Abstract
Objectives
Degeneration of articular cartilage is central to OA pathology; however, the molecular mechanisms leading to these irreversible changes are still poorly understood. Here, we investigated how changes in the chondrocyte translational apparatus may contribute to the pathology of OA.
Methods
Normal and OA human knee cartilage was used to analyze the activity of different components of the translational machinery. Chondrocytes isolated from lesional and non-lesional areas of OA cartilage were used to estimate relative rate of protein synthesis by metabolic labeling. Experimental OA was induced by transection of the anterior cruciate ligament in rats to investigate changes in the translational apparatus associated with OA. The role of IL-1β signaling was assessed in vitro using rat articular chondrocytes. Expression of mRNAs was analyzed by qPCR and protein levels by immunohistochemistry and Western blotting.
Results
We identified several novel traits of OA chondrocytes, including upregulation of the Serine/Threonine kinases AKT2 and AKT3 at the post-transcriptional level and increased rate of total protein synthesis, likely due to inactivation of 4E-BP1, a known repressor of cap-dependent translation. We found that 4E-BP1 inactivation is mTOR-dependent and crucial for upregulation of protein synthesis in general and in particular for MMP13 and ADAMTS5 expression. In addition, IL-1β treatment led to 4E-BP1 inactivation and upregulation of protein synthesis in articular chondrocytes.
Conclusions
Precise control of protein synthesis is vital for cartilage homeostasis and its dysregulation contributes to the molecular pathology of OA. Our study therefore identifies a novel set of potential therapeutic targets.
Keywords: osteoarthritis, chondrocytes, 4E-BP, protein synthesis, AKT/mTOR
INTRODUCTION
Osteoarthritis (OA) is one of the most prevalent chronic diseases, but the mechanisms leading to OA are still not well understood. However it’s well accepted that degeneration of the articular cartilage is a hallmark of OA [1, 2]. Recently several groups have shown the importance of proper regulation of mTOR (mammalian Target Of Rapamycin) kinase for cartilage homeostasis and investigated in great details its role in autophagy in articular chondrocytes [3–8]. However, protein synthesis, a vital cell function controlled by mTOR signaling, has not been investigated in OA pathophysiology. We therefore studied how the translation machinery is modulated to facilitate changes in the expression of established OA-associated catabolic and anabolic mediators.
Protein synthesis is a very complex process that proceeds through initiation, elongation and termination. Regulation of translation is a crucial component of gene expression control and is executed mostly at the initiation step [9], during which the small ribosomal subunit, bound to several translation initiation factors and initiator MettRNA, is recruited to mRNA. Ribosomal loading onto mRNAs is mediated by eukaryotic Initiation Factor 4F (eIF4F), a protein complex consisting of eIF4A, eIF4G and eIF4E (a cap-binding protein). By binding to the cap structure of mRNAs, eIF4E facilitates binding of the ribosomal complexes onto the mRNA 5’end and promotes protein synthesis [10]. One of the key mechanisms of translation regulation is modulation of eIF4E activity by eIF4E Binding Proteins (4E-BPs). There are three 4E-BPs isoforms with 4E-BP1 being the most abundant one. 4E-BP1 acts as a repressor of cap-dependent translation by sequestering eIF4E from the active eIF4F complexes. The ability of 4E-BP to bind eIF4E is restricted by 4E-BP phosphorylation, as the hyperphosphorylated form is unable to bind eIF4E [11]. 4E-BP phosphorylation/inactivation is mediated by mTORC1 (mTOR complex1) complexes comprised of mTOR kinase, Raptor and GβL [12].
We found that OA cartilage has accelerated protein synthesis, as a result of mTORC1-dependent 4E-BP1 inactivation mediated by AKT signaling. Using an animal model of OA we determined that changes in the activity of translational apparatus are prominent at early stages of the disease and persist to later stages.
Importantly, we found that modulation of 4E-BP1 activity in OA-like chondrocytes is correlated with the rate of protein synthesis in general and 4E-BP1 inactivation is required not only for efficient translation of extracellular matrix (ECM)-degrading enzymes, such as MMP13 and ADAMTS5, but for their transcription as well. We showed that IL-1β signaling, with a possible synergetic effect of other cytokines, is largely responsible for 4E-BP1 inactivation and upregulation of protein synthesis in chondrocytes. Inhibition of activity or expression of mTORC1 components in IL-1β treated articular chondrocytes not only diminished upregulation of (ECM)-degrading enzymes, but also annulled the cytokine effect on protein synthesis.
METHODS
Clinical Samples
Normal human knee cartilage samples from deceased subjects were procured from the National Disease Research Interchange (NDRI, Philadelphia, USA). Human OA cartilage was obtained from advanced OA patients who underwent knee replacement surgery. The surgically discarded human OA tissues were collected under the guidelines of the Institutional Review Board of NYU School of Medicine and total protein was extracted as described [13].
Human Chondrocyte Isolation
Chondrocytes were isolated as described [13]. Briefly, cartilage was dissected away from subchondral bone and digested with Pronase (Sigma-Aldrich) for 1h at 37°C followed by overnight digestion with Collagenase P (Roche Diagnostics) at 37°C. Cells were cultured in high-density monolayers.
Microarray analysis
Microarray analysis was done as described [14].
Experimental osteoarthritis in rats
All animal experiments were performed according to IACUC regulations at NYU School of Medicine. Experimental osteoarthritis was induced in 12 week old male Sprague Dawley rats by transection of the anterior cruciate ligament (ACLT) in the right knee as described in [15].
Rat chondrocyte isolation and culture
Articular cartilage was harvested from 12–15 week old Sprague Dawley rats for cytokines treatment and at 12 and 20 weeks post-surgery. Cartilage slices were incubated for 90min at 37°C with 1% pronase (w/v), then for 2h at 37°C with 0.4% (w/v) collagenase B (Sigma-Aldrich) in DMEM supplemented with 5% FBS and cultured in DMEM supplemented with 5% FBS. Once the cells reached confluence they were starved for 16h and treated with 10ng/ml of IL1-β and/or 10ng/ml of OSM or 100ng/ml IGF-1 for the indicated time. For the inhibitor experiments cells were pretreated with DMSO, Ku-063794 (1µM) or PI3K inhibitor IV (10µM) for 1h and then treated with IL1-β. Transfections with mTOR siRNA (nt2381; Sigma) were performed using Lipofectamine RNAiMax according to the manufacturer's protocol. MISSION siRNA Universal Negative Control was used as a control. Cells were starved for 16h in serum-free medium 24h post-transfection and then treated with IL1-β.
RT-qPCR
RNA was extracted from cultured chondrocytes or from cartilage isolated from rat knees immediately after Collagenase B digestion using the RNeasy Mini Kit (Life Technologies) according to the manufacturer’s protocol. 500ng of total RNA was used for cDNA synthesis with the iScript cDNA synthesis kit (Bio-Rad). qPCR was run in a MyiQ Single-Color Real-TimePCR Detection System (Bio-Rad) using iQ SYBR Green Supermix. Relative gene expression levels were calculated using the ΔΔCt method. Target mRNA levels were normalized to 18S rRNA. Primers used are listed in Supplemental Tables 1 and 2.
Immunohistochemistry (IHC)
IHC analysis for human samples was performed as described [14]. Rat knee joints from ACLT-operated (n=5) and sham-operated animals (n=5) were harvested and fixed in 4% PFA for 24h, decalcified in Formical2000 (Decal Chemical Corp) for 96h, followed by paraffin embedding. Serial sections (4µm) were also stained with Toluidine Blue, and examined for histopathological changes using a semiquantitative scoring system. Antibodies are listed in Supplemental Table 3. For antigen retrieval sections were incubated with Proteinase K for 7min at 37°C, followed by incubation with Triton-X100 for 15min. An avidin–biotin–peroxidase system (Vector) was used, and antigen-antibody complexes were visualized using diaminobenzidine as substrate. The tissues were counterstained with hematoxylin.
Metabolic 35S labeling
To analyze the rate of total protein synthesis an equal number of cells was used within each condition tested (100,000 cells). The assay was done as described in [16].
MMP13 Activity
MMP13 activity was analyzed using the Fluorokine E kit (R&D) following the manufacturer’s instructions. When 35S labeling was performed supernatant was collected just prior to labeling.
Statistics
The data in bars are expressed as mean±SD, while the data in dots show the distribution with the line depicting the mean. GraphPad Prism version 6.0 (www.graphpad.com) was used for statistical analysis, The level of significance was set at 0.05. More specifically, paired student t-test analyses were performed for lesional versus non lesional samples, for the quantification of positive IHC staining and for the RAC data when two sets of data were compared. In other cases a one way ANOVA analysis was performed with Bonferroni’s multiple comparisons correction. One sample t-test was performed for the 35S incorporation experiments comparing the median to the hypothetical value of 1, which is the value of the untreated control or the sham animal.
RESULTS
Aberrant activity and expression of components of translational machinery in human OA cartilage leads to increased rate of total protein synthesis
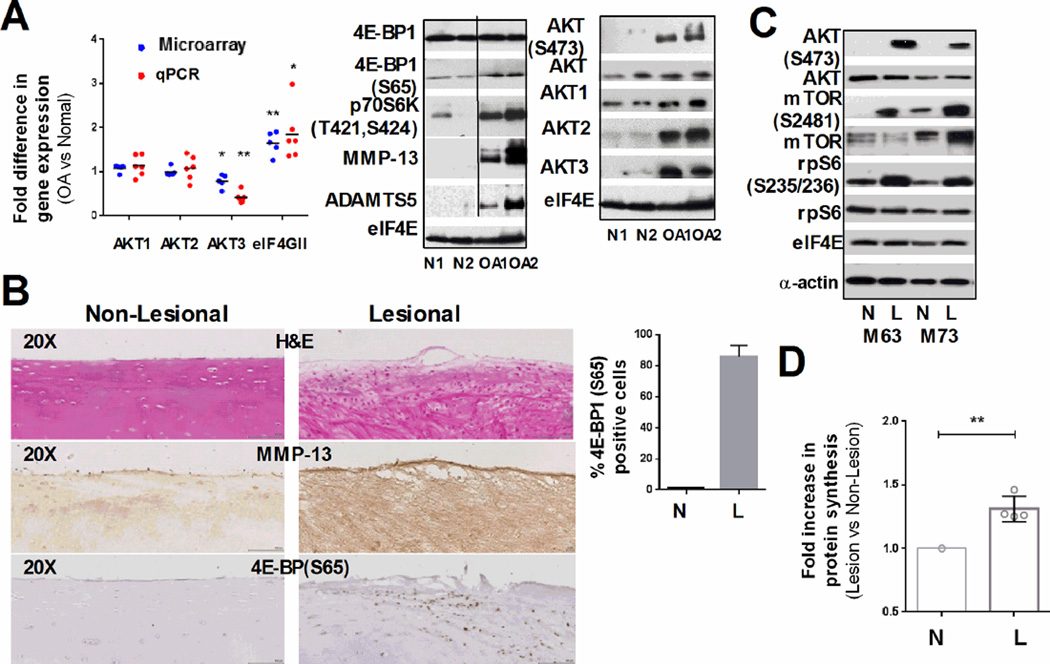
The mRNA expression of the components of eIF4F complexes (eIF4G, eIF4A and eIF4E) was examined in human OA and non-OA cartilage using Affymetrix U133A microarray data. This analysis revealed significant (p=0.0051) upregulation of eIF4GII in diseased tissue, with no significant changes in eIF4A and eIF4E expression (Fig. 1A and S1A). These data were further validated by qPCR analysis of individual cartilage samples (Fig. 1A, and S1A). Upregulation of eIF4G expression suggests a higher capacity of OA cartilage for accelerated protein synthesis, which is likely essential for the higher rates of cell turnover observed in OA tissue. We therefore analyzed abundance and activity of known effectors of protein synthesis, such as 4E-BP1, and components of the AKT/mTOR pathway by immunoblotting. To analyze changes in eIF4F activation we used eIF4E as a loading control, as we previously detected no changes in eIF4E mRNA level in OA cartilage. This loading control was consistently similar to actin in all the samples analyzed (Fig. 1C). As shown in Fig. 1A, there were no changes in total 4E-BP1 levels in OA cartilage relative to non-OA controls; however, we detected about a threefold increase in the hyperphosphorylated (Ser65 positive) form of 4E-BP1 in OA cartilage. Four known phosphorylation sites modulate 4E-BP1 activity: Thr37 and Thr46 are phosphorylated first, followed by Thr70 and Ser65. Ser65 phosphorylation by mTORC1 complexes precludes eIF4E binding and thus inactivates 4E-BP1 [17]. Therefore, our data indicate that the proportion of inactive 4E-BP1 is higher in OA cartilage that in normal tissue. OA samples had high protein and mRNA levels of MMP13 (matrix metalloproteinase 13) and ADAMTS5 (a disintegrin and metalloproteinase with thrombospondin-like motif 5) consistent with their well-established role in OA pathology [18, 19] (Figs. 1A and S1A).
Figure 1. Differential expression of proteins involved in translation in human OA.
(A) Graph: Expression of AKT isoforms and eIF4GII, in raw OA cartilage by microarray analysis (blue dots) and qPCR (red dots), (n=6, line at mean). Western blots (WB): Representative immunoblots of the raw OA cartilage samples (n=6) show changes in the phosphorylation and expression of the indicated proteins (20µg, eIF4E: loading control). (B) Representative hematoxylin and eosin (H&E) staining and IHC analysis of non-lesional and lesional cartilage areas of the same patient shows differential MMP13 expression and 4E-BP1 phosphorylation. The percentage of 4E-BP1(S65) positive cells is shown in the graph (mean ±SD, unpaired t-test). (C) Representative immunoblots of the indicated targets in cultured chondrocytes from lesional (L) and non-lesional (N) areas of the same patient (15µg of total protein, eIF4E and α-actin - loading controls) (D) Fold increase of protein synthesis in lesional and non-lesional chondrocytes of the same patient (n=4, values normalized to total cell number, * : P ≤ 0.05 and **: P ≤ 0.01).
4E-BP1 is a direct target of mTORC1 which has been recently implicated in autophagy defects in OA cartilage, and elevated mTOR mRNA level was reported in OA human cartilage [3, 20, 21]. We therefore characterized mTOR activity by using phospho-specific antibodies to p70S6K, its direct downstream target. As shown in Fig. 1A, p70S6K was robustly phosphorylated in human OA samples, but no significant changes in the p70S6K or mTOR mRNA expression was detected either by microarray or qPCR analysis (Fig. S1A).
We next examined the phosphorylation status of AKT, a positive regulator of mTORC1, in cartilage samples from OA patients. AKT is activated by phospholipid binding and phosphorylation on Thr308 and Ser473 [22]; therefore, Ser473 phosphorylation is commonly used as an indirect readout of AKT activity. As shown in Fig. 1A (right panel), AKT(S473) was present nearly exclusively in OA samples, showing that increased mTOR activity correlates with AKT(S473) phosphorylation. Surprisingly, while protein levels of AKT2 and AKT3 were upregulated in OA cartilage their mRNA levels were not increased (Fig. 1A). These data for the first time suggest that regulation of AKT2 and AKT3 expression at the post-transcriptional level might contribute to its increased phosphorylation/activity in OA cartilage.
To further validate our data we analyzed lesional and non-lesional areas of knee OA cartilage from the same patient, by IHC using 4E-BP1 (Ser65) antibodies. Both areas were located on the medial compartment within the closest possible proximity. Though this approach has some limitations, as non-lesional areas are not truly representative of non-OA cartilage, approximately 80% of the cells stained positively in lesional areas compared to 5% in non-lesional (Fig. 1B). The representative negative control is shown in Fig. S1C. This pattern co-localized with MMP13 immunostaining, and with the degree of tissue degeneration supporting our hypothesis that higher levels of inactive 4E-BP1 contribute to the degenerative changes in OA cartilage. Consistently, qPCR analysis showed upregulation of MMP13 and ADAMTS5 mRNAs in the lesional compared to the non-lesional cartilage from the same OA patient (Fig. S1B). In addition, Western blotting analysis showed increased phosphorylation of AKT(S473), mTOR(S2481) and its downstream target rpS6 in the lesion areas of all four samples (Fig. 1C). All of the phospho-sites analyzed are associated with increased activity of the corresponding proteins.
We next compared the rate of total protein synthesis in cells from lesional and non-lesional areas of cartilage obtained from total knee replacements (n=4). Total protein synthesis was measured by pulse-labeling with 35S Met/Cys. As shown in Fig. 1D, chondrocytes isolated from lesional areas incorporated 30–40% more 35S Met/Cys, than chondrocytes from non-lesional areas, validating our hypothesis that inactivation of 4E-BP1 leads to upregulation of translation in OA chondrocytes. As the lesional and non-lesional samples were obtained from OA joints at end-stage disease, there is a possibility that the changes in translational apparatus in advanced medial compartment knee OA tissue may not only represent direct effects of disease, but also by altered mechanical loading.
The rate of total protein synthesis is increased in rat articular cartilage after surgical induction of OA
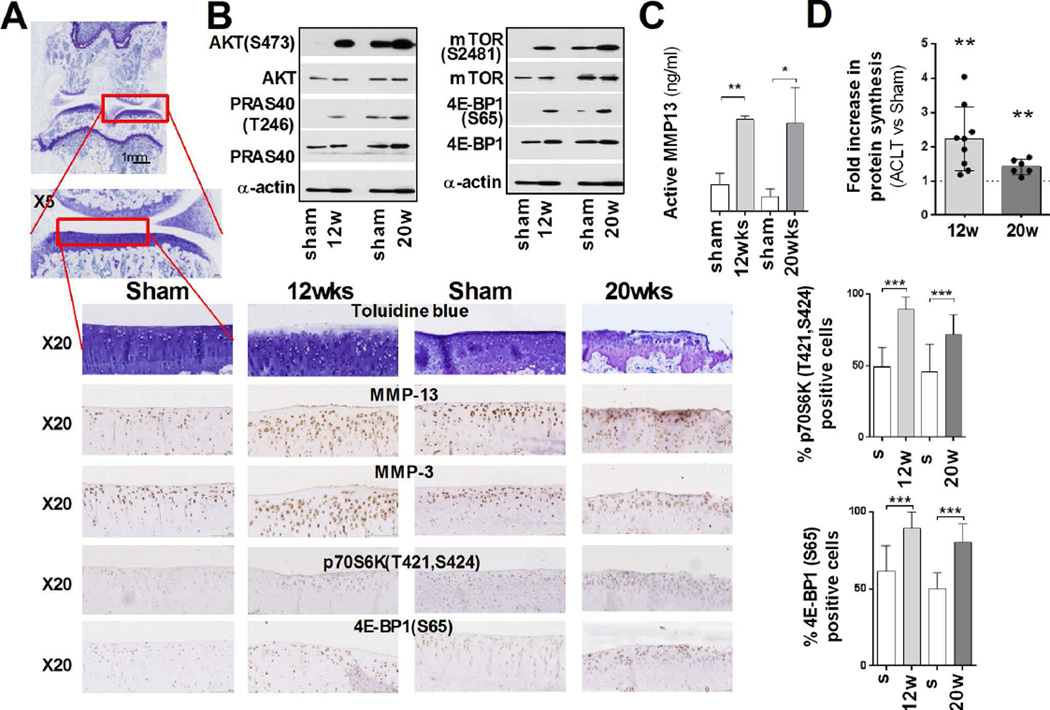
Since we observed increased activity of the translational apparatus in human OA cartilage, we further corroborated these findings using anterior cruciate ligament transection (ACLT) model of surgically induced (post-traumatic) OA in rats [15]. Articular cartilage harvested from ACLT operated animals was analyzed for MMP13, ADAMTS5, Aggrecan (ACAN) and Collagen Type II Alpha 1 (Col2a1) expression (Fig. S2A). Expression of MMP13, ADAMTS5 and Col2A1 mRNAs in the cartilage isolated from ACLT joints was upregulated at 12 weeks post-surgery and, as expect, higher upregulation was detected at 20 weeks, with no significant changes in ACAN mRNAs. At these times Toluidine Blue staining of sections of the samples showed loss of matrix and fibrillation of cartilage (Fig. 2A), and IHC analysis confirmed increased expression of MMP13 and MMP3 within the ACLT joint (Fig. 2A). While MMP13 localization was mostly at 12 weeks post-surgery (in agreement with other animal models [23, 24]), at 20 weeks prominent staining of matrix associated MMP13 was also detected.
Figure 2. Analysis of the translational apparatus in an ACLT rat model of OA.
(A) Representative Toluidine Blue staining of ACLT and sham operated joints (12 and 20 weeks post-surgery). IHC analysis of expression and phosphorylation of the indicated targets shows differences between ACLT and sham operated joints. The percentage of 4E-BP1(S65) and p70S6K (T421,S424) positive cells is shown in the graph (mean ±SD; unpaired t-test). (B) Representative immunoblots of the indicated proteins in cultured chondrocytes from ACLT and sham operated knee joints demonstrate (10µg of total protein, α-actin: loading control) (C) MMP13activity in the culture supernatant of chondrocytes from ACLT and sham operated joints. (n=3, mean ±SD, unpaired t-test) (D) Fold increase in the levels of protein synthesis in ACLT chondrocytes compared to chondrocytes from sham operated joints 12 (n=9) and 20 (n=6, normalized to total cell number, mean ±SD) weeks post-surgery, (*: P ≤ 0.05, **: P ≤ 0.01 and ***: P ≤ 0.001).
Increased MMP13 expression correlated with its catalytic activity in ACLT joints, as the amount of active enzyme was significantly higher in ACLT joints compared to sham operated joints (Fig. 2C). In line with our data from human OA cartilage, IHC of phospho-p70S6K, a direct mTORC1 target, and 4E-BP1(S65) indicated increased mTOR activity in OA cartilage (Fig 2A). Remarkably, 4E-BP(S65) positive cells showed similar increase at 12 and 20 weeks post-surgery, indicating that the proportion of inactive 4E-BP peaks as early as 12 weeks post-surgery. We obtained similar results when we analyzed chondrocytes, from ACLT and sham operated joints by immunoblotting (Fig. 2B). We also characterized phosphorylation of PRAS40, a direct AKT target [25], and the results were consistent with our analysis of AKT(S473) phosphorylation.
Chondrocytes isolated from ACLT joints at both 12 and 20 weeks post-surgery incorporated more 35S Meth/Cys than the control joints (Fig. 2D) indicating that the rate of total protein synthesis is higher in OA cartilage. Thus, our data showed that upregulation of mTOR signaling leads to inactivation of 4E-BP1 and correlates with an increased rate of protein synthesis in both human OA cartilage and a rodent model of post-traumatic OA. We also observed changes in the expression of autophagy genes affected by mTOR signaling as was previously reported [4] (Fig. S2B).
IL-1β signaling is largely responsible for changes in translational machinery leading to accelerated protein synthesis
Among the many pro-inflammatory cytokines implicated in OA pathology [26] IL-1β has a prominent role [27]. IL-1β treatment is routinely used to model OA-like changes in in vitro studies. We therefore adopted this system to investigate the changes in the chondrocyte translational machinery. We recognize the many limitations of this technique and therefore do not consider it as an OA model itself, but rather as an OA-like system which can help to decipher the complexity of this multifactorial, chronic disease.
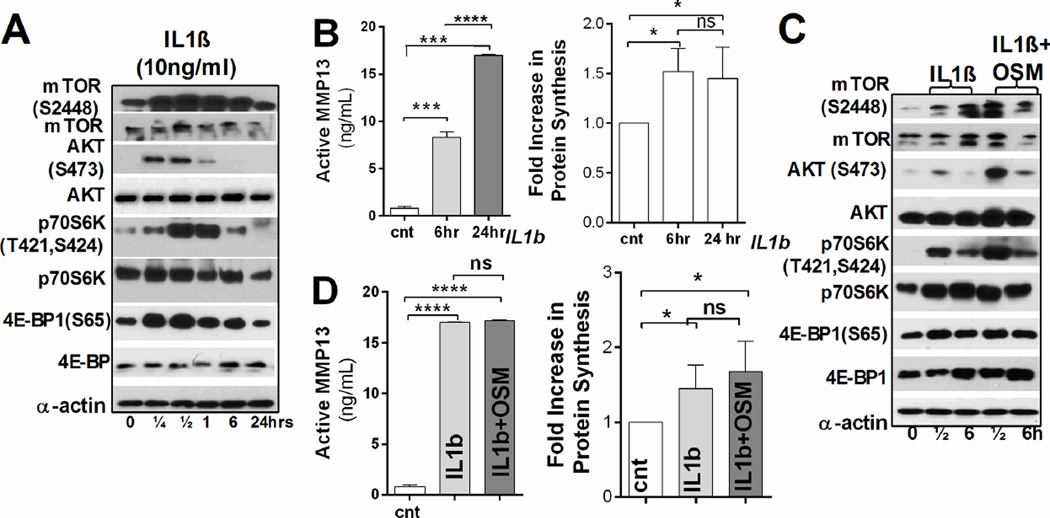
Consistent with previous reports, IL-1β treatment of articular chondrocytes (RAC) from 12-week old rats resulted in MAPKs (ERK, p38, and JNK) activation and upregulation of mRNA and protein expression of cartilage-specific targets (Fig. S3A, B and D) [28, 29], as well as in increased MMP13 expression and activity (Fig. 3B). We also observed activation of the AKT/mTOR pathway, and increased phosphorylation of 4E-BP1 and p70S6K (Fig. 3A). Importantly, total protein synthesis was increased as early as 6h of IL-1β treatment, implying that IL-1β signaling might be responsible for the higher translation rates observed in OA chondrocytes (Fig. 3B).
Figure 3. IL-1β signaling induces changes in the translational machinery of articular chondrocytes similar to the ones observed in OA cartilage.
RAC were treated with IL-1β (A, B) or with IL-1β and OSM (C, D) for the indicated times. (A, C) Representative immunoblots of proteins involved in the AKT/mTOR pathway (15µg of total protein, α-actin: loading control). (B, D) Left panel: Active MMP13 in treated and untreated RAC (n=3 independent experiments; mean ±SD, unpaired t-test). Right panel: Fold increase in protein synthesis in treated and untreated RAC (n=4 independent experiments, values normalized to total cell number, mean±SD, *: P ≤ 0.05, **: P ≤ 0.01, ***: P ≤ 0.001 and ****: P ≤ 0.0001).
We next investigated whether inclusion of an additional cytokine would have a cumulative or synergetic effect on protein synthesis. We chose Oncostatin M (OSM) as its expression is elevated in OA cartilage and it also stimulates the STAT3 pathway (Fig. S3C) [28, 30]. AKT(S473) phosphorylation was more robust when chondrocytes were treated with both cytokines than upon treatment with IL-1β alone (Fig. 3C). There were no significant changes in MMP13 and ADAMTS5 expression upon combined treatment (Fig. S3E), and MMP13 activity did not change (Fig. 3D). A similar effect was seen when total protein synthesis rate was compared in cells treated with both cytokines or with IL-1β alone (Fig. 3D, right panel). These data point out IL-1β signaling as one of the main modulators of the translational machinery in OA-like chondrocytes.
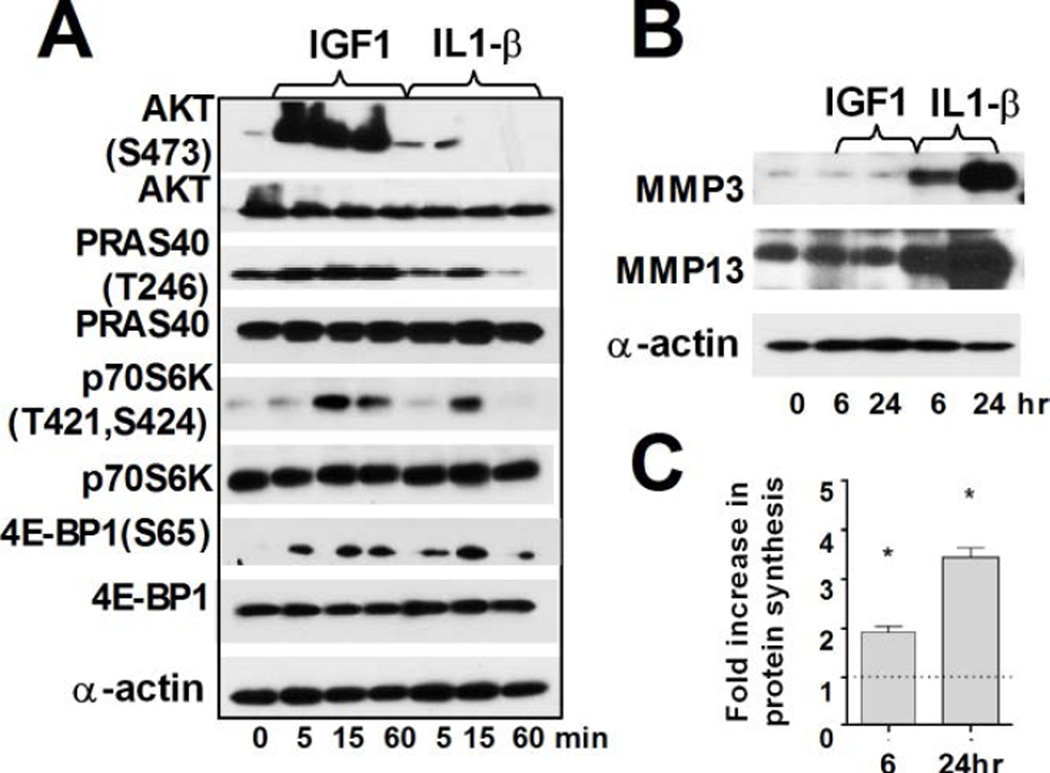
We next investigated whether the changes in the translation machinery induced by IL-1β treatment were comparable with those obtained by stimulation of proliferation by anabolic factors such as IGF1. As shown in Fig. 4A, both AKT(S473) and PRAS40 are phosphorylated more robustly by IGF treatment. While we observed an increase in 35S incorporation upon IGF1 treatment (Fig. 4C), this stimulation had no effect on the mRNA and protein levels of ECM-degrading enzymes (Fig. 4B, S4). Clearly, IL-1β signaling provides favorable environment for both mRNA and protein expression of genes implicated in OA pathogenesis and progression even if the increased rate of total protein synthesis is more modest.
Figure 4. IGF1 signaling induces protein synthesis in articular chondrocytes but does not affect expression of ECM-degrading proteases.
(A) Immunoblot analysis of the AKT/mTOR pathway in RAC treated with IGF1 or IL-1β for the times (15µg of total protein, α-actin-loading control). (B) Immunoblot analysis of MMP13 and MMP3 in treated and untreated RAC (15µg of total protein, α-actin: loading control). (C) Fold increase in protein synthesis in IGF1treated RAC compared to untreated control (n=3 independent experiments, values normalized to total cell number, mean±SD, *: P ≤ 0.05).
Expression of dominant negative (DN) 4E-BP1 decreases expression of ECM degrading enzymes and inhibits total protein synthesis upon IL-1β treatment
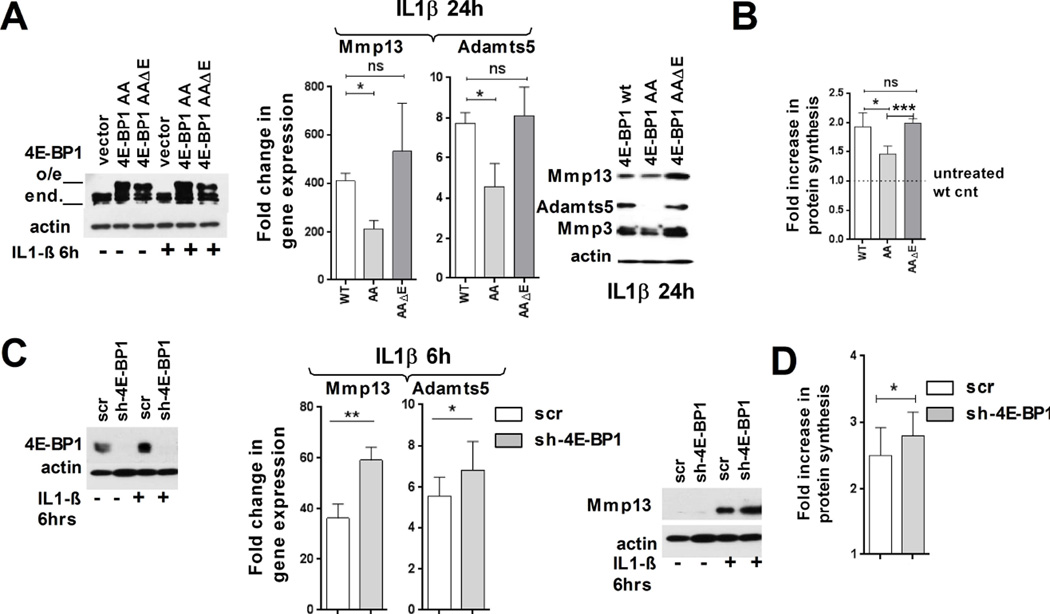
The changes in 4E-BP1 phosphorylation observed in OA chondrocytes prompted us to examine its potential role in the regulation of the expression of ECM-degrading enzymes. For this purpose, we used 4E-BP1 mutants that were previously characterized (Fig. 5A) [17]. 4E-BP1(AA) DN mutant has substitutions of Thr37 and Thr46 to Ala, and therefore cannot be phosphorylated/inactivated. 4E-BP1(AAΔ4E) mutant has the same mutations and deletion of eIF4E binding domain, and therefore cannot act as a DN mutant.
Figure 5. IGF1 signaling induces protein synthesis in articular chondrocytes but does not affect expression of ECM-degrading proteases.
(A, B) widl-type 4E-BP1, dominant negative (4EBP1-AA, DN) and mutant 4E-BP1 lacking the eIF4E-binding domain (4E-BP1-AAE) were expressed in IL-1β treated RAC. (A) Left panel: Immunoblot analysis of overexpression of the 4E-BP1 derivatives (α-actin: loading control). Middle and right panels: MMP13 and ADAMTS5 mRNA and protein expression in IL-1β treated RAC (n=3 independent experiments, mRNA expression normalized to 18S rRNA and untreated wt-control, mean ±SD; paired t-test; α-actin: loading control). (B) Fold increase in protein synthesis upon IL-1β treatment (n=3, mean ±SD). (C,D) 4E-BP1 expression was downregulated by shRNA using scrambled shRNA as a control. Left panel: Immunoblots of 4E-BP1 expression in RAC (α-actin: loading control). Middle and right panels: MMP13 and ADAMTS5 mRNA and protein expression in L-1β treated RAC (n=3, mRNA expression normalized to 18S rRNA and untreated scr-cnt, mean±SEM; α-actin used as a loading control) (D) Fold increase in protein synthesis upon IL-1β treatment (n=3, mean ±SD, *: P ≤ 0.05, **: P ≤ 0.01 and ***: P ≤ 0.001).
Expression of 4E-BP1 DN blocked the upregulation of MMP13, ADAMTS5 and MMP3 mRNA compared to the wt4E-BP1, while expression of 4E-BP1(AAΔ4E) had no significant effect (Figs. 5A, S5A). This effect was reflected at the protein level as well, with Adamts5 being surprisingly the most sensitive to the expression of DN mutant. Crucially, IL-1β-induced upregulation of total protein synthesis was suppressed by expression of DN 4E-BP1, but not by its deletion version (Fig. 5B). These data indicate that inactivation of 4E-BP1 is crucial for upregulation of total protein synthesis in general, and in particular, for expression of ECM-degrading enzymes.
Next, we examined if 4E-BP1 downregulation, which mimics its inactivation, would have an opposite effect. RAC were infected with a lentiviral vector expressing a control, scrambled shRNA or 4E-BP1 shRNA (Fig. 5C). Cells expressing 4E-BP1 shRNA showed stronger expression of MMP13 and ADAMTS5 upon IL-1β stimulation with nonsignificant changes in MMP3, Aggrecan and Col2A1 expression (Figs. 5C, S5B). Thus, these experiments established the importance of 4E-BP1 inactivation for upregulation of protein synthesis and efficient expression of MMP13 and ADAMTS5 in OA-like chondrocytes.
Activation of mTOR kinase is crucial for protein synthesis upregulation by IL-1β
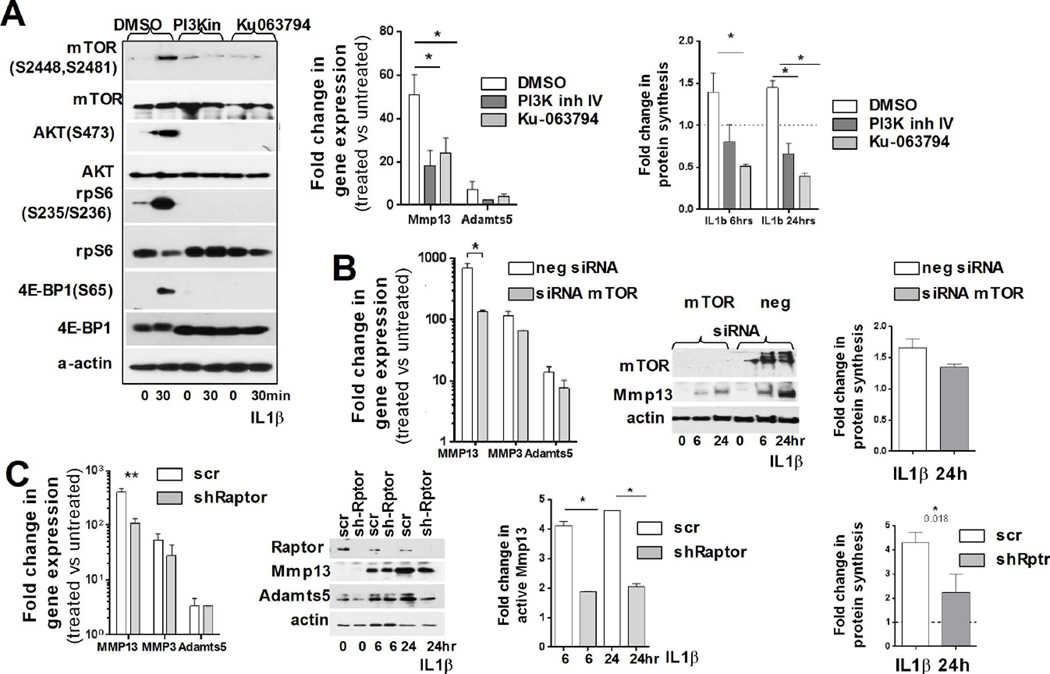
We next determined how mTOR inhibition affects IL-1β induced translation upregulation in RAC. Cells were pretreated with mTOR kinase inhibitor Ku-063794 [31] and then stimulated with IL-1β. We also used PI3K inhibitor IV. As shown in Fig. 6A, mTOR inhibition led to complete loss of AKT(S473), rpS6 and 4E-BP1(S65) phosphorylation) and significant reduction in MMP13 and ADAMTS5 expression was observed. Protein synthesis was considerably suppressed in the presence of Ku-063794 inhibitor (Fig. 6A, right graph).
Figure 6. mTORC1 activity is crucial for IL-1β induced changes in the translational machinery.
(A) Left panel: RAC pre-treated (1hr) with PI3K IV (10µM) or Ku-0063794 (1µM) inhibitor, were analyzed for activation of proteins of the AKT/mTOR pathway after IL-1β treatment (α-actin: loading control). Graphs: (Left) mTOR inhibition decreased MMP13 and ADAMTS5 mRNA upregulation compared to untreated control, (mean ±SD; values normalized to 18S rRNA). (Right) IL-1β mediated upregulation of total protein synthesis is abated when the PI3K/mTOR pathway was inhibited (mean ±SD). The data are representative of three independent experiments. (B) Downregulation of mTOR expression (by siRNA) significantly decreases MMP13 mRNA and protein upregulation compared to neg-si untreated control (values normalized to 18S rRNA). Right panel: Fold increase in protein synthesis upon IL-1β treatment (n=3, mean ±SD). (C) Specific downregulation of mTORC1 complexes by Raptor knockdown decreases MMP13 mRNA and protein upregulation compared to control cells (n=3, mean±SD; normalized to 18S rRNA). Right graphs: (1) Upregulation of total protein synthesis by IL-1β treatment is blocked by downregulation of Raptor expression (mean ±SD). (2) mTORC1 inhibition blocks the increase of active MMP13 (*: P ≤ 0.05).
We next analyzed the mechanism by which mTOR mediates the effect of IL-1β on translation in chondrocytes. RAC were pretreated with the mTOR kinase inhibitor Ku-063794 [31] or PI3K inhibitor IV and then stimulated with IL-1β. As shown in Fig. 6A, mTOR inhibition led to complete loss of AKT(S473), rpS6 and 4E-BP1(S65) phosphorylation and to significant reduction in MMP13 and ADAMTS5 expression. Protein synthesis was considerably suppressed in the presence of Ku-063794 (Fig. 6A, right graph).
Finally, we examined if mTOR or Raptor downregulation has similar effects. Downregulation of either mTOR by siRNA or Raptor by shRNA decreased the effect of IL-1β on MMP13, MMP3 and ADAMTS5 expression, with MMP13 and ADAMTS5 being particularly sensitive to Raptor downregulation, likely due to mTORC1 specific inhibition (Figs. 6B and C, Fig. S6). Thus, these results showed that the effect of IL-1β on protein synthesis in RAC is largely mediated by mTORC1 signaling and requires inactivation of 4E-BP1, the repressor of cap-dependent translation. This mechanism (with some additional tiers and players) is likely utilized in OA chondrocytes to accelerate the rate of total protein synthesis.
DISCUSSION
Currently there are no disease-modifying drugs to slow cartilage degeneration in OA, as the complex and heterogeneous nature of the disease complicates our understanding of the pathogenic mechanisms of OA. Therefore it is extremely important to identify new potential targets for diagnostics and treatment of OA at early stages. For this purpose understanding the role of translation control in OA progression is of particular importance, as it can allow us to modulate protein expression in a rapid and reversible way, when changes at transcriptional level are irreversible.
The importance of translational control has become apparent as correlations between changes at mRNA and protein levels are typically quite modest [32] and it is a challenge to correlate well described mRNA signatures for OA [33–35] with extensive proteomic data [36]. Clearly, post-transcriptional gene regulation must be taken into consideration alongside with mRNA expression. For example, Tew and colleagues have shown that the number of mRNAs with shortened half-lives is significantly higher in OA samples [37].
Our present results show that the rate of total protein synthesis is higher in human and rodent OA chondrocytes. Abnormal mTORC1 signaling resulting in reduced autophagy and increased cell death has been previously implicated in OA pathology [3, 20, 38]. These reports demonstrated that inhibition of mTOR activity by either its cartilage specific deletion or rapamycin protects mice from surgically induced OA. While this inhibition restores autophagy, it also downregulates the rate of total protein synthesis. Indeed, the translation rate of IL-1β treated chondrocytes is sensitive to mTORC1 inhibition (by siRNA, Ku063794 or inhibition of Raptor expression) indicating protein synthesis as an essential target of dysregulated mTORC1 signaling in OA cartilage. We established that in OA-like chondrocytes translation upregulation is mediated by mTORC1 signaling and requires inactivation of 4E-BP1, its direct substrate. It would be interesting to assess the rate of total protein synthesis in an animal model of OA in rapamycin-treated post-op animals compared to untreated ones.
Remarkably, the percentage of 4E-BP1(S65) positive cells was similar at 12 and 20 weeks post-surgery, indicating that 4E-BP1 activation likely reaches its maximum as early as at 12 weeks, when the morphological changes of the cartilage are still modest. We chose a later time point in order to see if we continue to observe similarity with human OA cartilage, as upregulation of protein synthesis in the ACLT model 20 weeks post-surgery was similar to that observed between lesional and non-lesional human chondrocytes. This finding, however, raises the important question of how early during OA progression we can detect an increase in the activity of the translational machinery, a point that warrants further investigation.
Our data also show that another mTORC1 substrate, p70S6K, is activated in human OA cartilage as well as in an animal model. Although we were not able to detect a significant increase in mTOR mRNA or protein expression in advanced human OA cartilage as reported before [3, 21], we consistently detected increased mTOR activity. Upregulation of AKT activity in OA cartilage that we observed is likely to be a key post-translational effector of mTORC1 activity. We found that AKT isoforms are differentially expressed in human OA chondrocytes and it would be interesting to investigate if there are any changes in expression/activation of these isoform during OA progression. PI3K/AKT signaling has been implicated in OA-like changes in vitro [28, 39], and its importance in vivo is indirectly supported by the notion that expression of mir-634, a negative regulator of PI3K, is decreased in OA cartilage [40].
We treated articular chondrocytes with IL-1β to mimic some changes observed in OA chondrocytes. While this system does not fully reflect the complexity of OA, our ACLT model corroborated our in vitro studies. We demonstrated that upregulation of protein synthesis requires mTORC1 activity and 4E-BP1 inactivation. While adding OSM simultaneously with IL-1β had minimal effect on the translational apparatus, upregulation of many age-related inflammatory cytokines should be also taken into account as expression of many of them, including leptin might affect translation machinery in a localized manner [41].
Interestingly, the increase in the rate of total protein synthesis was comparable between 6 and 24h of IL-1β treatment likely because it reaches its possible capacity. This level of upregulation might be used differentially by mRNAs depending on their 5’UTRs and relative abundance. Clearly this upregulation is adequate to promote efficient translation of MMP13, as its mRNA level is increased gradually.
The activation of the translational machinery in OA chondrocytes can serve several distinct purposes (Fig. S7). First, 4E-BP1 inactivation acts as a double-edge sword to accommodate increased production of proteins those mRNAs are upregulated in OA cartilage, such as matrix proteins and matrix-degrading enzymes. Second, 4E-BP1 inactivation, which results in a higher proportion of active eIF4F, might be crucial for translation of particular mRNAs with long and structured 5’UTR, such as, for example, ADAMTS5. This hypothesis is supported by the high sensitivity of ADAMTS5 translation (not transcription) to 4E-BP activity and CHX (the inhibitor of protein synthesis) treatment of IL-1β stimulated chondrocytes (Fig. S5C). Another potential, yet unidentified 4E-BP sensitive target seems to be crucial for MMP13 transcription, as MMP13 mRNA levels are sensitive to both 4E-BP activity and CHX treatment.
In conclusion, our data show the contribution of translational and non-translational effectors to chondrocyte homeostasis and offer a novel potential set of targets for OA therapy.
Supplementary Material
Fold difference in gene expression of indicated targets (A) in OA cartilage compared to normal cartilage, as determined by microarray analysis and qPCR, n=6 compared to average normal samples. (B) Fold increase in gene expression of MMP13 and ADAMTS5 of lesional versus non lesional samples of the same donor (n=4). (C) Representative IHC of 4EBP1(S65) and negative control of lesional versus non lesional cartilage of the same donor.
Expression of different genes in OA rat model: (A) Fold change in gene expression of MMP13, ADAMTS5, COL2A1 and ACAN 12 and 20 weeks post-surgery in cartilage isolated from ACLT joined compared to sham operated joint (n=3,18S rRNA used for normalization). (B) Expression of LC3b, a central player in the autophagy pathway, in the chondrocytes, isolated from ACLT joint (12 weeks post-surgery) compared to sham operated animals as assayed by qPCR (18S rRNA used for normalization).
Treatment of rat articular chondrocytes (RAC) with IL-1β and IL-1β/OSM (A) Fold difference in gene expression of degradative enzymes and extracellular matrix genes 6 and 24hrs post IL-1β treatment (each dot represents an individual biological sample, compared to untreated control and normalized to 18S rRNA, line at mean) (B,C) Representative immunoblot analysis of IL-1β and L1β+OSM treatment respectively, shows activation of MAPK and STAT3 pathways (α-actin used as a loading control, 15µg of total protein was loaded). (D) Representative immunoblots of MMP13 and MMP3, 6 and 24hrs post IL-1β treatment (α-actin was used as a loading control, 15µg of total protein was loaded). (E) Comparison of the levels of expression of MMP13 and ADAMTS5 gene in IL-1β and combined L1β+OSM treatment of RAC (n=3, compared to untreated control and normalized to 18S rRNA, mean ±SD).
IGF-1 signaling in RAC: Fold difference in gene expression of degradative enzymes and extracellular matrix genes 24hrs post IGF-1 treatment (n=3, compared to untreated control and normalized to 18S rRNA, mean ±SD).
Modulation of 4E-BP1 activity: Gene expression levels of MMP3, COL2A1 and ACAN in (A) DN 4EBP-AA and 4E-BP-AAE mutants and (B) upon 4E-BP1 knockdown post IL-1β treatment (n=3 independent experiments, mRNA expression was normalized to 18S rRNA and untreated wt-control or scr-control respectively, mean ±SD). (C) Gene and protein expression levels of ECM-degrading enzymes and extracellular matrix genes upon IL-1β treatment in the presence of CHX (cycloheximide), the inhibitor of protein synthesis (n=3 independent experiments, mRNA expression was normalized to 18S rRNA and untreated-control, mean ±SD; α-actin used as a loading control, 15µg loaded).
Inhibition of mTORC1 activity in rat articular chondrocytes: (A) mRNA expression levels of COL2A1 and ACAN upon IL-1β treatment in the cells transfected with mTOR siRNA (B) mRNA expression levels of COL2A1 and ACAN upon IL-1β treatment in the cells infected with lentivirus containing shRNA against Raptor.
A model for the changes in the translational apparatus in normal compared to OA cartilage.
Acknowledgments
We are grateful to Dr. Paolo Mignatti for the critical reviewing of the manuscript and Dr. Basilico for his comments. We are thankful to Dr. Kennedy for sharing his animal model for our preliminary experiments. This work was supported by NIH grants R01 AR063128 (to V.K.) and in part R01-AR054817 (to S.B.A.).
REFERENCES
- 1.Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum. 2012 Jun;64(6):1697–1707. doi: 10.1002/art.34453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krasnokutsky S, Attur M, Palmer G, Samuels J, Abramson SB. Current concepts in the pathogenesis of osteoarthritis. Osteoarthritis Cartilage. 2008;16(Suppl 3):S1–S3. doi: 10.1016/j.joca.2008.06.025. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Vasheghani F, Li YH, Blati M, Simeone K, Fahmi H, et al. Cartilage-specific deletion of mTOR upregulates autophagy and protects mice from osteoarthritis. Ann Rheum Dis. 2015 Jul;74(7):1432–1440. doi: 10.1136/annrheumdis-2013-204599. [DOI] [PubMed] [Google Scholar]
- 4.Carames B, Taniguchi N, Otsuki S, Blanco FJ, Lotz M. Autophagy is a protective mechanism in normal cartilage, and its aging-related loss is linked with cell death and osteoarthritis. Arthritis Rheum. 2010 Mar;62(3):791–801. doi: 10.1002/art.27305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasheghani F, Zhang Y, Li YH, Blati M, Fahmi H, Lussier B, et al. PPARgamma deficiency results in severe, accelerated osteoarthritis associated with aberrant mTOR signalling in the articular cartilage. Ann Rheum Dis. 2015 Mar;74(3):569–578. doi: 10.1136/annrheumdis-2014-205743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Adamo S, Alvarez-Garcia O, Muramatsu Y, Flamigni F, Lotz MK. MicroRNA-155 suppresses autophagy in chondrocytes by modulating expression of autophagy proteins. Osteoarthritis Cartilage. 2016 Jan 19; doi: 10.1016/j.joca.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duarte JH. Osteoarthritis: Autophagy prevents age-related OA. Nat Rev Rheumatol. 2015 Dec;11(12):683. doi: 10.1038/nrrheum.2015.145. [DOI] [PubMed] [Google Scholar]
- 8.Bouderlique T, Vuppalapati KK, Newton PT, Li L, Barenius B, Chagin AS. Targeted deletion of Atg5 in chondrocytes promotes age-related osteoarthritis. Ann Rheum Dis. 2016 Mar;75(3):627–631. doi: 10.1136/annrheumdis-2015-207742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hershey JW, Sonenberg N, Mathews MB. Principles of translational control: an overview. Cold Spring Harb Perspect Biol. 2012 Dec;4(12) doi: 10.1101/cshperspect.a011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010 Feb;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, et al. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999 Jun 1;13(11):1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009 May;10(5):307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 13.Attur M, Yang Q, Shimada K, Tachida Y, Nagase H, Mignatti P, et al. Elevated expression of periostin in human osteoarthritic cartilage and its potential role in matrix degradation via matrix metalloproteinase-13. FASEB J. 2015 Oct;29(10):4107–4121. doi: 10.1096/fj.15-272427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Attur MG, Palmer GD, Al-Mussawir HE, Dave M, Teixeira CC, Rifkin DB, et al. F-spondin, a neuroregulatory protein, is up-regulated in osteoarthritis and regulates cartilage metabolism via TGF-beta activation. FASEB J. 2009 Jan;23(1):79–89. doi: 10.1096/fj.08-114363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams JM, Felten DL, Peterson RG, O'Connor BL. Effects of surgically induced instability on rat knee articular cartilage. J Anat. 1982 Jan;134(Pt 1):103–109. [PMC free article] [PubMed] [Google Scholar]
- 16.Ruoff R, Katsara O, Kolupaeva V. Cell type-specific control of protein synthesis and proliferation by FGF-dependent signaling to the translation repressor 4E-BP. Proc Natl Acad Sci U S A. 2016 Jun 16; doi: 10.1073/pnas.1605451113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001 Nov 1;15(21):2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell PG, Magna HA, Reeves LM, Lopresti-Morrow LL, Yocum SA, Rosner PJ, et al. Cloning, expression, and type II collagenolytic activity of matrix metalloproteinase-13 from human osteoarthritic cartilage. J Clin Invest. 1996 Feb 1;97(3):761–768. doi: 10.1172/JCI118475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glasson SS, Askew R, Sheppard B, Carito B, Blanchet T, Ma HL, et al. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005 Mar 31;434(7033):644–648. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 20.Carames B, Hasegawa A, Taniguchi N, Miyaki S, Blanco FJ, Lotz M. Autophagy activation by rapamycin reduces severity of experimental osteoarthritis. Ann Rheum Dis. 2012 Apr;71(4):575–581. doi: 10.1136/annrheumdis-2011-200557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tchetina EV, Poole AR, Zaitseva EM, Sharapova EP, Kashevarova NG, Taskina EA, et al. Differences in Mammalian target of rapamycin gene expression in the peripheral blood and articular cartilages of osteoarthritic patients and disease activity. Arthritis. 2013;2013:461486. doi: 10.1155/2013/461486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franke TF, Yang SI, Chan TO, Datta K, Kazlauskas A, Morrison DK, et al. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995 Jun 2;81(5):727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 23.Xu W, Xie Y, Wang Q, Wang X, Luo F, Zhou S, et al. A novel fibroblast growth factor receptor 1 inhibitor protects against cartilage degradation in a murine model of osteoarthritis. Sci Rep. 2016;6:24042. doi: 10.1038/srep24042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu L, Polur I, Lim C, Servais JM, Dobeck J, Li Y, et al. Early-onset osteoarthritis of mouse temporomandibular joint induced by partial discectomy. Osteoarthritis Cartilage. 2009 Jul;17(7):917–922. doi: 10.1016/j.joca.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovacina KS, Park GY, Bae SS, Guzzetta AW, Schaefer E, Birnbaum MJ, et al. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J Biol Chem. 2003 Mar 21;278(12):10189–10194. doi: 10.1074/jbc.M210837200. [DOI] [PubMed] [Google Scholar]
- 26.Goldring SR, Goldring MB. The role of cytokines in cartilage matrix degeneration in osteoarthritis. Clin Orthop Relat Res. 2004 Oct;(427 Suppl):S27–S36. doi: 10.1097/01.blo.0000144854.66565.8f. [DOI] [PubMed] [Google Scholar]
- 27.Goldring MB, Otero M. Inflammation in osteoarthritis. Curr Opin Rheumatol. 2011 Sep;23(5):471–478. doi: 10.1097/BOR.0b013e328349c2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greene MA, Loeser RF. Function of the chondrocyte PI-3 kinase-Akt signaling pathway is stimulus dependent. Osteoarthritis Cartilage. 2015 Jun;23(6):949–956. doi: 10.1016/j.joca.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gouze JN, Bianchi A, Becuwe P, Dauca M, Netter P, Magdalou J, et al. Glucosamine modulates IL-1-induced activation of rat chondrocytes at a receptor level, and by inhibiting the NF-kappa B pathway. FEBS Lett. 2002 Jan 16;510(3):166–170. doi: 10.1016/s0014-5793(01)03255-0. [DOI] [PubMed] [Google Scholar]
- 30.Ni J, Yuan XM, Yao Q, Peng LB. OSM is overexpressed in knee osteoarthritis and Notch signaling is involved in the effects of OSM on MC3T3-E1 cell proliferation and differentiation. Int J Mol Med. 2015 Jun;35(6):1755–1760. doi: 10.3892/ijmm.2015.2168. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Martinez JM, Moran J, Clarke RG, Gray A, Cosulich SC, Chresta CM, et al. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem J. 2009 Jul 1;421(1):29–42. doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012 Apr;13(4):227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loeser RF, Olex AL, McNulty MA, Carlson CS, Callahan MF, Ferguson CM, et al. Microarray analysis reveals age-related differences in gene expression during the development of osteoarthritis in mice. Arthritis Rheum. 2012 Mar;64(3):705–717. doi: 10.1002/art.33388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Appleton CT, Pitelka V, Henry J, Beier F. Global analyses of gene expression in early experimental osteoarthritis. Arthritis Rheum. 2007 Jun;56(6):1854–1868. doi: 10.1002/art.22711. [DOI] [PubMed] [Google Scholar]
- 35.Poulet B, Ulici V, Stone TC, Pead M, Gburcik V, Constantinou E, et al. Time-series transcriptional profiling yields new perspectives on susceptibility to murine osteoarthritis. Arthritis Rheum. 2012 Oct;64(10):3256–3266. doi: 10.1002/art.34572. [DOI] [PubMed] [Google Scholar]
- 36.Lourido L, Calamia V, Mateos J, Fernandez-Puente P, Fernandez-Tajes J, Blanco FJ, et al. Quantitative proteomic profiling of human articular cartilage degradation in osteoarthritis. J Proteome Res. 2014 Dec 5;13(12):6096–6106. doi: 10.1021/pr501024p. [DOI] [PubMed] [Google Scholar]
- 37.Tew SR, McDermott BT, Fentem RB, Peffers MJ, Clegg PD. Transcriptome-wide analysis of messenger RNA decay in normal and osteoarthritic human articular chondrocytes. Arthritis Rheumatol. 2014 Nov;66(11):3052–3061. doi: 10.1002/art.38849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takayama K, Kawakami Y, Kobayashi M, Greco N, Cummins JH, Matsushita T, et al. Local intra-articular injection of rapamycin delays articular cartilage degeneration in a murine model of osteoarthritis. Arthritis Res Ther. 2014;16(6):482. doi: 10.1186/s13075-014-0482-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shakibaei M, John T, Schulze-Tanzil G, Lehmann I, Mobasheri A. Suppression of NF-kappaB activation by curcumin leads to inhibition of expression of cyclo-oxygenase-2 and matrix metalloproteinase-9 in human articular chondrocytes: Implications for the treatment of osteoarthritis. Biochem Pharmacol. 2007 May 1;73(9):1434–1445. doi: 10.1016/j.bcp.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Cui X, Wang S, Cai H, Lin Y, Zheng X, Zhang B, et al. Overexpression of microRNA-634 suppresses survival and matrix synthesis of human osteoarthritis chondrocytes by targeting PIK3R1. Sci Rep. 2016;6:23117. doi: 10.1038/srep23117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greene MA, Loeser RF. Aging-related inflammation in osteoarthritis. Osteoarthritis Cartilage. 2015 Nov;23(11):1966–1971. doi: 10.1016/j.joca.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fold difference in gene expression of indicated targets (A) in OA cartilage compared to normal cartilage, as determined by microarray analysis and qPCR, n=6 compared to average normal samples. (B) Fold increase in gene expression of MMP13 and ADAMTS5 of lesional versus non lesional samples of the same donor (n=4). (C) Representative IHC of 4EBP1(S65) and negative control of lesional versus non lesional cartilage of the same donor.
Expression of different genes in OA rat model: (A) Fold change in gene expression of MMP13, ADAMTS5, COL2A1 and ACAN 12 and 20 weeks post-surgery in cartilage isolated from ACLT joined compared to sham operated joint (n=3,18S rRNA used for normalization). (B) Expression of LC3b, a central player in the autophagy pathway, in the chondrocytes, isolated from ACLT joint (12 weeks post-surgery) compared to sham operated animals as assayed by qPCR (18S rRNA used for normalization).
Treatment of rat articular chondrocytes (RAC) with IL-1β and IL-1β/OSM (A) Fold difference in gene expression of degradative enzymes and extracellular matrix genes 6 and 24hrs post IL-1β treatment (each dot represents an individual biological sample, compared to untreated control and normalized to 18S rRNA, line at mean) (B,C) Representative immunoblot analysis of IL-1β and L1β+OSM treatment respectively, shows activation of MAPK and STAT3 pathways (α-actin used as a loading control, 15µg of total protein was loaded). (D) Representative immunoblots of MMP13 and MMP3, 6 and 24hrs post IL-1β treatment (α-actin was used as a loading control, 15µg of total protein was loaded). (E) Comparison of the levels of expression of MMP13 and ADAMTS5 gene in IL-1β and combined L1β+OSM treatment of RAC (n=3, compared to untreated control and normalized to 18S rRNA, mean ±SD).
IGF-1 signaling in RAC: Fold difference in gene expression of degradative enzymes and extracellular matrix genes 24hrs post IGF-1 treatment (n=3, compared to untreated control and normalized to 18S rRNA, mean ±SD).
Modulation of 4E-BP1 activity: Gene expression levels of MMP3, COL2A1 and ACAN in (A) DN 4EBP-AA and 4E-BP-AAE mutants and (B) upon 4E-BP1 knockdown post IL-1β treatment (n=3 independent experiments, mRNA expression was normalized to 18S rRNA and untreated wt-control or scr-control respectively, mean ±SD). (C) Gene and protein expression levels of ECM-degrading enzymes and extracellular matrix genes upon IL-1β treatment in the presence of CHX (cycloheximide), the inhibitor of protein synthesis (n=3 independent experiments, mRNA expression was normalized to 18S rRNA and untreated-control, mean ±SD; α-actin used as a loading control, 15µg loaded).
Inhibition of mTORC1 activity in rat articular chondrocytes: (A) mRNA expression levels of COL2A1 and ACAN upon IL-1β treatment in the cells transfected with mTOR siRNA (B) mRNA expression levels of COL2A1 and ACAN upon IL-1β treatment in the cells infected with lentivirus containing shRNA against Raptor.
A model for the changes in the translational apparatus in normal compared to OA cartilage.