Abstract
Detection of evolutionary conserved molecules on microbial pathogens by host immune sensors represents the initial trigger of the immune response against infection. Cytosolic receptors sense viral and intracellular bacterial genomes, as well as nucleic acids produced during replication. Once activated, these sensors trigger multiple signaling cascades, converging on the production of type I interferons and proinflammatory cytokines. Although distinct classes of receptors are responsible for the RNA and DNA sensing, the downstream signaling components are physically and functionally interconnected. This review will highlight the importance of the crosstalk between RIG-I-MAVS RNA sensing and the cGAS-STING DNA sensing pathways in potentiating efficient antiviral responses. The potential of cGAS-STING manipulation as a component of cancer immunotherapy will also be reviewed.
Innate Sensing of Pathogens
Scattered throughout the body and particularly concentrated in mucous membranes and skin, the cells of the innate immune system provide a first line of defense against the potential pathogens. Granulocytes, macrophages, dendritic and natural killer cells recognize microbes using a relatively small set of evolutionarily ancient, germline-encoded sensors called pattern recognition receptors (PRRs). These receptors identify conserved structures, such as cell wall components, that are usually unique to microbes and often essential for microbial existence, collectively referred to as pathogen-associated molecular patterns (PAMPs) [1]. Pathogen-PRR association drives the initial innate immune response and forms the basis for the generation of an effective adaptive immune response. Extracellular microorganisms and viruses are detected by PRRs strategically located at the cell surface, including members of the Toll-like receptor family, as well as the C-Type lectin receptors, whose biochemical and physiological functions have been reviewed extensively [2–4].
Viruses represent a unique challenge to innate immune receptors, because they possess few unique features that could serve as PAMPs. Therefore, innate immune detection of viruses relies on the recognition of viral genomes that differs from host nucleic acids in some unusual biochemical features; for example, viral RNAs can be tri- or di- phosphorylated, and lack a 7-methylguanosine cap. In DNA virus-infected cells, the crucial discrimination between self and non-self is achieved by specific localization of PRRs in the endosomal or cytosolic compartments, to which DNA has limited access in healthy cells (for a more exhaustive review of self/non-self nucleic acid discrimination, see [5]). Most eukaryotic DNA is enclosed in the nucleus or in mitochondria of cells; the appearance of DNA in the cytoplasm during an infection can lead to its detection by specific PRRs and the initiation of downstream signaling pathways. Studies performed in recent years have uncovered the components, structures and mechanisms involved in cytosolic nucleic acid detection (Figure 1). This review will focus on the innate immune responses triggered by pathogenic viral genomes or endogenous damaged DNA, and will address the interrelationship between cytoplasmic RIG-I MAVS RNA sensing and cGAS-STING DNA sensing pathways.
Figure 1. Schematic Representation of the Intracellular Viral Sensing Pathways.
DNA and RNA viruses release their genomes in the cytoplasm, where innate sensors for nucleic acids reside. Upon ss/dsRNA binding, RIG-I engages the adaptor protein MAVS on the mitochondria outer membrane. The cGAS receptor in contrast recognizes dsDNA and the RNA:DNA hybrids generated during retroviral replication, and catalyzes the synthesis of cGAMP, which is the paramount agonist of the adaptor protein STING. Another sensor, IFI16 can recruit STING in response to cytoplasmic DNA, through a molecular mechanism yet to be described. Both STING and MAVS stimulate downstream signaling cascades that involve multiple kinases and finally lead to IRF3 phosphorylation and nuclear translocation. The primary consequence of these virus sensing pathways is the induction of type I IFN and IFN stimulated genes. Abbreviations used: ss/dsRNA, single-stranded/double-stranded RNA, vRNA/DNA, viral RNA/DNA; RIG-I, retinoic acid inducible gene-I; MAVS, mitochondrial antiviral-signaling protein; cGAS, cyclic GMP-AMP synthase; cGAMP, 2’3’ guanosine-adenosine monophosphate; STING, stimulator of interferon genes; TANK, TRAF-associated NFκB activator; TBK1, TANK binding kinase 1; IKK, IκB kinase; IFI16, Interferon Gamma Inducible Protein 16; IRF3, interferon regulatory factor 3.
Cytosolic Nucleic Acid Sensing Pathways
RIG-I Mediated Sensing of Cytosolic RNA
Retinoic acid inducible gene-I (RIG-I)-like receptors (RLRs) are responsible for the detection of single stranded (ss) and double-stranded (ds) RNA generated in the course of a virus infection [6]. This receptor family contains three members: RIG-I, melanoma differentiation associated gene 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2). All RLRs are characterized by a central DEAD box helicase/ATPase domain and a C-terminal regulatory domain (CTD), essential for RNA binding and for the autorepression of RLR activity. RIG-I and MDA5 possess, moreover, two N-terminal tandemly linked caspase activation and recruitment domains (CARDs), that mediate signaling to downstream adaptor proteins [7]. These two RLRs are activated by specific RNA motifs: RIG-I preferentially detects 5’ end-di/triphosphorylated (5′pp/5’ppp) RNA sequences rich in poly-U or poly-UC tracts, whereas MDA5 responds to high-molecular weight viral RNAs and to the synthetic dsRNA poly(I:C). Accordingly, the involvement of the two receptors in different virus infections is variable [8].
In uninfected cells, RIG-I is maintained in a closed, autoinhibited state. Binding of 5′ ppp-RNA to the CTD/helicase region triggers dephosphorylation, ubiquitination and ATP-dependent conformational changes that promote RIG-I oligomerization; the resulting tetramer, resembling a lock-washer in its 3D structure, serves as a scaffold for binding to the adaptor mitochondrial antiviral-signaling protein (MAVS) on the surface of mitochondria [9, 10]. Once activated by RIG-I via CARD-CARD homotypic interactions, MAVS forms aggregates, described as prion-like structures, that self-perpetuate the conversion of inactive MAVS into functional multimeric filaments to which numerous adapter proteins and kinases are recruited [11, 12]. From this point, RIG-I signaling bifurcates into two molecular cascades: one involves the Tank binding kinase-1 (TBK1) and IκB kinase epsilon (IKK ɛ), that directly phosphorylate the interferon (IFN) regulatory factors 3 and 7 (IRF3, IRF7), to promote the expression of type I and type III IFNs; the other pathway engages the IKKα/β/γ complex, and leads to the NF-κB dependent upregulation of proinflammatory genes [7]. The IFN response induced by RIG-I stimulation effectively inhibits human pathogenic RNA virus infection, as well as DNA virus infection (Figure 2).
Figure 2. STING is a RIG-I Inducible Gene.
In cells infected by JEV, SeV or treated with 5’pppRNA, RIG-I signaling is strongly activated; through the NF-κB and IRF3 cascades this pathway induces the production of IFNα/β and proinflammatory cytokines such as TNFα through an autocrine/paracrine manner that synergistically upregulates STING expression. By a positive feedback mechanism, induction of STING contributes to a sustained inflammatory response that protects treated cells from infection, including DNA virus infection. Abbreviations used: JEV, Japanese encephalitis virus; SeV, Sendai virus; VSV, vesicular stomatitis virus; HCV hepatitis C virus; 5’pppRNA, 5’-triphosphorylated RNA; TRAF, TNF receptor associated factor; MAVS, mitochondrial antiviral-signaling protein; RIG-I, Retinoic acid inducible gene-I; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; IκB, inhibitor of kappa B; IKK, IκB kinase; IRF, interferon regulatory factor; IFNα/β, Interferon α/β; IFNAR: IFNα receptor; JAK, Janus kinase; TYK: tyrosine kinase; STAT, signal transducer and activator of transcription; ISGF3, IFN-stimulated gene factor 3; STING, stimulator of interferon genes; HSV-1, Herpes simplex virus type 1.
Because of the potential to develop new broadly acting antiviral strategies based on stimulation of these evolutionarily conserved innate immune pathways, considerable effort has been directed toward the development of small molecules that bind to RIG-I or MDA5, activate the downstream antiviral response and protect cells against a range of RNA viruses [13]. A recently characterized sequence-optimized immunostimulatory RNA (termed M8) that maintains specificity for RIG-I engagement was identified as a potent antiviral agent that blocked influenza, chikungunya and dengue infection in vitro and in vivo in animal models of viral pathogenesis [14, 15]. At picomolar concentrations, M8 stimulated in MoDCs greater IFN and inflammatory responses compared to other RLR binding sequences; this immune modulation required RIG-I, but was not affected by knockdown of other RNA receptors, a clear indication of M8 selectivity for RIG-I. The adjuvant properties of M8 were also examined in vaccination studies in combination with influenza viruslike particles (VLP). In combination with VLP, M8 increased the antibody response to VLP immunization, provided VLP antigen sparing, and protected mice from a lethal challenge with influenza H5N1. M8-VLP immunization also led to long term protective responses against influenza infection in mice [14, 15]. Collectively, these data argue that stimulation of the conserved RIG-I pathway may be an additional target for antiviral and vaccine development.
cGAS-STING Mediated Sensing of Cytosolic DNA
While the cytosolic recognition of viral RNA is mediated almost exclusively by RLRs, many different sensors have been implicated in the detection of non-self DNA, present at aberrant concentrations or locations within the cell (for a more exhaustive review of this topic, see [16] and [17]). A central regulator of cytosolic DNA sensing is cyclic GMP-AMP (cGAMP) synthase (cGAS), a nucleotidyl transferase previously described to be upregulated by IFN and to possess a broad antiviral activity [18, 19]. An activation loop within this molecule binds dsDNA in a sequence-independent manner and undergoes rearrangements that promote cGAS dimerization [20]; once activated, cGAS catalyzes the synthesis of two phosphodiester bonds between a GMP and a AMP, thus resulting in the generation of 2’3’cGAMP [21–24].
With a higher affinity than the bacterial cyclic dinucleotides (CDNs), mammalian cGAMP binds to an essential cytosolic sensor which was independently identified by several groups: stimulator of interferon genes (STING), also known as mitochondrial mediator of IRF3 activation (MITA), N-terminal methionine-proline-tyrosine-serine plasma membrane tetraspanner (MPYS) or endoplasmic reticulum IFN stimulator (ERIS) [25–28]. In resting conditions, STING is anchored as a homodimer within the endoplasmic reticulum (ER) membrane, at the interface between the mitochondrion and the ER [29]. When CDNs bind STING within a groove between two monomers, the autoinhibited state caused by intramolecular interaction between the C-terminal tail and the CDN binding domain is relieved, and an E3 ubiquitin ligase complex conjugates K27-linked polyubiquitin polymers to STING [30, 31]. In this conformation, STING is able to bind TBK1, and together they translocate from the ER via the Golgi to perinuclear endosomes. In the trans-Golgi network, STING undergoes palmitoylation on Cys88 and Cys91, an essential event for the subsequent activation of TBK1 [32]. Phosphorylated on Ser366 by TBK1, STING interacts with and activates IRF3, thus inducing IFNβ upregulation; STING also drives NF-κB phosphorylation, nuclear translocation, and target gene expression [33]. As a negative regulatory feedback mechanism to prevent the overstimulation of innate immune genes, E3 ligases are responsible for ubiquitin-dependent STING degradation in viral infections [34, 35].
A unique aspect of the cGAS-STING response is that cGAMP is able to move between infected and bystander cells via gap junction communication; cGAMP can also be incorporated during the assembly of new virions and therefore transferred to newly infected cells [36–38]. The cell-cell transmission of cGAMP into neighbouring, uninfected cells thus propagates an IFN and cytokine-independent spread of antiviral immunity through STING activation, leading to the production of antiviral effector ISGs that limited virus propagation (for a more detailed review on the cGAS-STING pathway, see the recent review [39]).
Cross-Talk Between RNA and DNA sensing pathways
Cytoplasmic nucleic acids lead to the generation of additional ligands for RIG-I-MAVS and cGAS-STING
While the RNA and DNA pathways rely on different receptors and adaptors, it is clear that they at least partially overlap, as highlighted by the fact that the IFN response to poly(dA:dT) was reduced by >99% in STING−/−, MAVS−/− macrophages and dendritic cells, but not in phagocytes deficient in STING alone [40]. A molecular mechanism linking DNA sensing with the RIG-I pathway was first identified by Chiu et al, who demonstrated that cytosolic DNA could be used as a template for RNA polymerase (pol) III-driven synthesis of dsRNA [41]. Newly synthesized dsRNA was shown to bind RIG-I, activate MAVS and induce IFNβ production (Figure 3). RNA pol III could thus be considered a cytosolic sensor for bacterial and viral DNA, specifically recognizing poly(dA:dT) sequences [41].
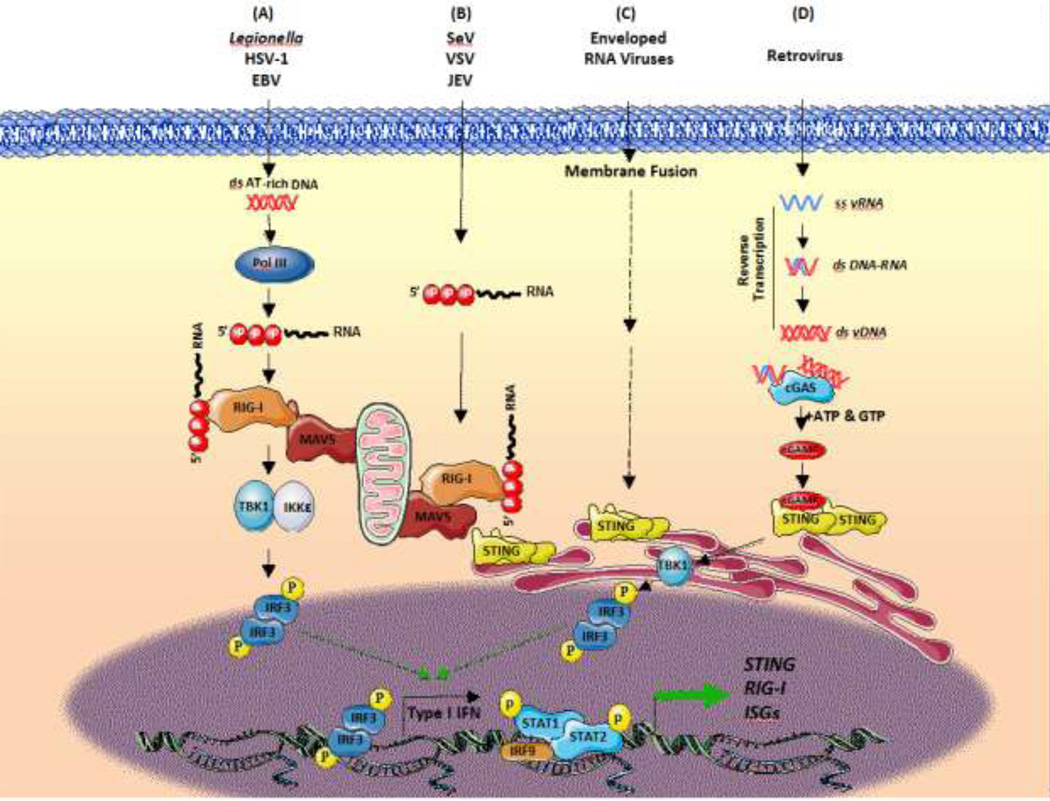
Figure 3. Cross-talk between the Viral RNA and DNA Sensing Pathways.
(A) AT-rich dsDNA from bacteria (Legionella) or DNA viruses (EBV, HSV-1) can be used as a template for RNA polymerase (Pol) III-driven synthesis of dsRNA bearing a 5’ triphosphate end group. Newly synthesized dsRNA binds RIG-I, activates MAVS and induces IFNβ production through TBK1-induced IRF3 phosporylation. (B) STING interacts with RIG-I and MAVS to facilitate the triggering of the antiviral response in a complex that is stabilized upon RNA virus infection. (C) Enveloped RNA viruses such as influenza A activate the STING-IFN axis independently of cGAS through a membrane fusion process. (D) The RNA genome of retroviruses is reverse transcribed in a multistep reaction that generates DNA:RNA duplexes and ssDNA as intermediates, and dsDNA molecules as final products. The cGAS-STING axis is stimulated by retroviral replicative intermediates to counteract infection and limit proviral integration via induction of ISG. Abbreviations used: HSV, herpes simplex virus; EBV, Epstein-Barr virus; SeV, Sendai virus; VSV, vesicular stomatitis virus; JEV, Japanese encephalitis virus; RIG-I, retinoic acid inducible gene-I; MAVS, mitochondrial antiviral-signaling protein; cGAS, cyclic GMP-AMP synthase; cGAMP, 2’3’ guanosine-adenosine monophosphate; STING, stimulator of interferon genes; TBK1, TANK binding kinase 1; IKK, IκB kinase; IRF3, interferon regulatory factor 3; STAT, signal transducer and activator of transcription; ISGs, interferon stimulated genes.
Significant cross-talk between DNA and RNA sensing mechanisms was also implied from studies with the human retroviruses HIV-1 and HTLV-1 (Figure 3). The RNA genome of retroviruses is reverse transcribed in a multistep reaction that generates DNA:RNA duplexes and ssDNA as intermediates, and dsDNA molecules as final products [42–44]. In HIV and HTLV-1 infected T lymphocytes and myeloid cells, DNA replicative intermediates activate the IFI16/cGAS-STING axis to counteract retroviral infection and proviral integration via induction of ISG and apoptosis [42, 44–47]. Unlike the sequence independent binding of dsDNA, cGAS exclusively recognizes particular structures within ssDNA, characterized by short double-stranded loops (12–20bp) flanked by unpaired guanosine residues [48] (Table 1).
Table 1.
Mechanisms of cGAS-STING and RIG-I-MAVS Crosstalk
| Nucleic Acid | Mechanism | References |
|---|---|---|
| Bacterial/viral DNA | RNA pol III-mediated transcription in dsRNA, RIG-I activation |
[41] |
| ssRNA retroviruses | Reverse transcriptase mediated generation of RNA:DNA hybrids and ss/dsDNA molecules; cGAS activation |
[47, 48] |
| RNA viruses | Assembly of the RIG-I/MAVS/STING complex | [26, 27, 51] |
| ssDNA | Upregulation of RIG-I mRNA | [58] |
| RNA viruses, Poly(I:C), M8 | Upregulation of STING mRNA | [51, 59, 60] |
| RNA viruses | Coactivation of RIG-I and STING pathways; potentiated IFN response |
[53, 62, 63] |
STING can directly transmit RIG-I-MAVS mediated signals
A number of additional mechanisms demonstrating the interplay between DNA and RNA sensing have been found, with STING identified as the central player in this crosstalk (Figure 2). STING deficiency in cell lines, primary cultures and mouse models significantly increased permissivity to RNA virus infection, while reducing type I IFN production [25, 27, 49]. In human embryonic stem cells, the absence of STING expression correlated with the inability of RIG-I to produce IFN in response to cytoplasmic dsRNA [50]. Evidence for a role for STING in potentiating RIG-I-mediated antiviral signaling also came from studies with Japanese encephalitis virus (JEV), a flavivirus inducing neuroinflammation in humans. The ssRNA genome of JEV is recognized by RIG-I, which then recruits STING to initiate a downstream cascade leading to the antiviral response. Genetic ablation of STING inhibited activation of the IRF/IFN pathway, reduced the expression of IFN-related genes and inhibited cytokine and chemokine release, leading to increased intracellular viral load [51].
In several studies STING appeared to interact with RIG-I and MAVS, in a complex that was stabilized upon virus infection [26, 27, 51] (Figure 3 and Table 1). The involvement of STING in transmitting RIG-I signaling is so fundamental that many RNA viruses have evolved strategies to block STING-dependent innate immunity. The serine protease NS4B of hepatitis C virus competed with MAVS for binding to STING on mitochondria-associated membranes; this interaction resulted in displacement of MAVS from the RIG-I/MAVS/STING complex and impeded activation of downstream effectors [52]. Also, enveloped RNA viruses such as influenza A activated the STING-IFN axis independently of cGAS, and this mechanism was blocked by STING interaction with the viral hemagglutinin [53]. Dengue virus was shown to antagonize STING-dependent IFN production via the NS2B3 protease that physically interacted with and cleaved STING. Variants of STING with mutations in the cleavage site induced stronger IFN responses to dengue [54, 55].
Conversely, a potentiating role for RNA sensing mechanisms in the activation of host responses against non-self DNA has also been identified. In HeLa or HepG2 cells, TBK-1 phosphorylation after DNA transfection or DNA virus infection relied on MAVS-TBK1 interaction. Knockdown of MAVS in these cells markedly reduced p-TBK1 and IFN-β levels induced by cytoplasmic DNA [56]. Recently, a small molecule termed G10 capable of activating type I and type III IFN in human cells was identified [57]. Although not a direct ligand for STING, the synthetic G10 compound stimulated the STING-IRF3 axis and protected cells against alphavirus infection without targeting the RIG-I pathway; nonetheless, G10-induced IFN secretion was strongly reduced in follicular helper CD4 T cells lacking MAVS [57].
Expression levels of cytoplasmic nucleic acid sensing pathways are co-regulated
Crosstalk by the DNA and RNA cytosolic sensing pathways occurs not only at the level of physical interaction or transactivation between the mediators of the two pathways, but also by coordinate regulation of STING and RIG-I expression levels (Figure 3). Chronic stimulation of the DNA immune sensing, mimicked by γ-irradiation or treatment with the DNA damaging agent etoposide, induced not only various components of the DNA sensing pathway, including cGAS, IFI16 and STING, but also RIG-I in bone marrow derived macrophages (BMDMs) [58]. As a consequence, when challenged with a RNA virus, BMDMs treated with etoposide mounted a high IFNβ response that suppressed virus replication [58]. Conversely, RNA viruses, poly(I:C) and 5′pppRNA upregulated STING in rat and mouse models, thus demonstrating a contribution of RIG-I signaling to STING-mediated protection against DNA viruses [51, 59, 60]. A synergistic interplay between DNA and RNA sensing pathways was also observed in studies examining the off-target immunostimulatory effects of siRNAs. siRNA treatment enhanced, in a target-independent manner, IFN-λ1 production triggered by DNA transfection or DNA virus infection; RIG-I, IFI16, STING, TBK-1 and IRF3 were indispensable for IFN-λ1 production [61].
To date, no direct role for cytosolic DNA receptors in the recognition and response to foreign RNA has been described [17, 18]. One study did report cGAS binding to a synthetic 50mer dsRNA, although this interaction did not result in the generation of cGAMP [24]. West Nile virus (WNV) infection induced a significantly higher mortality in cGAS−/− mice than in wild type animals, and higher viral titers were quantified in cGAS−/− macrophages compared to controls [62]. The effect was not related to a direct sensing of viral RNA genome by cGAS, but rather to a cGAS-mediated modulation of IFN-stimulated genes (ISGs). cGAS deficient macrophages exhibited in fact low ISG expression level following infection which provided an advantage for WNV replication [62]. Similarly, silencing of the DNA sensor IFI16 attenuated IFNα/β responses to RNA viruses or synthetic 5′-ppp RNA [63]. It was suggested that IFI16 could regulate expression by directly binding to the IFN promoter and facilitating the recruitment of RNA polymerase II to activate gene transcription [63].
Implications of STING and RIG-I Agonists in Cancer Therapy
In addition to its essential function in the detection of invading microbe genomes, STING also has an equally important function as a sensor of self DNA, released from the nucleus into the cytoplasm after DNA damage, a scenario characteristic of autoimmune diseases. In fact, the cGAS-STING pathway contributes to the high levels of circulating cytokines found in inflammation-related disorders [64]. Furthermore, numerous studies have highlighted the importance of STING in antitumor immunity. DNA derived from dying tumor cells can enter the cytosol of DCs as a consequence of TLR9 ligation, phagocytosis or cell-cell contact, leading to the induction of STING signaling. The ensuing type I IFN production activates DCs in an autocrine manner, resulting in cross-priming of tumor-specific CD8+ T cells [65]. This model was corroborated by the observation that in STING −/− mice bearing an F16 melanoma, spontaneous priming of antitumor T cells was severely reduced and tumor rejection suppressed [66]. The absence of STING also abrogated the effects of radiotherapy on tumor growth, IFNβ production and DC-mediated cross-priming [67]. The role of STING in cancer immunosurveillance is not limited to DC activation; STING expression in tumor and host immune cells also inhibited melanoma growth by generating a chemokine profile in the tumor microenvironment that was favorable to cytotoxic NK cell infiltration [68].
These observations provided a rationale for the utilization of CDNs as molecular adjuvants, together with tumor-derived antigens to initiate an efficient cellular and humoral response against malignancies [69]. c-di-GMP was shown to be a powerful adjuvant for anticancer vaccines, regulating lymphocyte infiltration and inflammation in tumor microenvironment, as well as triggering apoptotic pathways in tumor cells [70–72]. Another molecule - 5,6-dimethyllxanthenone-4-acetic acid (DMXAA), a selective agonist for murine STING, was shown to possess significant anticancer properties in animal studies; however, the inability of DMXAA to interact with human STING limited its potential use in human cancer immunotherapy [73].
The search for new synthetic human STING agonists to potentiate antitumor immunity has intensified. Corrales et al. modified natural CDNs to develop ligands for both mouse and human STING; intratumoral injection of these synthetic compounds triggered a systemic, tumor specific T-cell response that caused regression of the primary tumor, as well as distant metastases [74]. In an alternate strategy, Fu et al. introduced chemical modifications into natural CDNs that conferred resistance to phosphodiesterase degradation and a higher affinity to human STING [70]. The resulting compounds were used as adjuvants in granulocyte-macrophage colony-stimulating factor secreting cells to generate a potent cancer vaccine, termed STINGVAX. In vivo experiments in melanoma, colon carcinoma and pancreatic carcinoma demonstrated the ability of STINGVAX to reduce tumor growth. STINGVAX was also directly responsible for lymphocyte tumor infiltration and for high-level expression of PD-L1 in the tumor microenvironment. Combination treatment with PD-1-blocking antibody and STINGVAX induced regression of established tumors and blocked the development of new tumors after a second inoculation [75].
Not all studies support the anti-tumor potential of STING agonists. In some tumor models, cGAMP in the tumor microenvironment actually appears to favor tumor progression. In the brain, metastatic tumor growth and chemoresistance are supported by glia: cancer cells were shown to associate with normal astrocytes and to transfer cGAMP via gap junctions. As a consequence of STING stimulation, astrocytes produced and secreted IFNα and TNF, two paracrine factors that activated survival pathways in tumor cells [76]. The potential of DMXAA as a chemotherapeutic agent has also been questioned; when used as an adjuvant in combination with a model antigen, DMXAA preferentially induced a Th2 response [77], which is considered antagonistic to the antitumor immunity mediated by Th1 cells.
An unexpected role for STING in promoting cancer was observed in epidermal carcinogenesis, chemically induced by the polycyclic aromatic hydrocarbon 7,12-dimethylbenz[α]anthracene (DMBA); the DMBA-damaged DNA was released into the cytosol and activated STING to produce proinflammatory cytokines that attracted inflammatory cells. After phagocytosis, DNA from engulfed keratinocytes further activated STING to propagate inflammation and facilitate skin tumorigenesis. STING−/− mice treated with DMBA failed to produce pro-inflammatory cytokines or develop tumors, evidencing that STING was required for the carcinogenic activity of DMBA [78]. These STING mediated effects reflect the dual role of the immune system in protecting from malignant transformation, or generating chronic inflammatory conditions that promote tumorigenesis.
Another crucial protein involved in the STING-IFNα/β pathway in hematopoietic cells is the immunosuppressor indoleamine 2,3-dioxygenase (IDO) [79]. Upon sensing of DNA from dying tumor cells, STING elevated IDO activity in murine, tumor-driven node-resident DCs. The activated DCs inhibited effector T cells and stimulated Foxp3+ regulatory CD4 T cells to suppress tumor immunity. In STING−/− mice, higher levels of tumor infiltrating lymphocytes were detected, whereas the number of myeloid suppressor cells decreased, as did levels of the immunoregulatory cytokine IL10, and tumor growth was attenuated. These observations are not universally true, and antigenicity is probably a key factor influencing these immune responses. In particular, the above described tolerogenic induction was observed in mice bearing Lewis lung carcinoma, a tumor with low antigenicity. In tumors with higher antigenicity, the STING pathway likely promoted immunogenic responses instead of tumorigenesis and IDO activation [80]. Altogether, the important considerations of microenvironment, cell specificity of response, tumor immunogenicity and nature of the agonists are all likely to impact the efficacy of STING-mediated immunotherapy.
RIG-I and MDA5 also represent potential targets for cancer therapy. Cancer cell lines and tumors are highly susceptible to apoptotic pathways induced by RLR ligands [81–83]; this event has the characteristics of immunogenic cell death, resulting in IFN production, DC activation and antigen cross-presentation [81, 84]. In malignant conditions, RIG-I expression can be regulated at the level of promoter activity [85] or via miRNA targeting its 3′ untranslated region [86, 87]. Because of its significant downregulation in human and mouse models of disease, RIG-I represents a prognostic biomarker for hepatocellular carcinoma (HCC) [88]; when overexpressed in HCC, melanoma and cervical cancer cells, RIG-I inhibited proliferation and cell cycle progression by blocking both mitogenic MAPKs and survival PI3K/AKT pathways [86, 87, 89]. RIG-I was recently shown to be essential for radio/chemotherapy sensitivity; the RNA sensing pathway was activated in tumor and host irradiated cells by small non-coding RNAs translocated from the nucleus to the cytoplasm; the resultant RIG-I-RNA complexes activated a signaling cascade leading to the cytotoxic IFNβ response [90].
RIG-I agonists have also been successfully used as vaccine adjuvants: M8 and 5’-pppRNA, in combination with influenza hemagglutinin as antigen, protected mice against challenge with a lethal inoculation of influenza, an effect dependent on antibody production and cytotoxic T lymphocyte activation [14, 91]. With their ability to induce tumor cell death and lymphocyte cross-priming, RIG-I ligands are among the most promising molecules for the development of new immune-stimulatory adjuvants in cancer vaccines [69]. A model, based in part on recent data, illustrates how RIG-I stimulation by 5’pppRNA, coupled with potentiation of the response by STING, could impact adaptive immune responses in cancer immunotherapy and/or vaccination (Figure 4).
Figure 4. A Model of RIG-I Mediated Stimulation of Adaptive Immune Responses.
1. RIG-I agonist activates cGAS-STING in DC (dendritic cells). Stimulation of the RIG-I cytosolic sensor by 5’pppRNA engages the MAVS adapter, resulting in release of reactive oxygen species (ROS) and mitochondrial DNA release that in turn stimulates cGAS-dependent production of cGAMP, activation of the STING sensor on the endoplasmic reticulum, downstream activation of IRF-3 and production of type I interferon (IFN). 2. Activated RIG-I and STING lead to type I IFN-dependent DC activation. 5’pppRNA in combination with type I IFN induces DC maturation, based on enhanced surface marker expression and increased antigen presenting capacity. 3. Activated DC drive adaptive immune responses. Activated DC engage CD4 + T cells and stimulate a Th1 skewed T cell response that includes IFNγ release, maturation of B lymphocytes to antibody producing plasma cells and the production of antibodies directed against viral or tumor antigens (14,15). Abbreviations used: RIG-I, retinoic acid inducible gene-I; MAVS, mitochondrial antiviral-signaling protein; IFN, interferon; cGAS, cyclic GMP-AMP synthase; cGAMP, 2’3’ guanosine-adenosine monophosphate; STING, stimulator of interferon genes
Concluding Remarks
Innate cytosolic sensing of foreign nucleic acids represents an important trigger of innate immune responses in several pathologic conditions, ranging from microbial and viral infections to inflammatory diseases and cancer. The pathways that converge to MAVS and STING-dependent signaling are involved in RNA and DNA sensing, respectively, but there is also a high degree of interconnection between those two pathways. These interactions rely on the conversion of nucleic acid ligands by the host during the viral cycle, direct interactions between components of these sensing pathways, as well as co-regulation of their expression. These mechanisms of cross-talk between the cytosolic RIG-I-MAVS and cGAS-STING nucleic acid sensing pathways amplify the innate antiviral responses against both RNA and DNA viral pathogens (Figure 3 and Table 1). The importance of these regulatory networks in the generation of an effective immune response has prompted the characterization of immunostimulatory RNA and small molecule agonists of the RIG-I pathway, as well as the derivation of natural and synthetic CDN activators of the cGAS-STING pathway. These experimental therapies have been used successfully as antivirals, vaccine adjuvants, and as cancer therapeutics against highly immunogenic tumors in pre-clinical models. Several mechanisms have been proposed to explain the antitumoral effects of CDN, including STING-dependent induction of apoptosis in tumor cells and immune cell activation. However, contradictory studies have shown that STING may contribute to tumorigenicity in weakly immunogenic tumors. Further elucidation of the complete range of events triggered by cytosolic nucleic acid sensing mechanisms in tumors will be essential to elaborate new rational anticancer strategies (see Outstanding Questions).
Outstanding questions.
DNA and RNA sensing pathways are composed of a multitude of receptors and adaptor proteins. Do they all converge on the same responses via the activation of a common set of host response genes, or is there a selective divergence among them?
What are the mechanisms downstream of RIG-MAVS and/or cGAS-STING that govern the decision between antiviral response and cell death?
Is the MAVS-STING association triggered by cytosolic DNA or cyclic di-nucleotide stimuli?
Have DNA viruses evolved mechanisms to counteract the RIG-I pathway?
Do other mediators of RNA and/or DNA sensing remain to be discovered?
What is the relationship between the multiple DNA sensors and the cGAS-STING pathway? Do they all converge at the level of STING?
In tumor studies, would targeting both RIG-I and STING pathways simultaneously potentiate or interfere with response? Could the efficacy of STING agonist treatment depend on the RIG-I status of the tumor?
Does the combination of stimuli a cancer cell receives determine its fate (death vs survival)? Or is it rather the intensity of STING activation and the time of exposure to STING agonists that influence cell responses?
Could STING agonists have a tumor-specific role? Understanding the complex functions of STING in tumors could help generate new anticancer therapies. Could RIG-I and STING agonists act in a synergistic way against virus infection and tumor development?
Highlights.
cGAS-STING and RIG-I-MAVS pathways are critical cytosolic pattern recognition receptors that recognize and respond to DNA and RNA nucleic acids respectively from invading microbial pathogens.
Although activated by distinct nucleic acids, cGAS-STING and RIG-I-MAVS signaling are functionally interconnected; STING and MAVS physically interact, and coordinate their expression levels through positive feedback mechanisms.
Natural and synthetic RIG-I and STING agonists are being explored as antiviral or anti-cancer agents, given their broad spectrum efficacy in in vitro and in vivo models of infection. These agonists can trigger cell death in malignant cells, recruit immune cells into the tumor microenvironment, and represent promising tools in cancer immunotherapies.
In some types of cancer, STING may facilitate tumor development by promoting chronic inflammation, and by promoting immunosuppression rather than cytotoxicity.
Acknowledgments
This research was supported by grants from Fondazione Cenci Bolognetti, NIH grant 7R21CA192185-02, and the Italian Association for Cancer Research (IG16901). David Olagnier was supported by a Carlsberg Foundation International Research Fellowship. The figures of the review were illustrated using the Servier Medical Art library http://www.servier.com/Powerpoint-image-bank (CC-BY 3.0 license). The authors would like to acknowledge all the contributions in the field that could not be included in the review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Chow J, Franz KM, Kagan JC. PRRs are watching you: Localization of innate sensing and signaling regulators. Virology. 2015;479–480:104–109. doi: 10.1016/j.virol.2015.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Neill LA, Golenbock D, Bowie AG. The history of Toll-like receptors - redefining innate immunity. Nat Rev Immunol. 2013;13(6):453–460. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- 4.Hoving JC, Wilson GJ, Brown GD. Signalling C-type lectin receptors. microbial recognition and immunity. Cell Microbiol. 2014;16(2):185–194. doi: 10.1111/cmi.12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlee M, Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol. 2016;16(9):566–580. doi: 10.1038/nri.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 7.Belgnaoui SM, Paz S, Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol. 2011;23(5):564–572. doi: 10.1016/j.coi.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Yoneyama M, et al. Viral RNA detection by RIG-I-like receptors. Curr Opin Immunol. 2015;32:48–53. doi: 10.1016/j.coi.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Gack MU. Mechanisms of RIG-I-like receptor activation and manipulation by viral pathogens. J Virol. 2014;88(10):5213–5216. doi: 10.1128/JVI.03370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peisley A, et al. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509(7498):110–114. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou F, et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146(3):448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs JL, Coyne CB. Mechanisms of MAVS regulation at the mitochondrial membrane. J Mol Biol. 2013;425(24):5009–5019. doi: 10.1016/j.jmb.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pattabhi S, et al. Targeting Innate Immunity for Antiviral Therapy through Small Molecule Agonists of the RLR Pathway. J Virol. 2016;90(5):2372–2387. doi: 10.1128/JVI.02202-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beljanski V, et al. Enhanced Influenza Virus-Like Particle Vaccination with a Structurally Optimized RIG-I Agonist as Adjuvant. J Virol. 2015;89(20):10612–10624. doi: 10.1128/JVI.01526-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiang C, et al. Sequence-Specific Modifications Enhance the Broad-Spectrum Antiviral Response Activated by RIG-I Agonists. J Virol. 2015;89(15):8011–8025. doi: 10.1128/JVI.00845-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38(5):870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Unterholzner L. The interferon response to intracellular DNA: why so many receptors? Immunobiology. 2013;218(11):1312–1321. doi: 10.1016/j.imbio.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Sun L, et al. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472(7344):481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, et al. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 2014;6(3):421–430. doi: 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51(2):226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ablasser A, et al. cGAS produces a 2’-5’-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498(7454):380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao P, et al. Cyclic [G(2’,5’)pA(3’,5’)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153(5):1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Civril F, et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498(7454):332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun W, et al. ERIS an endoplasmic reticulum IFN stimulator. activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A. 2009;106(21):8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29(4):538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin L, et al. MPYS a novel membrane tetraspanner. is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28(16):5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchi S, Patergnani S, Pinton P. The endoplasmic reticulum-mitochondria connection: one touch. multiple functions. Biochim Biophys Acta. 2014;1837(4):461–469. doi: 10.1016/j.bbabio.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 30.Yin Q, et al. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol Cell. 2012;46(6):735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q, et al. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity. 2014;41(6):919–933. doi: 10.1016/j.immuni.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 32.Mukai K, et al. Activation of STING requires palmitoylation at the Golgi. Nat Commun. 2016;7:11932. doi: 10.1038/ncomms11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abe T, Barber GN. Cytosolic-DNA-mediated. STING-dependent proinflammatory gene induction necessitates canonical NF-kappaB activation through TBK1. J Virol. 2014;88(10):5328–5341. doi: 10.1128/JVI.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong B, et al. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30(3):397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, et al. TRIM30alpha Is a Negative-Feedback Regulator of the Intracellular DNA and DNA Virus-Triggered Response by Targeting STING. PLoS Pathog. 2015;11(6):e1005012. doi: 10.1371/journal.ppat.1005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bridgeman A, et al. Viruses transfer the antiviral second messenger cGAMP between cells. Science. 2015;349(6253):1228–1232. doi: 10.1126/science.aab3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gentili M, et al. Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science. 2015;349(6253):1232–1236. doi: 10.1126/science.aab3628. [DOI] [PubMed] [Google Scholar]
- 38.Ablasser A, et al. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503(7477):530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17(10):1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 40.Brunette RL, et al. Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J Exp Med. 2012;209(11):1969–1983. doi: 10.1084/jem.20121960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138(3):576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Montfoort N, Olagnier D, Hiscott J. Unmasking immune sensing of retroviruses: interplay between innate sensors and host effectors. Cytokine Growth Factor Rev. 2014;25(6):657–668. doi: 10.1016/j.cytogfr.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 43.Doitsh G, Greene WC. Dissecting How CD4 T Cells Are Lost During HIV Infection. Cell Host Microbe. 2016;19(3):280–291. doi: 10.1016/j.chom.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jakobsen MR, Olagnier D, Hiscott J. Innate immune sensing of HIV-1 infection. Curr Opin HIV AIDS. 2015;10(2):96–102. doi: 10.1097/COH.0000000000000129. [DOI] [PubMed] [Google Scholar]
- 45.Sze A, et al. Host restriction factor SAMHD1 limits human T cell leukemia virus type 1 infection of monocytes via STING-mediated apoptosis. Cell Host Microbe. 2013;14(4):422–434. doi: 10.1016/j.chom.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 46.Gao D, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341(6148):903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mankan AK, et al. Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. EMBO J. 2014;33(24):2937–2946. doi: 10.15252/embj.201488726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herzner AM, et al. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat Immunol. 2015;16(10):1025–1033. doi: 10.1038/ni.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated. type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen LL, Yang L, Carmichael GG. Molecular basis for an attenuated cytoplasmic dsRNA response in human embryonic stem cells. Cell Cycle. 2010;9(17):3552–3564. doi: 10.4161/cc.9.17.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nazmi A, et al. STING mediates neuronal innate immune response following Japanese encephalitis virus infection. Sci Rep. 2012;2:347. doi: 10.1038/srep00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nitta S, et al. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology. 2013;57(1):46–58. doi: 10.1002/hep.26017. [DOI] [PubMed] [Google Scholar]
- 53.Holm CK, et al. Influenza A virus targets a cGAS-independent STING pathway that controls enveloped RNA viruses. Nat Commun. 2016;7:10680. doi: 10.1038/ncomms10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu CY, et al. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 2012;8(6):e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aguirre S, et al. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012;8(10):e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzuki T, et al. Cell type-specific subcellular localization of phospho-TBK1 in response to cytoplasmic viral DNA. PLoS One. 2013;8(12):e83639. doi: 10.1371/journal.pone.0083639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sali TM, et al. Characterization of a Novel Human-Specific STING Agonist that Elicits Antiviral Activity Against Emerging Alphaviruses. PLoS Pathog. 2015;11(12):e1005324. doi: 10.1371/journal.ppat.1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hartlova A, et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 2015;42(2):332–343. doi: 10.1016/j.immuni.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 59.Liu Y, et al. RIG-I-Mediated STING Upregulation Restricts Herpes Simplex Virus 1 Infection. J Virol. 2016;90(20):9406–9419. doi: 10.1128/JVI.00748-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang Z, et al. Cloning and functional characterization of rat stimulator of interferon genes (STING) regulated by miR-24. Dev Comp Immunol. 2012;37(3–4):414–420. doi: 10.1016/j.dci.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 61.Sui H, et al. siRNA enhances DNA-mediated interferon lambda-1 response through crosstalk between RIG-I and IFI16 signalling pathway. Nucleic Acids Res. 2014;42(1):583–598. doi: 10.1093/nar/gkt844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schoggins JW, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505(7485):691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson MR, et al. Interferon gamma-inducible protein (IFI) 16 transcriptionally regulates type i interferons and other interferon-stimulated genes and controls the interferon response to both DNA and RNA viruses. J Biol Chem. 2014;289(34):23568–23581. doi: 10.1074/jbc.M114.554147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahn J, Barber GN. Self-DNA. STING-dependent signaling and the origins of autoinflammatory disease. Curr Opin Immunol. 2014;31:121–126. doi: 10.1016/j.coi.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 65.Klarquist J, et al. STING-mediated DNA sensing promotes antitumor and autoimmune responses to dying cells. J Immunol. 2014;193(12):6124–6134. doi: 10.4049/jimmunol.1401869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Woo SR, et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41(5):830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deng L, et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity. 2014;41(5):843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takashima K, et al. STING in tumor and host cells cooperatively work for NK cell-mediated tumor growth retardation. Biochem Biophys Res Commun. 2016 doi: 10.1016/j.bbrc.2016.09.021. [DOI] [PubMed] [Google Scholar]
- 69.Gutjahr A, et al. Triggering Intracellular Receptors for Vaccine Adjuvantation. Trends Immunol. 2016;37(10):716. doi: 10.1016/j.it.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 70.Chandra D, et al. STING ligand c-di-GMP improves cancer vaccination against metastatic breast cancer. Cancer Immunol Res. 2014;2(9):901–910. doi: 10.1158/2326-6066.CIR-13-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Z, Celis E. STING activator c-di-GMP enhances the anti-tumor effects of peptide vaccines in melanoma-bearing mice. Cancer Immunol Immunother. 2015;64(8):1057–1066. doi: 10.1007/s00262-015-1713-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakamura T, et al. Liposomes loaded with a STING pathway ligand cyclic di-GMP. enhance cancer immunotherapy against metastatic melanoma. J Control Release. 2015;216:149–157. doi: 10.1016/j.jconrel.2015.08.026. [DOI] [PubMed] [Google Scholar]
- 73.Conlon J, et al. Mouse but not human STING. binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. J Immunol. 2013;190(10):5216–5225. doi: 10.4049/jimmunol.1300097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Corrales L, et al. Direct Activation of STING in the Tumor Microenvironment Leads to Potent and Systemic Tumor Regression and Immunity. Cell Rep. 2015;11(7):1018–1030. doi: 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fu J, et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci Transl Med. 2015;7(283):283ra52. doi: 10.1126/scitranslmed.aaa4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen Q, et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature. 2016;533(7604):493–498. doi: 10.1038/nature18268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tang CK, et al. The chemotherapeutic agent DMXAA as a unique IRF3-dependent type-2 vaccine adjuvant. PLoS One. 2013;8(3):e60038. doi: 10.1371/journal.pone.0060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ahn J, et al. Inflammation-driven carcinogenesis is mediated through STING. Nat Commun. 2014;5:5166. doi: 10.1038/ncomms6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang L, et al. Cutting edge: DNA sensing via the STING adaptor in myeloid dendritic cells induces potent tolerogenic responses. J Immunol. 2013;191(7):3509–3513. doi: 10.4049/jimmunol.1301419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lemos H, et al. STING Promotes the Growth of Tumors Characterized by Low Antigenicity via IDO Activation. Cancer Res. 2016;76(8):2076–2081. doi: 10.1158/0008-5472.CAN-15-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Poeck H, et al. 5’-Triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma. Nat Med. 2008;14(11):1256–1263. doi: 10.1038/nm.1887. [DOI] [PubMed] [Google Scholar]
- 82.Van DN, et al. Innate immune agonist dsRNA. induces apoptosis in ovarian cancer cells and enhances the potency of cytotoxic chemotherapeutics. FASEB J. 2012;26(8):3188–3198. doi: 10.1096/fj.11-202333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Besch R, et al. Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. J Clin Invest. 2009;119(8):2399–2411. doi: 10.1172/JCI37155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ellermeier J, et al. Therapeutic efficacy of bifunctional siRNA combining TGF-beta1 silencing with RIG-I activation in pancreatic cancer. Cancer Res. 2013;73(6):1709–1720. doi: 10.1158/0008-5472.CAN-11-3850. [DOI] [PubMed] [Google Scholar]
- 85.Su ZZ, et al. Central role of interferon regulatory factor-1 (IRF-1) in controlling retinoic acid inducible gene-I (RIG-I) expression. J Cell Physiol. 2007;213(2):502–510. doi: 10.1002/jcp.21128. [DOI] [PubMed] [Google Scholar]
- 86.Liu Z, et al. Ftx non coding RNA-derived miR-545 promotes cell proliferation by targeting RIG-I in hepatocellular carcinoma. Oncotarget. 2016;7(18):25350–25365. doi: 10.18632/oncotarget.8129. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 87.Wang JH, et al. microRNA-34a–Upregulated Retinoic Acid-Inducible Gene-I Promotes Apoptosis and Delays Cell Cycle Transition in Cervical Cancer Cells. DNA Cell Biol. 2016;35(6):267–279. doi: 10.1089/dna.2015.3130. [DOI] [PubMed] [Google Scholar]
- 88.Hou J, et al. Hepatic RIG-I predicts survival and interferon-alpha therapeutic response in hepatocellular carcinoma. Cancer Cell. 2014;25(1):49–63. doi: 10.1016/j.ccr.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 89.Szabo A, et al. RIG-I inhibits the MAPK-dependent proliferation of BRAF mutant melanoma cells via MKP-1. Cell Signal. 2016;28(5):335–347. doi: 10.1016/j.cellsig.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 90.Ranoa DR, et al. Cancer therapies activate RIG-I-like receptor pathway through endogenous non-coding RNAs. Oncotarget. 2016;7(18):26496–26515. doi: 10.18632/oncotarget.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hochheiser K, et al. Cutting Edge: The RIG-I Ligand 3pRNA Potently Improves CTL Cross-Priming and Facilitates Antiviral Vaccination. J Immunol. 2016;196(6):2439–2443. doi: 10.4049/jimmunol.1501958. [DOI] [PubMed] [Google Scholar]