.11.017. [DOI] [PubMed] [Google Scholar]
- Meissner G. Adenine nucleotide stimulation of Ca2+-induced Ca2+ release in sarcoplasmic reticulum. The Journal of biological chemistry. 1984;259:2365–2374. [PubMed] [Google Scholar]
- Mosbah A, Kharrat R, Fajloun Z, Renisio JG, Blanc E, Sabatier JM, El Ayeb M, Darbon H. A new fold in the scorpion toxin family, associated with an activity on a ryanodine-sensitive calcium channel. Proteins. 2000;40:436–442. doi: 10.1002/1097-0134(20000815)40:3<436::aid-prot90>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Narasimhan L, Singh J, Humblet C, Guruprasad K, Blundell T. Snail and spider toxins share a similar tertiary structure and ‘cystine motif’. Nature structural biology. 1994;1:850–852. doi: 10.1038/nsb1294-850. [DOI] [PubMed] [Google Scholar]
- Pallaghy PK, Nielsen KJ, Craik DJ, Norton RS. A common structural motif incorporating a cystine knot and a triple-stranded beta-sheet in toxic and inhibitory polypeptides. Protein science : a publication of the Protein Society. 1994;3:1833–1839. doi: 10.1002/pro.5560031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possani LD, Becerril B, Delepierre M, Tytgat J. Scorpion toxins specific for Na+-channels. European journal of biochemistry / FEBS. 1999;264:287–300. doi: 10.1046/j.1432-1327.1999.00625.x. [DOI] [PubMed] [Google Scholar]
- Quintero-Hernandez V, Jimenez-Vargas JM, Gurrola GB, Valdivia HH, Possani LD. Scorpion venom components that affect ion-channels function. Toxicon. 2013;76:328–342. doi: 10.1016/j.toxicon.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero-Hernandez V, Ramirez-Carreto S, Romero-Gutierrez MT, Valdez-Velazquez LL, Becerril B, Possani LD, Ortiz E. Transcriptome analysis of scorpion species belonging to the Vaejovis genus. PloS one. 2015;10:e0117188. doi: 10.1371/journal.pone.0117188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Franco J, Fill M. Approaching ryanodine receptor therapeutics from the calcin angle. The Journal of general physiology. 2016;147:369–373. doi: 10.1085/jgp.201611599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remijsen Q, Verdonck F, Willems J. Parabutoporin, a cationic amphipathic peptide from scorpion venom: much more than an antibiotic. Toxicon. 2010;55:180–185. doi: 10.1016/j.toxicon.2009.10.027. [DOI] [PubMed] [Google Scholar]
- Santibáñez-López CE, Francke OF, Ureta C, Possani LD. Scorpions from Mexico: From species diversity to venom complexity. Toxins (Basel) 2015 doi: 10.3390/toxins8010002. doi:10.3390/toxins8010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsó M, Trujillo R, Gurrola GB, Valdivia HH, Wagenknecht T. Three-dimensional location of the imperatoxin A binding site on the ryanodine receptor. J. Cell Biol. 1999;146:493–499. doi: 10.1083/jcb.146.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo AL, del Rio-Portilla F, Picco C, Estrada G, Prestipino G, Possani LD, Delepierre M, Corzo G. Solution structure of native and recombinant expressed toxin CssII from the venom of the scorpion Centruroides suffusus suffusus, and their effects on Nav1.5 sodium channels. Biochimica et biophysica acta. 2012;1824:478–487. doi: 10.1016/j.bbapap.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Schwartz EF, Capes EM, Diego-Garcia E, Zamudio FZ, Fuentes O, Possani LD, Valdivia HH. Characterization of hadrucalcin, a peptide from Hadrurus gertschi scorpion venom with pharmacological activity on ryanodine receptors. British journal of pharmacology. 2009;157:392–403. doi: 10.1111/j.1476-5381.2009.00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo IR, Choi MR, Park CS, Kim do H. Effects of recombinant imperatoxin A (IpTxa) mutants on the rabbit ryanodine receptor. Molecules and cells. 2006;22:328–335. [PubMed] [Google Scholar]
- Shahbazzadeh D, Srairi-Abid N, Feng W, Ram N, Borchani L, Ronjat M, Akbari A, Pessah IN, De Waard M, El Ayeb M. Hemicalcin, a new toxin from the Iranian scorpion Hemiscorpius lepturus which is active on ryanodine-sensitive Ca2+ channels. The Biochemical journal. 2007;404:89–96. doi: 10.1042/BJ20061404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Larios A, Gurrola GB, Zamudio FZ, Possani LD. Hadrurin, a new antimicrobial peptide from the venom of the scorpion Hadrurus aztecus. European journal of biochemistry / FEBS. 2000;267:5023–5031. doi: 10.1046/j.1432-1327.2000.01556.x. [DOI] [PubMed] [Google Scholar]
- Tripathy A, Resch W, Xu L, Valdivia HH, Meissner G. Imperatoxin A induces subconductance states in Ca2+ release channels (ryanodine receptors) of cardiac and skeletal muscle. The Journal of general physiology. 1998;111:679–690. doi: 10.1085/jgp.111.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia HH, Kirby MS, Lederer WJ, Coronado R. Scorpion toxins targeted against the sarcoplasmic reticulum Ca(2+)-release channel of skeletal and cardiac muscle. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:12185–12189. doi: 10.1073/pnas.89.24.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z, Gurrola-Briones G, Papp F, Rodríguez de la Vega RC, Pedraza-Alva G, Tajhya RB, Gaspar R, Cardenas L, Rosenstein Y, Beeton C, Possani LD, Panyi G. Vm24, a natural immunosuppressive peptide, potently and selectively blocks Kv1.3 potassium channels of human T cells. Mol. Pharmacol. 2012;82:372–82. doi: 10.1124/mol.112.078006. doi:10.1124/mol.112.078006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CG, He XL, Shao F, Liu W, Ling MH, Wang DC, Chi CW. Molecular characterization of an anti-epilepsy peptide from the scorpion Buthus martensi Karsch. European journal of biochemistry / FEBS. 2001;268:2480–2485. doi: 10.1046/j.1432-1327.2001.02132.x. [DOI] [PubMed] [Google Scholar]
- Xiao L, Gurrola GB, Zhang J, Valdivia CR, SanMartin M, Zamudio FZ, Zhang L, Possani LD, Valdivia HH. Structure-function relationships of peptides forming the calcin family of ryanodine receptor ligands. The Journal of general physiology. 2016;147:375–394. doi: 10.1085/jgp.201511499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Zhao Z, He Y, Wu L, Cai D, Hong W, Wu Y, Cao Z, Zheng C, Li W. A new natural alpha-helical peptide from the venom of the scorpion Heterometrus petersii kills HCV. Peptides. 2011;32:11–19. doi: 10.1016/j.peptides.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Zamudio FZ, Gurrola GB, Arevalo C, Sreekumar R, Walker JW, Valdivia HH, Possani LD. Primary structure and synthesis of Imperatoxin A (IpTx(a)), a peptide activator of Ca2+ release channels/ryanodine receptors. FEBS letters. 1997;405:385–389. doi: 10.1016/s0014-5793(97)00227-5. [DOI] [PubMed] [Google Scholar]
- Zhu S, Darbon H, Dyason K, Verdonck F, Tytgat J. Evolutionary origin of inhibitor cystine knot peptides. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2003;17:1765–1767. doi: 10.1096/fj.02-1044fje. [DOI] [PubMed] [Google Scholar]