Abstract
Polyamines are multivalent and organic cations essential for cellular growth, proliferation, differentiation, and apoptosis. Increased levels of polyamines are closely associated with numerous forms of cancer. An autoregulatory circuit composed of ornithine decarboxylase (ODC), antizyme (AZ) and antizyme inhibitor (AZI) govern the intracellular level of polyamines. Antizyme binds with ODC to inhibit ODC activity and to promote the ubiquitin‐independent degradation of ODC. Antizyme inhibitor binds to AZ with a higher affinity than ODC. Consequently, ODC is released from the ODC–AZ complex to rescue its activity. Antizyme inhibitor increases the ODC activity to accelerate the formation of intracellular polyamines, triggering gastric and breast carcinogenesis as well as hepatocellular carcinoma and esophageal squamous cell carcinoma development. Antizyme inhibitor 1 (AZIN1), a primary member of the AZI family, has aroused more attention because of its contribution to cancer. Even though its conformation is changed by adenosine‐to‐inosine (A→I) RNA editing, it plays an important role in tumorigenesis through regulating intracellular polyamines. Encouragingly, AZIN1 has been revealed to have an additional function outside the polyamine pathway so as to bypass the deficiency of targeting the polyamine biosynthetic pathway, promising to become a critical target for cancer therapy. Here, we review the latest research advances into AZIN1 and its potential contribution to carcinogenesis.
Keywords: Antizyme, antizyme inhibitor 1, carcinogenesis, ornithine decarboxylase, polyamine
Polyamines are multivalent, organic cations that play important roles in cell growth, proliferation, differentiation, and apoptosis. Ornithine decarboxylase (ODC) is the first rate‐limiting enzyme in the polyamine biosynthesis pathway. It is a pyridoxal 5‐phosphate (PLP)‐dependent enzyme that catalyzes decarboxylation of ornithine into putrescine, then into spermidine and spermine by a concerted action of downstream enzymes.1 Ornithine decarboxylase functions only as a homodimeric complex. Hence, its monomers have no enzymatic activity.2 Individual ODC monomers bind to each other, appearing in a relatively low affinity. A balance is rapidly formed between ODC dimers and inactive ODC monomers under physiological conditions.
Different studies have revealed that ODC and polyamines serve as key regulators in the development of cancer.3 Being such a key regulator of cellular proliferation, it is not surprising that intracellular ODC is tightly regulated at multiple levels, especially by regulation of its stability through a polyamine‐mediated autoregulatory circuit. Small proteins termed antizymes are in the center of this autoregulatory circuit. Antizyme‐1 (AZ1), the most highly characterized member of these proteins, was originally described as a polyamine‐induced ODC inhibitory activity. As the affinity of antizymes (AZs) towards ODC subunits is higher than that of ODC subunits for each other, AZs trap transient ODC monomers to form inactive ODC–AZ heterodimers that are recognized by the 26S proteasome, resulting in ubiquitin‐independent degradation of ODC. Antizyme reduces cellular polyamine pools not only by stimulating ODC degradation, but also by interfering with the uptake of external polyamines through a mechanism not yet elucidated. Until now, despite the fact that all AZ isoforms are able to inhibit ODC and the polyamine uptake, only AZ1 clearly induces ODC proteasomal degradation. In fact, there are controversial data on the capacity of AZ2 for targeting ODC to degradation in vivo and in vitro,4 and recently it has been reported that AZ3, a testis‐specific isoform, is unable to target ODC to degradation.5
Intriguingly, another ODC‐related protein, an antizyme inhibitor, efficiently negates the activities of AZ. The recent development of the relationship between antizyme inhibitor 1 (AZIN1) and cancer draws upon more than three decades of basic research into understanding the biological function of this mysterious protein molecule. The protein was originally found in rat liver extract as an antizyme inhibitor (AZI). Until now, AZI has been reported to have two isoforms, of which the most predominant is AZIN1. The latter is ubiquitously expressed at a high level so as to be highly cared. Antizyme inhibitor 1 was shown to be a heat‐labile molecule with a molecular weight similar to ODC, but with a higher affinity for antizyme 1 than ODC. In addition, AZIN1 has a highly identical sequence and structure to ODC.6, 7 Therefore, AZIN1 was originally considered to be a derivative of ODC, but it has now been reported to be a distinct protein from different aspects, including the lack of ornithine decarboxylating activity and ubiquitin‐dependent degradation. With some significant findings, researchers gradually realized that AZIN1 may play an important role in tumorigenesis in many different tumors (Fig. 1). However, in contrast to considerable interest in ODC and AZ in terms of cell proliferation and tumorigenesis, AZI has been less well studied. Antizyme may act as a tumor suppressor to negatively regulate tumor cell proliferation and transformation.8 Antizyme inhibitor is combined with AZ to suppress its function, thus resulting in enhanced polyamine synthesis with concomitant cell proliferation, transformation, and tumorigenesis. Recent reports show that A→I RNA editing of AZIN1 is intimately associated with augmented tumor‐initiating potential and tumor‐aggressive behaviors.9, 10 The growing evidence suggests that AZIN1 might be a desired target for anticancer therapy. Here, we highlight the biological properties and degradation of AZIN1, and its contributions to carcinogenesis.
Figure 1.
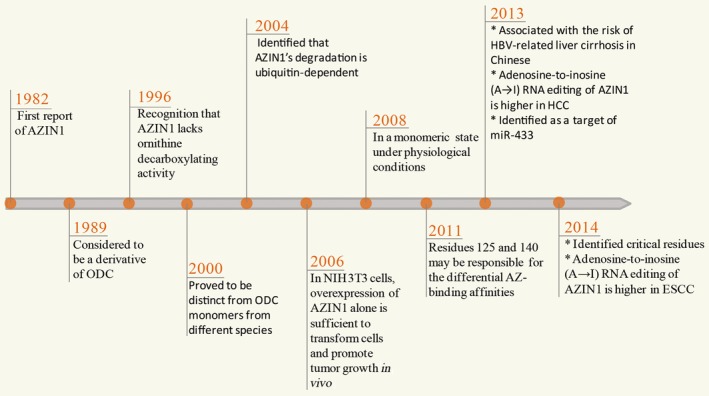
Historic process from basic research into understanding the biological function of antizyme inhibitor 1 (AZIN1). The protein, originally found in rat liver extract, binds antizyme (AZ) with higher affinity than ornithine decarboxylase (ODC). First thought to be an ODC derivative, AZIN1 was then proved to be a distinct protein from different aspects, including the lack of an ornithine decarboxylating activity and ubiquitin‐dependent degradation. Researchers have gradually realized that AZIN1 is associated with the risk of hepatitis B virus (HBV)‐related liver cirrhosis, or may play an important role in tumorigenesis in many different tumors, such as hepatocellular carcinoma (HCC) and esophageal squamous cell carcinoma (ESCC). miR, microRNA.
AZIN 1 Binds More Strongly to AZ Than to ODC
Like ODC, AZIN1 binds to AZ with a higher affinity than ODC. Thus, it can release ODC from the ODC–AZ complex to rescue ODC activity (Fig. 2). The dissociation constant (Kd) of the AZ–ODC complex is 0.21 mM, whereas that of the AZ–AZIN1 complex is 0.02 mM. As a result, there is an approximately 10‐fold difference between the binding affinities of AZ–AZIN1 and AZ–ODC.11 This suggests that AZIN1 can restore ODC activity by forming a tight complex with AZ, thereby releasing ODC from AZIN1. Although AZIN1 is crystallized as a dimer, it has been detected in a monomeric state under physiological conditions. The fact that AZIN1 is a monomer increases its availability for interaction with AZ, partially contributing to its most important feature, namely its higher affinity for AZ compared to ODC. This higher affinity provides the basis for the ability of AZIN1 to rescue ODC subunits from interaction with AZ, thus protecting them from degradation (Fig. 2).12, 13 As increased accessibility is unlikely to fully account for the higher affinity to AZ, it must be assumed that the sequence of the relevant binding segments may also be of importance. Initial studies, based on the differential affinities for AZ between murine and trypanosomal ODC, led to the identification of the AZ‐binding element (AZBE), as a sequence spanning the residues from 117 to 140 in mouse ODC. Antizyme inhibitor 1 binds to AZ through the AZBE region, but in this case the binding to AZ makes AZIN1 more resistant against proteasomal degradation, probably because AZ inhibits its ubiquitylation. Subsequently, the analysis of the tertiary structure of ODC revealed that this region is present in two α‐helices included in a TIM‐like α/β‐barrel domain, in which several basic residues are exposed toward the surface. The interaction between ODC and AZIN1 with AZ through the AZBE region has been computationally predicted by docking, being quite similar in both cases.14 Although the amino acid sequences of the AZBE region of ODC and AZIN1 are quite similar, recent data have indicated that residues 125 and 140 are the critical residues contributing to the differential AZ‐binding affinities.15, 16 In addition, another study showed that N327 and Y331 of ODC are replaced by A325 and that S329 in AZIN1, which may be another important factor contributing to the differential AZ‐binding affinities (Table 1).17 Therefore, AZIN1 may positively regulate the polyamine pathway through binding to AZ, finally preventing the degradation of ODC mediated by AZ.
Figure 2.
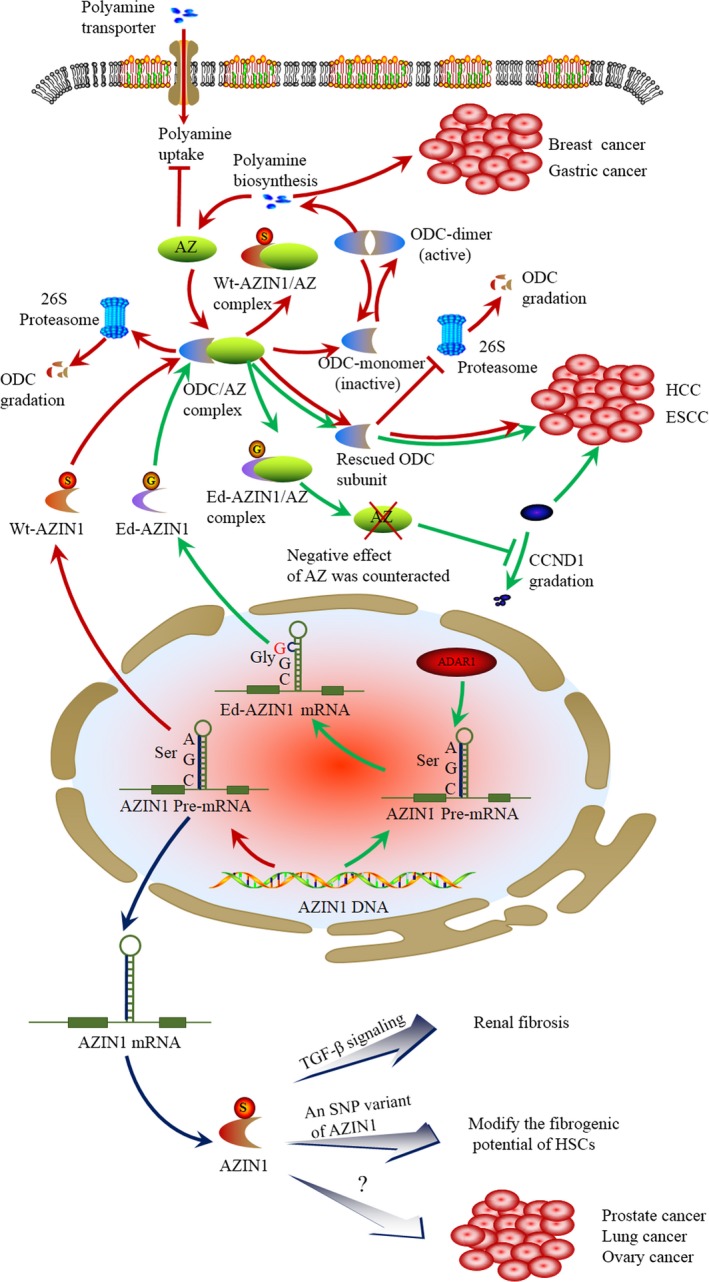
RNA‐edited antizyme inhibitor 1 (AZIN1) interferes with the formation of the antizyme–ornithine decarboxylase (AZ–ODC) complex, contributing to cancer development. AZIN1 and AZ regulate polyamine biosynthesis through ornithine decarboxylase (red arrows). ODC catalyzes the first step in polyamine synthesis. AZ disrupts ODC homodimers, targets ODC for degradation by the 26S proteasome in a ubiquitin‐independent manner, and inhibits polyamine uptake. Activity of AZ is further regulated by AZIN1. AZIN1 can displace ODC from the ODC–AZ complex, restoring polyamine biosynthesis by ODC. The latter is degraded by the 26S proteasome in a ubiquitin‐dependent manner. The overexpression of AZIN1 leads to a concomitant increase in ODC activity to accelerate the formation of polyamine, triggering gastric and breast carcinogenesis, and development of hepatocellular carcinoma (HCC) and esophageal squamous cell carcinoma (ESCC). RNA editing and its contribution to AZIN1 (green arrows). Adenosine‐to‐inosine editing of AZIN1 transcripts, specifically regulated by adenosine deaminase 1 acting on double‐stranded RNA (ADAR1), leads to a serine (Ser) to glycine (Gly) substitution at residue 367 of AZIN1, which is predicted to cause a conformational alteration. Compared with wild‐type (Wt‐) AZIN1 protein, the edited (Ed‐) AZIN1 binds to AZ with a higher affinity toward AZ, thus the Ed‐AZIN1 protein sequestrates AZ to block the degradation of ODC and cyclin D1 (CCND1), thus promoting cell proliferation possibly with cancer onset. Other potential roles of AZIN1 (dark blue arrows). Overexpression of AZIN1 reduces the expression levels of transforming growth factor (TGF)‐β1, TGF‐β receptor type I, and Smad3, and phosphorylation of Smad3, suppressing TGF‐β signaling and the fibrotic response. A single nucleotide polymorphism (SNP) variant in the AZIN1 gene leads to the enhanced generation of a novel alternative splice form that modifies the fibrogenic potential of hepatic stellate cells (HSCs). In addition, AZIN1 is highly expressed in cancer of the prostate, lung, and ovary. But its detailed mechanism is completely unclear.
Table 1.
Comparison of human ornithine decarboxylase (ODC) and human antizyme inhibitor 1 (AZIN1) from different biological characteristics
| Biological characteristics | ODC | AZIN1 | Reference |
|---|---|---|---|
| State under physiological conditions | Dimeric | Monomeric | 13 |
| Dissociation constants of ODC‐AZ and AZIN1‐AZ complexes | 0.21 mM | 0.02 mM | 19 |
| Dissociation constants of ODC‐ODC and AZIN1‐ AZIN1 complexes | 0.18 mM | 84 mM | 11 |
| Critical amino acid residues of contributing to differential AZ‐binding affinities | N125, M140, N327, Y331 | K125, K140, A325, S329 | 17, 20 |
| Critical amino acid residues contributing to determination of ornithine decarboxylating activities | Y331, Y389 | S331, D389 | 14 |
| Degradation | Ubiquitin‐independent | Ubiquitin‐dependent | 16 |
AZ, antizyme.
AZIN 1 has No Ornithine Decarboxylating Activity
Antizyme inhibitor also crystallizes as a dimer like ODC. The latter is a stable dimer, with a Kd of 0.18 mM. However, AZIN1 can form a rapid balance between the monomer and the dimer, with a Kd value of 84 mM. Therefore, the self‐association of both protein subunits differs by >400‐fold.18 However, a smaller buried surface area, fewer interactions at the dimer interface, and lack of symmetry of the interactions between residues from the two monomers suggest that AZIN1 does not dimerize under physiological conditions. Indeed, biochemical studies revealed that AZIN1 remains monomeric in solution, whereas ODC is dimeric.19 As the active site of ODC is formed at the interface between the two monomers, the lack of physiological dimerization of AZIN1 is sufficient to explain the lack of ornithine decarboxylating activity. The observation that AZIN1 is unable to bind PLP provides an additional and independent explanation for the lack of enzymatic activity.19 What are the essential amino acid residues that determine why AZIN1 exists as a monomer and is unable to bind PLP? The structure of human ODC indicates that some amino acid residues in the dimer interface may play significant roles in either dimer formation or enzyme activity. Multiple sequence alignments of ODC and AZIN1 from various species have shown that residues 277, 331, 332, and 389 are not conserved between ODC and AZIN1. Recent data suggest that residue 331 is the most important residue governing the dimerization of human AZIN1 protein. Tyr389 may play a role not only in PLP binding, but also in dimer formation and structural stability (Table 1).14 The size‐distribution of the ODC interface mutants shows that the dimer formation results from the cooperative behavior of the interface residues.18
Degradation of AZIN1
Similar to ODC, AZIN1 is a rapidly degraded protein. However, in contrast to ODC, AZIN1 degradation is in a ubiquitin‐dependent manner (Table 1).16 It does not need interaction with AZ or the C‐terminal segment20 which, for ODC, acts as a proteasome recognition signal. Interaction between AZ and ODC greatly stimulates ODC degradation. In contrast, interaction with AZ actually stabilizes AZIN1 by interfering with its ubiquitination.20 Therefore, it is tempting to hypothesize that, similar to the conformational alteration, AZ imposes on ODC resulting in exposure of the C‐terminal degradation signal. The interaction with AZ imposes a conformational change on AZIN1, masking its as‐yet unidentified degradation signal. As AZ is also stabilized when binding to AZIN1, it seems that AZIN1 buffers AZ by maintaining it in a stable complex. The behavior and destiny of this AZIN1–AZ complex is presently unclear. It is possible that increased intracellular polyamine levels may disrupt the interaction between AZIN1 and AZ, releasing AZ to fulfill its original role.
Evidence of a Role for AZIN1 as a Regulator of Human Cancer
Much recent evidence suggests that elevated levels of ODC and polyamines are tightly involved in various types of neoplastic transformation, including prostate cancer and other tumors.21 Although high levels of ODC and polyamines are responsible for the development of many tumors, the role of AZIN1 in this process is just emerging. The AZIN1 gene was located at chromosome 8q22.3 in human cells, which was previously considered to be a frequently amplified hotspot that contributes to decreased survival and accelerated distant metastases in breast tumor.22 Antizyme inhibitor 1 has also been shown to be highly expressed in stomach, lung, prostate, and ovary tumors,23, 24, 25 and especially in liver, where AZIN1 has been found to be associated with the risk of hepatitis B virus‐related liver cirrhosis in Chinese patients.26, 27
By binding to AZ, AZIN1 can prevent the effects of AZ on ODC degradation and cellular uptake of polyamines. Thus, polyamine homeostasis is highly regulated and AZIN1 appears to be a potential carcinogenic molecule in this regulation. Keren‐Paz et al. reported that NIH3T3 cells overexpressing AZIN1 resulted in elevation of ODC activity and polyamine uptake, and showed improved cell proliferation. Moreover, these cells gave rise to tumors when injected into nude mice, whereas knockdown of AZIN1 using siRNA or shRNA decreased cell proliferation both in vitro and in vivo.28 Another study has also shown that downregulated expression of AZIN1 in A549 cells resulted in decreased polyamine levels with the inhibition of cell proliferation. However, the ability of these cells to form tumor in vivo was not determined.29 Similar findings were obtained by Silva et al. using human breast carcinoma cell line JIMT‐1. By diluting cells in fresh medium, the cellular content of AZIN1 increased transiently after induction of cell proliferation medium and inhibition of polyamine biosynthesis induced an even larger increase in the cellular AZIN1 content. Moreover, induced overexpression of AZIN1 caused increased cell proliferation with a concomitant increase in ODC activity and putrescine amount.22 Using AZIN1‐targeting siRNA to knock down the azin1 gene in both human and rat prostate cancer cells, smaller tumors occurred in vivo.30 In rodent tissues and human cell lines, differential expressions of AZIN and AZ were also observed.31 These results indicate that the increasing level of AZIN1 within the cells has a significant promoting effect on tumor growth.
Although AZIN1 plays a major role in the polyamine pathway, the precise cell signaling pathways remain an enigma. An exciting study recently published in Nature by Li et al.32 provides important clues and evidence to reveal the molecular mechanism of AZIN1 involved in polyamine pathway regulation. It showed that microRNA (miR)‐433 acted as an important component of transforming growth factor‐β (TGF‐β)/Smad3‐driven renal fibrosis, and AZIN1 was identified as a target of miR‐433. During kidney disease, injury activates TGF‐β signaling to induce miR‐433 expression. Upregulation of miR‐433 then suppresses AZIN1 expression to derepress AZ. Then polyamine levels decline, which triggers TGF‐β signaling, causing the activation of transcription of extracellular matrix (ECM) genes and tubulointerstitial fibrosis. Another study also indicated that a variant of single nucleotide polymorphisms in the AZIN1 gene results in the enhanced generation of a novel alternative splice that alters the fibrogenicity of hepatic stellate cells.33 Thus, AZIN1 may be a potential therapeutic target in tissue fibrosis.
Recoding RNA Editing of AZIN1 Predisposes to Carcinoma
These results indicate that overexpression of AZIN1 has a significant promoting effect on tumor growth by fulfilling an essential regulatory function in polyamine homeostasis and cell proliferation. Interestingly, from an epigenetic aspect, A→I RNA editing of AZIN1 has also been reported to cause cancer through the same mechanisms.
Recently, two independent studies have identified that A→I RNA editing of AZIN1 is more significant in hepatocellular carcinoma (HCC)9 and esophageal squamous cell carcinoma10 compared with their corresponding normal specimens. Moreover, its process conferred a “gain‐of‐function” phenotype involving augmented tumor‐initiating potential and more aggressive behaviors.9, 10 In HCC,A→I editing of AZIN1 transcripts leads to a serine‐to‐glycine substitution at residue 367, influencing protein conformation and inducing a cytoplasmic‐to‐nuclear translocation. Compared with wild‐type AZIN1, the edited AZIN1 has a stronger affinity to AZ with resultant higher protein stability, promoting cell proliferation and tumor progression through restraining AZ‐mediated degradation of the ODC and cyclin D1 (CCND1) oncoproteins.9 Therefore, A→I (G) AZIN1 editing is increased in HCC tumor, resulting in the inhibition of AZ tumor suppressor, and the appearance of a gain‐of‐function phenotype involving tumor‐aggressive behaviors (Fig. 2). In the field of cancer stem cells, increased RNA editing of AZIN1 is detected in human leukemia stem cells from patients with blast crisis chronic myeloid leukemia by a sensitive RNA editing fingerprint assay.34 Given the oncogenic role of the edited AZIN1, it could become a promising target for tumor therapy in the future. Further studies will be required to confirm the importance and significance of RNA editing of AZIN1 during carcinogenesis.
Could AZIN1 Become a Critical Target for Anticancer Therapy?
Thus far, we have primarily focused on AZIN1 in tumor progression through increasing ODC activity to accelerate the formation of intracellular polyamine. However, as expected, several recent studies put forward an alternative possibility, suggesting that AZIN1 might also be an important molecule for modulating cell proliferation and oncogenesis through a polyamine‐independent mechanism.
Cyclin D1 serves as an active switch in the regulation of cell cycle progression and can function as a transcriptional co‐regulator, overexpression of CCND1 and deregulation of CCND1 degradation are thought to be associated with the development and progression of cancer.35, 36 A previous study confirmed AZ‐mediated degradation of CCND1 in a ubiquitin‐independent reaction.37 However, another study reported that not only AZ can regulate the level of CCND1, but AZIN1 can also directly bind to CCND1 and prevent its degradation.38 Silencing AZIN1 expression leads to diminished levels of CCND1 and reduces cell proliferation. Overexpression of the non‐AZ‐binding mutant AZIN1117‐140 can interact with CCND1 to lead to increased cell proliferation. This suggests that both AZ and AZIN1 may contribute in separate ways to the control of CCND1 levels. However, it is still not clear how, and under what physiological status, this AZ‐stimulated degradation and/or AZIN1‐preventing degradation in an AZ‐independent mechanism operates. Therefore, it deserves to be further calculated what the binding constants between AZIN1 and CCND1 owe to the binding affinity of AZ–CCND1 complex that are already known.19 Thus, it would greatly enhance our understanding of the relative importance of these interactions within the cells.
In addition to the direct interaction between AZIN1 and CCND1, a recent study suggests that AZIN1 is involved in the degradation of anti‐apoptotic DeltaNp73 by a c‐Jun‐dependent mechanism in response to genotoxic stress through the AZ‐mediated pathway.39 Coexpression of AZIN1 and DNp73 prior to doxorubicin treatment can block the decrease of DNp73 levels. Similarly, coexpression of AZIN1 with c‐Jun and DNp73 prevented the reduction of DNp73 levels by c‐Jun, suggesting that AZIN1 is indeed required for preventing c‐Jun‐mediated DNp73 degradation. Because DNp73 expression is elevated in many human cancers,28 and its overexpression has been shown to inhibit apoptosis,40 the role of AZIN1 in preventing the reduction of DNp73 may be critical to tumorigenesis.
Centrosome amplification is a common feature of numerous solid tumors and seems to be an early event in tumorigenesis.41 Silencing of AZIN1 results in a decrease of numerical centrosome abnormalities, whereas overexpression of AZIN1 leads to centrosome over‐duplication.42 This provides a novel mechanism by which changes in the level of AZIN1 affect additional compounds related to tumor behavior. In addition, AZ has also been found to modulate degradation of numerous other proteins associated with the cell cycle, including Smad1,43 Aurora A,44 and Mps145, 46 that regulate centrosome duplication and upregulate loricrin (LOR) to promote differentiation of SCC15 cells.47 In view of the fact that AZIN1 can bind and inactivate AZ, AZIN1 may involve the regulation of these proteins, which remains to be further studied.
To identify whether or not there are other candidate loci, we used AZIN1 as a query to obtain protein–protein interactions through analysis of The European Molecular Biology Laboratory's Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database. The STRING database, containing known and predicted physical and functional protein–protein interactions, identifies interaction partners according to a variety of criteria, and only interactions with high confidence levels (>0.7) are kept.48 As expected, polyamine metabolism‐related regulatory proteins, including ornithine decarboxylase antizyme family proteins (OAZ1, OAZ2, and OAZ3), ornithine decarboxylase 1, ubiquitin C, adenosylmethionine decarboxylase 1, spermidine synthase, threonine synthase‐like 1, and spermine synthase, have a strong connection with AZIN1. These proteins have been reported in published works and some of them have been discussed above. Unexpectedly, the protein interaction network in STRING also identified that fibroblast growth factor 2, a known angiogenic factor, is highly correlated with AZIN1, which has not been mentioned in previous reports. It remains to be further studied. This analysis revealed that AZIN1 has polyamine‐independent functions, perhaps mediated by preventing degradation of CCND1 and other cell cycle regulators or by interacting with fibroblast growth factor 2 (Fig. 3).
Figure 3.
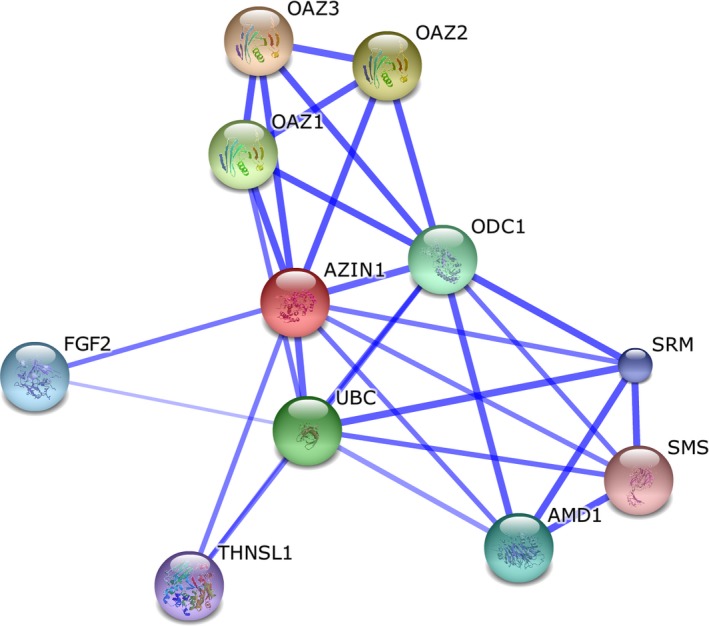
Antizyme inhibitor 1 (AZIN1) interactome network. The interaction network with high confidence levels (>0.7) is shown as displayed by The European Molecular Biology Laboratory's Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) for genetically interacting proteins possibly related in function to antizyme inhibitor 1 (AZIN1). Stronger associations are represented by thicker lines. In addition to polyamine metabolism‐related regulatory proteins, including ornithine decarboxylase antizyme (OAZ)1, OAZ2, OAZ3, ubiquitin C (UBC), ornithine decarboxylase 1 (ODC1), adenosylmethionine decarboxylase 1 (AMD1), threonine synthase‐like 1 (THNSL1), and spermine synthase (SMS), fibroblast growth factor 2 (FGF2) is also correlated highly with AZIN1. SRM, spermidine synthase.
Based on the importance of polyamines for cellular growth and transformation, there has been long‐term interest in targeting polyamine metabolism as a target for therapeutic efforts in battling cancer and other hyperproliferative diseases.49 However, targeting the polyamine pathway through enzyme inhibitors and polyamine analogs proves rather ineffective due to the ability of cells to enhance polyamine uptake from the microenvironment to overcome the effect of the disturbance of the polyamine biosynthetic pathway. Moreover, enzyme inhibitors are reported to be generally well tolerated with mostly reversible toxicities including thrombocytopenia, gastrointestinal anomalies, and hearing loss.49 Some analogs also exert their cytotoxic effect by negating the function of the physiological polyamines or stimulating antizyme production.50
As previously described, AZIN1 has been shown to have additional functions outside the polyamine pathway. Therefore, we have sufficient reasons to believe that targeting AZIN1 for cancer therapy may actually be a better strategy than targeting the polyamine biosynthetic pathway, because such a treatment would affect cell proliferation not only through the polyamine pathway but also through additional mediators. Regardless, AZIN1 opens up an entirely new approach for polyamine‐dependent or polyamine‐independent antitumor drug development.
Disclosure Statement
The authors have no conflict of interest.
Acknowledgments
This project was supported by the National Natural Science Foundation of China (Grant No. 81172824) to F.Y. Xing.
Cancer Sci 108 (2017) 163–169
Funding Information
National Natural Science Foundation of China (Grant No. 81172824).
Contributor Information
Jing Liu, Email: tjliu@jnu.edu.cn.
Feiyue Xing, Email: tfyxing@jnu.edu.cn.
References
- 1. Pegg AE. Regulation of ornithine decarboxylase. J Biol Chem 2006; 281: 14529–32. [DOI] [PubMed] [Google Scholar]
- 2. Kahana C, Asher G, Shaul Y. Mechanisms of protein degradation: an odyssey with ODC. Cell Cycle 2005; 4: 1461–4. [DOI] [PubMed] [Google Scholar]
- 3. Gerner EW, Meyskens FL. Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer 2004; 4: 781–92. [DOI] [PubMed] [Google Scholar]
- 4. Ramos‐Molina B, Lambertos A, Lopez‐Contreras AJ et al Structural and degradative aspects of ornithine decarboxylase antizyme inhibitor 2. FEBS Open Bio 2014; 4: 510–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Snapir Z, Keren‐Paz A, Bercovich Z, Kahana C. Antizyme 3 inhibits polyamine uptake and ornithine decarboxylase (ODC) activity, but does not stimulate ODC degradation. Biochem J 2009; 419: 99–103, 101 p following 103. [DOI] [PubMed] [Google Scholar]
- 6. Nilsson J, Grahn B, Heby O. Antizyme inhibitor is rapidly induced in growth‐stimulated mouse fibroblasts and releases ornithine decarboxylase from antizyme suppression. Biochem J 2000; 346(Pt 3): 699–704. [PMC free article] [PubMed] [Google Scholar]
- 7. Jackson LK, Brooks HB, Osterman AL, Goldsmith EJ, Phillips MA. Altering the reaction specificity of eukaryotic ornithine decarboxylase. Biochemistry 2000; 39: 11247–57. [DOI] [PubMed] [Google Scholar]
- 8. Wang X, Jiang L. Effects of ornithine decarboxylase antizyme 1 on the proliferation and differentiation of human oral cancer cells. Int J Mol Med 2014; 34: 1606–12. [DOI] [PubMed] [Google Scholar]
- 9. Chen L, Li Y, Lin CH et al Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med 2013; 19: 209–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qin YR, Qiao JJ, Chan TH et al Adenosine‐to‐inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer Res 2014; 74: 840–51. [DOI] [PubMed] [Google Scholar]
- 11. Kahana C. Antizyme and antizyme inhibitor, a regulatory tango. Cell Mol Life Sci 2009; 66: 2479–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Su KL, Liao YF, Hung HC, Liu GY. Critical factors determining dimerization of human antizyme inhibitor. J Biol Chem 2009; 284: 26768–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Albeck S, Dym O, Unger T, Snapir Z, Bercovich Z, Kahana C. Crystallographic and biochemical studies revealing the structural basis for antizyme inhibitor function. Protein Sci 2008; 17: 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee CY, Liu YL, Lin CL, Liu GY, Hung HC. Functional roles of the dimer‐interface residues in human ornithine decarboxylase. PLoS One 2014; 9: e104865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cohavi O, Tobi D, Schreiber G. Docking of antizyme to ornithine decarboxylase and antizyme inhibitor using experimental mutant and double‐mutant cycle data. J Mol Biol 2009; 390: 503–15. [DOI] [PubMed] [Google Scholar]
- 16. Bercovich Z, Kahana C. Degradation of antizyme inhibitor, an ornithine decarboxylase homologous protein, is ubiquitin‐dependent and is inhibited by antizyme. J Biol Chem 2004; 279: 54097–102. [DOI] [PubMed] [Google Scholar]
- 17. Liu YC, Liu YL, Su JY, Liu GY, Hung HC. Critical factors governing the difference in antizyme‐binding affinities between human ornithine decarboxylase and antizyme inhibitor. PLoS One 2011; 6: e19253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lopez‐Contreras AJ, Ramos‐Molina B, Cremades A, Penafiel R. Antizyme inhibitor 2: molecular, cellular and physiological aspects. Amino Acids 2010; 38: 603–11. [DOI] [PubMed] [Google Scholar]
- 19. Liu YC, Lee CY, Lin CL, Chen HY, Liu GY, Hung HC. Multifaceted interactions and regulation between antizyme and its interacting proteins cyclin D1, ornithine decarboxylase and antizyme inhibitor. Oncotarget 2015; 6: 23917–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu HY, Chen SF, Hsieh JY et al Structural basis of antizyme‐mediated regulation of polyamine homeostasis. Proc Natl Acad Sci USA 2015; 112: 11229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hogarty MD, Norris MD, Davis K et al ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res 2008; 68: 9735–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Silva TM, Cirenajwis H, Wallace HM, Oredsson S, Persson L. A role for antizyme inhibitor in cell proliferation. Amino Acids 2015; 47: 1341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jung MH, Kim SC, Jeon GA et al Identification of differentially expressed genes in normal and tumor human gastric tissue. Genomics 2000; 69: 281–6. [DOI] [PubMed] [Google Scholar]
- 24. Schaner ME, Ross DT, Ciaravino G et al Gene expression patterns in ovarian carcinomas. Mol Biol Cell 2003; 14: 4376–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Duin M, van Marion R, Vissers K et al High‐resolution array comparative genomic hybridization of chromosome arm 8q: evaluation of genetic progression markers for prostate cancer. Genes Chromosomes Cancer 2005; 44: 438–49. [DOI] [PubMed] [Google Scholar]
- 26. Peng L, Guo J, Zhang Z et al A candidate gene study for the association of host single nucleotide polymorphisms with liver cirrhosis risk in Chinese hepatitis B patients. Genet Test Mol Biomarkers 2013; 17: 681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peng LJ, Guo JS, Zhang Z et al Polymorphism of ornithine decarboxylase antizyme inhibitor 1 gene is associated with liver cirrhosis in Chinese hepatitis B patients. Zhonghua Gan Zang Bing Za Zhi 2011; 19: 169–73. [DOI] [PubMed] [Google Scholar]
- 28. Keren‐Paz A, Bercovich Z, Porat Z, Erez O, Brener O, Kahana C. Overexpression of antizyme‐inhibitor in NIH3T3 fibroblasts provides growth advantage through neutralization of antizyme functions. Oncogene 2006; 25: 5163–72. [DOI] [PubMed] [Google Scholar]
- 29. Choi KS, Suh YH, Kim WH, Lee TH, Jung MH. Stable siRNA‐mediated silencing of antizyme inhibitor: regulation of ornithine decarboxylase activity. Biochem Biophys Res Commun 2005; 328: 206–12. [DOI] [PubMed] [Google Scholar]
- 30. Olsen RR, Chung I, Zetter BR. Knockdown of antizyme inhibitor decreases prostate tumor growth in vivo. Amino Acids 2012; 42: 549–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramos‐Molina B, Lopez‐Contreras AJ, Cremades A, Penafiel R. Differential expression of ornithine decarboxylase antizyme inhibitors and antizymes in rodent tissues and human cell lines. Amino Acids 2012; 42: 539–47. [DOI] [PubMed] [Google Scholar]
- 32. Li R, Chung AC, Dong Y, Yang W, Zhong X, Lan HY. The microRNA miR‐433 promotes renal fibrosis by amplifying the TGF‐beta/Smad3‐Azin1 pathway. Kidney Int 2013; 84: 1129–44. [DOI] [PubMed] [Google Scholar]
- 33. Paris AJ, Snapir Z, Christopherson CD et al A polymorphism that delays fibrosis in hepatitis C promotes alternative splicing of AZIN1, reducing fibrogenesis. Hepatology 2011; 54: 2198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Crews LA, Jiang Q, Zipeto MA et al An RNA editing fingerprint of cancer stem cell reprogramming. J Transl Med 2015; 13: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Casimiro MC, Velasco‐Velazquez M, Aguirre‐Alvarado C, Pestell RG. Overview of cyclins D1 function in cancer and the CDK inhibitor landscape: past and present. Expert Opin Investig Drugs 2014; 23: 295–304. [DOI] [PubMed] [Google Scholar]
- 36. Finn RS, Aleshin A, Slamon DJ. Targeting the cyclin‐dependent kinases (CDK) 4/6 in estrogen receptor‐positive breast cancers. Breast Cancer Res 2016; 18: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Newman RM, Mobascher A, Mangold U et al Antizyme targets cyclin D1 for degradation. A novel mechanism for cell growth repression. J Biol Chem 2004; 279: 41504–11. [DOI] [PubMed] [Google Scholar]
- 38. Kim SW, Mangold U, Waghorne C et al Regulation of cell proliferation by the antizyme inhibitor: evidence for an antizyme‐independent mechanism. J Cell Sci 2006; 119: 2583–91. [DOI] [PubMed] [Google Scholar]
- 39. Dulloo I, Gopalan G, Melino G, Sabapathy K. The antiapoptotic DeltaNp73 is degraded in a c‐Jun‐dependent manner upon genotoxic stress through the antizyme‐mediated pathway. Proc Natl Acad Sci USA 2010; 107: 4902–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Belloni L, Moretti F, Merlo P et al DNp73alpha protects myogenic cells from apoptosis. Oncogene 2006; 25: 3606–12. [DOI] [PubMed] [Google Scholar]
- 41. Meraldi P, Nigg EA. The centrosome cycle. FEBS Lett 2002; 521: 9–13. [DOI] [PubMed] [Google Scholar]
- 42. Mangold U, Hayakawa H, Coughlin M, Munger K, Zetter BR. Antizyme, a mediator of ubiquitin‐independent proteasomal degradation and its inhibitor localize to centrosomes and modulate centriole amplification. Oncogene 2008; 27: 604–13. [DOI] [PubMed] [Google Scholar]
- 43. Gruendler C, Lin Y, Farley J, Wang T. Proteasomal degradation of Smad1 induced by bone morphogenetic proteins. J Biol Chem 2001; 276: 46533–43. [DOI] [PubMed] [Google Scholar]
- 44. Lim SK, Gopalan G. Antizyme1 mediates AURKAIP1‐dependent degradation of Aurora‐A. Oncogene 2007; 26: 6593–603. [DOI] [PubMed] [Google Scholar]
- 45. Kasbek C, Yang CH, Fisk HA. Antizyme restrains centrosome amplification by regulating the accumulation of Mps1 at centrosomes. Mol Biol Cell 2010; 21: 3878–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kasbek C, Yang CH, Yusof AM, Chapman HM, Winey M, Fisk HA. Preventing the degradation of mps1 at centrosomes is sufficient to cause centrosome reduplication in human cells. Mol Biol Cell 2007; 18: 4457–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xiong A, Zhang Y, Chen X et al Ornithine decarboxylase antizyme 1 upregulate LOR to promote differentiation of SCC15 cells by binding CBP/p300 in promoter region. Int J Clin Exp Med 2016; 9: 2359–66. [Google Scholar]
- 48. Szklarczyk D, Franceschini A, Wyder S et al STRING v10: protein‐protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015; 43: D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Casero RA Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discovery 2007; 6: 373–90. [DOI] [PubMed] [Google Scholar]
- 50. Mitchell JL, Thane TK, Sequeira JM, Marton LJ, Thokala R. Antizyme and antizyme inhibitor activities influence cellular responses to polyamine analogs. Amino Acids 2007; 33: 291–7. [DOI] [PubMed] [Google Scholar]


