Abstract
Aberrant sialylation profiles on the cell surface have been recognized for their potential diagnostic value in identifying the regulation of tumor properties in several cancers, including hepatocellular carcinoma (HCC). Recently, increasing evidence has suggested that the deregulation of microRNA (miRNA) is a common feature in human cancers. In this study, we found obvious upregulation of sialyltransferase ST3GAL6 both in HCC cell lines and in tissue samples. The altered expression of ST3GAL6 was found to correlate with cell proliferation, migration, and invasion ability in HCC. Further investigation showed that miR‐26a negatively regulated ST3GAL6, inducing the suppression of cell proliferation, migration, and invasion in vitro. Moreover, we identified the protein kinase B/mammalian target of rapamycin (Akt/mTOR) pathway as the target of ST3GAL6 based on Western blot analysis. Analysis of a xenograft mouse model showed that miR‐26a significantly reduced tumor growth by suppressing activation of the Akt/mTOR pathway by directly targeting ST3GAL6. In conclusion, these data indicate that ST3GAL6 promotes cell growth, migration, and invasion and mediates the effect of miR‐26a through the Akt/mTOR signaling pathway in HCC.
Keywords: Akt/mTOR pathway; growth, invasion and migration; hepatocellular carcinoma; miR‐26a; ST3GAL6
Primary liver cancer, with HCC representing the major histological subtype, is the fifth most frequently diagnosed cancer worldwide but the second most frequent cause of cancer death in men.1 Currently, surgical resection and liver transplantation remain the most appropriate treatments and are regarded as the only options for recovery. However, only approximately 20–30% of HCC patients are cured after surgical resection, and the reported 5‐year survival rate is 40–50%, with a high rate of cancer recurrence and metastasis.2, 3 This is considered an arduous clinical challenge. Recently, more attention has been paid to identification of the molecular mechanisms possibly underlying metastasis and tumor angiogenesis in HCC. In addition, this might be a promising strategy for improving the diagnosis and treatment of HCC.
Sialyltransferases (STs) are a group of enzymes that specifically conjugate sialic acids (Neu5Ac) to other sugar molecules in glycans. The ST family contains a total of 20 members and is divided into three subfamilies based on the linkage form: alpha‐2,3‐sialyltransferases (ST3Gal I–VI), alpha‐2,6‐sialyltransferases (ST6Gal I–II and ST6GalNAc I–VI), and alpha‐2,8‐sialyltransferases (ST8Sia I–VI).4 Abnormal expression of ST family members has been discovered in various types of cancers, including colon carcinoma, breast cancer, and ovarian carcinoma.5, 6, 7 For the ST3Gal gene family, overexpression of ST3GAL1 has been confirmed to contribute to poor prognosis in clear cell renal cell carcinoma, glioblastoma, and HCC.8, 9, 10 ST3GAL3 was also found to modulate pancreatic cancer cell motility and adhesion in vitro and enhance its metastatic potential in vivo.11 In our study, we found that ST3GAL6 was significantly upregulated in both HCC cells and tissues and plays important roles in the progression of several cancers. However, the mechanism through which ST3GAL6 becomes upregulated in HCC cells and its specific molecular function remain unclear, prompting further research.
MicroRNAs (miRNAs) are short, non‐protein coding RNAs that are approximately 20 nt in length and have been confirmed to alter gene expression at the post‐transcriptional level.12, 13 MicroRNAs mainly bind to the 3′‐UTRs of target mRNAs, negatively regulating gene expression through mRNA degradation or the suppression of mRNA translation.14, 15 Numerous studies have proved that miRNAs function as critical tumor suppressors or promoters, participating in the development of most cancers. For instance, the downregulation of miR‐31 is associated with poor prognosis in gastric cancer, and its restoration suppresses malignant tumor cell phenotypes by inhibiting E2F2.16 MicroRNA‐224 (miR‐224) was recently reported to promote colorectal cancer metastasis by regulating SMAD4.17 In this study, we identified a group of miRNAs that are possible regulators of ST3GAL6 using bioinformatic analysis. Among the predicted miRNAs, miR‐26a, which has been reported to have a correlation with HCC, was identified as the most likely regulator. Thus, we sought to detect the effect of miR‐26a on HCC cells specifically and its potential correlation with ST3GAL6 in further research.
In general, we sought to determine whether ST3GAL6 could mediate the effects of miR‐26a on cell growth, migration, and invasion and the detailed mechanisms involved in this regulation. Our results suggest that ST3GAL6 could be a possible therapeutic target for HCC.
Materials and Methods
Cell culture
The MHCC97‐H, MHCC97‐L, and HCCLM3 human HCC cell lines were obtained from KeyGEN (Nanjing, China). Compared with MHCC97‐L, MHCC97‐H has a high metastasis rate.18 Two HCC clones with the same genetic background were established from the MHCC parental hepatocellular carcinoma line. HEK293T cells were obtained from ATCC (Manassas, VA, USA). Cell lines were cultured in 90% DMEM (Gibco, Grand Island, NY, USA) supplemented with antibiotics (13 penicillin/streptomycin 100 U/mL; HyClone Laboratories, Logan, UT, USA) and 10% heat‐inactivated FBS (Gibco). Cells were incubated at 37°C in a humidified atmosphere containing 5% CO2.
Patient tissue samples
Twenty‐eight pairs of HCC tissues and their adjacent non‐cancerous liver tissues were collected from patients who underwent surgical resection at The Second Affiliated Hospital of Dalian Medical University (Dalian, China) between September 2013 and August 2015 after obtaining informed consent. The study and informed consent were approved by the Ethics Committee of The Second Affiliated Hospital of Dalian Medical University. The extracted specimens were confirmed to be HCC tissue with pathological diagnosis using Union for International Cancer Control guidelines. The specimens were immediately frozen in liquid nitrogen and then stored at −80°C for analysis.
Real‐time PCR analysis
Quantitative real‐time PCR (qRT‐PCR) was used to measure the gene expression level of ST3GAL6 and miRNAs. The total RNA was extracted from cell lines using an RNeasy Mini Kit (Qiagen, Valencia, CA, USA), and cDNA was synthesized using a QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer's instructions. To quantify miRNAs, total RNA was polyadenylated and then reversed transcribed using an NCode miRNA First‐Strand cDNA Synthesis Kit (Invitrogen, Grand Island, NY, USA). The relative expression of miRNAs was determined using a mirVana qRT‐PCR microRNA Detection Kit according to the manufacturer's protocol (Ambion, Austin, TX, USA) and was normalized to U6‐small nuclear RNA using the method. ST3GAL6 mRNA levels were quantified with SYBR Green qRT‐PCR (Takara, Otsu, Shiga, Japan) and normalized to GAPDH. Real‐time PCR was carried out on an ABI Prism 7500 fast real‐time PCR system (Applied Biosystems, Foster City, CA, USA). The sequences of the upstream and downstream primers were as follows: ST3GAL6, 5′‐ATGTCTATTGGGTGGCAC‐3′ and 5′‐CGCACACAGAAAAGGGTG‐3′, respectively; and GAPDH, 5′‐CTCCTCCACCTTTGACGCTG‐3′ and 5′‐TCCTCTTGTGCTCTTGCTGG‐3′, respectively. All samples were normalized to internal controls, and fold‐changes were calculated by relative quantification ().
Western blot analysis
Whole cell proteins were electrophoresed under reducing conditions in 10% polyacrylamide gels. The separated proteins were transferred to a PVDF membrane. After blocking with 5% non‐fat milk in PBS containing 0.1% Tween 20 (PBST), the membrane was incubated with antibodies (anti‐ST3GAL6, 1:500 dilution; anti‐mammalian target of rapamycin [mTOR] and anti‐p‐mTOR, 1:500 dilution [Thermo Fisher Scientific, Rockford, IL, USA]; and anti‐protein kinase B [Akt] and anti‐p‐Akt, 1:1000 dilution [Thermo Fisher Scientific]) overnight at 4°C and then with peroxidase‐conjugated anti‐rabbit IgG (1:10 000 dilution; GE Healthcare, Little Chalfont, UK). A GAPDH antibody (1:200 dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used as a control. All bands were detected using an ECL Western blot kit (Amersham Biosciences, Little Chalfont, UK), and the bands were analyzed with LabWorks (version 4.6; UVP, BioImaging Systems, Upland, CA, USA).
Oligonucleotides, plasmid construction, and cell transfection
ST3GAL6 coding sequences obtained from Takara (Dalian, China) were cloned into the pcDNA3.1 vector (Invitrogen) to generate the pcDNA3.1/ST3GAL6 expression vector. We used siST3GAL6 (100 nM; GenePharma, Shanghai, China) and scrambled siRNA (siSCR) to transfect MHCC97‐H and MHCC97‐L cells. MicroRNA‐26a mimics or negative control miRNA (miR‐NC) and miR‐26a inhibitor (anti‐miR‐26a) or negative control miRNA (anti‐miR‐NC) were obtained from RiboBio (GenePharma, Shanghai, China). Lipofectamine 2000 (Invitrogen) was used for all transfection experiments according to the manufacturer's instructions. Forty‐eight hours after transfection, cells were harvested for further study.
Dual‐luciferase assay
The wild‐type ST3GAL6 3′‐UTR was cloned into the pGL3‐control vector (Promega, Madison, WI, USA) downstream of the firefly luciferase gene, and the mutant 3′‐UTR plasmid was subsequently created by site‐directed mutagenesis. HEK293T cells were seeded (5 × 104 cells per well) into a 24‐well plate and were incubated overnight. For the 3′‐UTR luciferase assay, cells were cotransfected with the miR‐26a mimics and the wild‐type or mutant target sequence using Lipofectamine 2000. Forty‐eight hours after transfection, lysates were collected, and the activities of firefly and Renilla luciferase were measured using the Dual‐Luciferase Reporter Assay System (Promega) and normalized to Renilla luciferase activity. The mean of the results from the cells transfected with the miR‐control was set at 1.0. The data are presented as the mean value ± SD for triplicate experiments.
Transwell invasion assay
Cell invasion was investigated in vitro using 24‐well Transwell units (Corning, New York, NY, USA) with 8‐μm pore size polycarbonate filters coated with a continuous thin layer of ECMatrix gel (Chemicon, Billerica, Massachusetts, USA). MHCC97‐H and MHCC97‐L cells (3 × 105) were harvested in serum‐free medium containing 0.1% BSA and added to the upper chamber. The lower chamber contained 500 μL DMEM. The cells were incubated for 24 h at 37°C with 5% CO2. At the end of the incubation, the cells on the upper surface of the filter were completely removed by wiping with a cotton swab. The filters were then fixed in methanol and stained with Wright Giemsa. The cells that had invaded the Matrigel and reached the lower surface of the filter were counted under a light microscope at a magnification of ×400.
Wound‐healing assay
Cell migration was measured using a wound‐healing assay. Fully confluent cells were seeded into 12‐well plates. A cell‐free area was created by scraping using a pipette tip. Wound closure was measured at 0 and 24h.
Cell proliferation assay
Cell Counting Kit‐8 (CCK‐8; Biotool, Houston, TX, USA) was used to measure cell proliferation. Cells (2 × 103/well) were seeded into 96‐well plates containing complete DMEM (100 μL) in triplicate for each condition and were maintained in an incubator at 37°C with 5% CO2. Then CCK‐8 solution (10 μL) was added to each well and incubated for 4 h. Optical density values were measured with a water‐soluble tetrazolium salt assay using microplate computer software (Bio‐Rad Laboratories, Hercules, CA, USA) according to the protocol of the CCK‐8 assay kit (Biotool). The absorbance at 450 nM (A450) was read on a microplate reader (168–1000 Model 680; Bio‐Rad Laboratories), and proliferation curves were plotted.
Colony formation assays
Cells were suspended as a single cell suspension, seeded into six‐well plates at 500–1000 cells/well and cultured in high‐glucose‐DMEM containing 10% FBS at 37°C and 5% CO2 for 2–3 weeks. The cell colonies were then fixed with 10% formaldehyde for 40 min and stained with 0.1% crystal violet solution for 20 min. Colonies with more than 50 cells were counted using ImagePro Plus 6.0 software (Media Cybernetics, Silver Spring, MD, USA).
Xenograft mouse model and lentivirus infection
Four‐week‐old male BALB/c athymic nude mice were purchased from the Animal Facility of Dalian Medical University. All animal experiments were approved by the Committee on the Ethics of Animal Experiments of Dalian Medical University. Lentiviral constructs containing pre‐miR‐26a (LV‐miR‐26a) were purchased from GeneChem (Shanghai, China). As a control, we also generated a lentiviral vector that expressed green fluorescent protein alone (LV‐miR‐NC). Approximately 1 × 107 MHCC97‐H cells were injected s.c. into the right flank of each nude mouse. After the development of palpable tumors (approximately 1 week after tumor cell injection), mice were treated with intratumoral injections of 1 × 107 pfu/mL LV‐miR‐26a or LV‐miR‐NC lentiviral construct, and the length and width of tumors were measured every week. Tumor volume was calculated using the following formula: tumor volume = 1/2 (length × width2). Five to six weeks after injection, mice were killed and tumors were harvested and weighed.
Immunohistochemical staining analysis
After the xenograft tumor was harvested, it was immediately fixed in 4% paraformaldehyde, dehydrated in a graded series of alcohol, and then embedded in paraffin. Slices (0.5‐mm thick) were cut, dried, deparaffinized, rehydrated, and then immersed in 3% hydrogen peroxide for 10 min to quench endogenous peroxidase activity. After consecutive washes with PBS, the slides were labeled with antibodies (Abcam, Cambridge, UK) at a dilution of 1:200 at 4°C overnight. Staining with the secondary streptavidin–HRP‐conjugated antibody (Santa Cruz Biotechnology) was carried out at room temperature for 60 min. Finally, the sections were counterstained with hematoxylin and coverslipped. ImagePro Plus 4.5 software (Media Cybernetics, Silver Spring, MD, USA) was used to analyze the expression of proteins.
Statistical analysis
Each experiment was carried out in at least triplicate. Data are displayed as the mean ± SD and were analyzed using SPSS 13.0 (SPSS, Chicago, IL, USA). The significance of the differences in multiple comparisons was determined using Student's t‐test. The association between miR‐26a and ST3GAL6 expression was determined using Spearman's correlation analysis. P < 0.05 was considered statistically significant.
Results
ST3GAL6 is upregulated in human HCC tissues and cell lines
In our previous research, ST3GAL6 was found to be overexpressed in MHCC97‐H cells relative to MHCC97‐L cells but attracted little attention. To further confirm our previous data, we measured the expression levels of ST3GAL6 in three types of HCC cell lines (MHCC97‐H, MHCC97‐L, and HCCLM3) and normal cell lines (LO2) using qRT‐PCR. As expected, ST3GAL6 expression was higher in HCCLM3 and MHCC97‐H cells than in MHCC97‐L cells, and the same result was also found in MHCC97‐L cells compared with the normal LO2 cell line (Fig. 1a). Next, the expression level of the ST3GAL6 gene in 28 pairs of HCC tissues and corresponding adjacent non‐cancerous tissues was measured. Similarly, ST3GAL6 was significantly upregulated in HCC tissues compared with adjacent non‐tumor tissues (Fig. 1b). Taken together, these data indicate that ST3GAL6 may contribute to the malignant phenotype of HCC.
Figure 1.
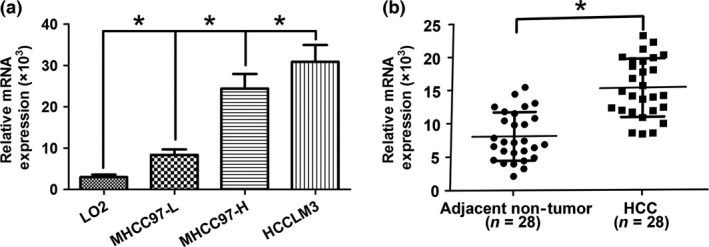
Sialyltransferase ST3GAL6 was upregulated in human hepatocellular carcinoma (HCC) tissues and cell lines. (a) Differences in expression in HCC cell lines (MHCC97‐H, MHCC97‐L, and HCCLM3) and a normal human hepatic cell line (LO2). (b) ST3GAL6 (measured by SYBR Green quantitative real‐time PCR) was upregulated in HCC tissues compared with the matched adjacent non‐tumorous liver tissues. The central horizontal line represents the mean value. *P < 0.05.
ST3GAL6 regulates the invasion, migration, and proliferation of HCC cells
To elucidate the potential biological functions of ST3GAL6 in the progression of HCC, we overexpressed ST3GAL6 by transfecting the ST3GAL6 expression vector into MHCC97‐L cell lines. Quantitative RT‐PCR and Western blot analysis were carried out, and a significant increase in ST3GAL6 expression was found in the transfected cells relative to control cells (Fig. 2a). Next, cell metastatic ability was measured with a Transwell assay and a wound‐healing assay; cell proliferation was investigated using a colony formation assay and a CCK‐8 assay. As shown in Figure 2, overexpression of ST3GAL6 significantly promoted cell migration, invasion, and proliferation compared to control cells. To further confirm our hypothesis, we knocked down the expression of ST3GAL6 in MHCC97‐H cells by transfecting these cells with siST3GAL6 or siSCR. As expected, when ST3GAL6 levels decreased, MHCC97‐H cells showed decreased migration, invasion, and proliferation compared to control cells. Moreover, inhibiting the expression of ST3GAL6 in HCCLM3 cells showed a trend similar to that in MHCC97‐H cells (Fig. S1). Together, these results indicated that ST3GAL6 was involved in the regulation of cell migration, invasion, and proliferation.
Figure 2.
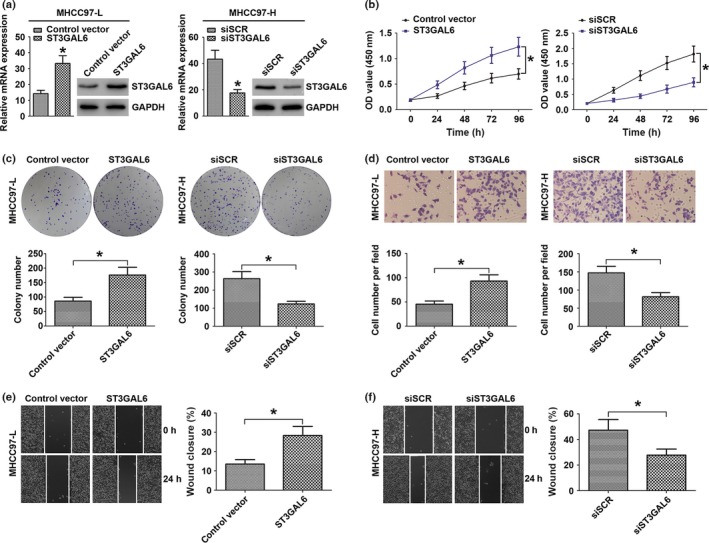
Sialyltransferase ST3GAL6 regulated invasion, migration, and proliferation of hepatocellular carcinoma (HCC) cells. (a–f) MHCC97‐L cells were transfected with ST3GAL6 or control vector; MHCC97‐H cells were transfected with siST3GAL6 or scrambled siRNA (siSCR). (a) Relative expression of ST3GAL6 was detected by quantitative real‐time PCR and Western blot analysis in the two hepatoma cell lines after transfection. Proliferation curves according to the results of CCK‐8 assay (b) and representative results of the colony formation assay (c) for MHCC97‐L and MHCC97‐H cells with different transfections. Representative results of the Transwell invasion assay (d) and wounding healing assay (e, f) for MHCC97‐L transfected with ST3GAL6 (or control vector) and MHCC97‐H cells transfected with siST3GAL6 (or siSCR). Each experiment was independently repeated at least three times. Data are presented as the mean ± SD. *P < 0.05 compared with the control.
MicroRNA‐26a is a negative regulator of ST3GAL6
Given that miRNA is acknowledged as an important upstream regulator involved in most types of cancers, we questioned whether there was any miRNA modulating the expression of ST3GAL6. Twenty miRNAs were selected as possible regulators using miRNA target prediction software (TargetScan, www.targetscan.org). Quantitative RT‐PCR was then undertaken to measure the different expression levels of these selected miRNAs in the MHCC97‐H and MHCC97‐L cell lines. Finally, we identified miR‐26a as a hypothetical negative regulator of ST3GAL6 due to its most obvious decrease in expression level in MHCC97‐H cells compared with MHCC97‐L cells among these selected miRNAs (Fig. 3a). Subsequently, we measured the expression level of miR‐26a in 28 pairs of HCC tissues and the corresponding adjacent non‐cancerous tissues. Consistent with the above results, decreased expression of miR‐26a was also observed in HCC tissues compared to non‐cancerous tissues (Fig. 3b), and analysis of miR‐26a and ST3GAL6 expression in 28 HCC specimens with Spearman's correlation analysis revealed a significant inverse correlation (Fig. 3c). For the luciferase reporter assay, HEK293T cells were cotransfected with miR‐26a mimics and the wild‐type or mutant ST3GAL6 target sequence. The cotransfection of miR‐26a dramatically suppressed the activity of the luciferase reporter containing the wild‐type 3′‐UTR of ST3GAL6, whereas this effect was abrogated when the predicted 3′‐UTR binding site was mutated (Fig. 3d,e). Moreover, upregulating miR‐26a in MHCC97‐H cells dramatically suppressed the expression of ST3GAL6, whereas downregulating miR‐26a in MHCC97‐L cells had the opposite effect (Fig. 3f). However, altering the expression of ST3GAL6 had little effect on miR‐26a (Fig. 3g).
Figure 3.
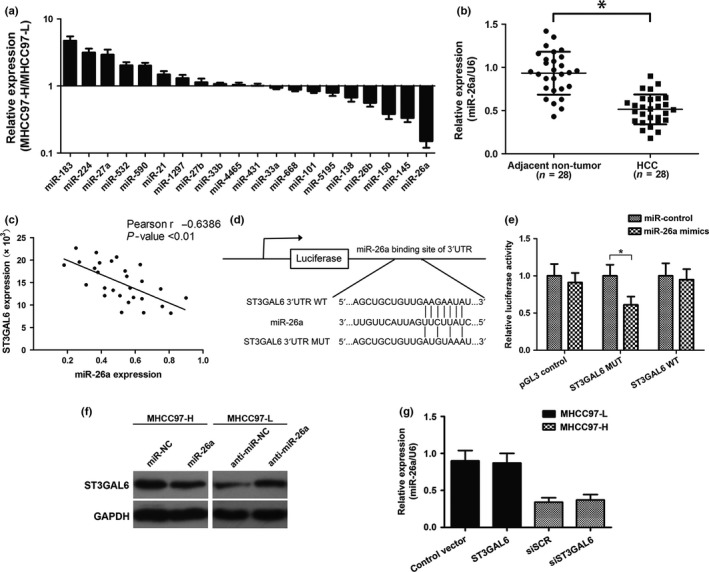
MicroRNA‐26a (miR‐26a) was a negative regulator of sialyltransferase ST3GAL6. (a) Relative expression of 20 types of predicted miRNAs in MHCC97‐H hepatocellular carcinoma (HCC) cells compared with MHCC97‐L cells. (b) Relative expression level of miR‐26a in HCC tissues and adjacent non‐tumor tissues (28 pairs). (c) Correlation of miR‐26a to ST3GAL6 mRNA levels in 28 pairs of HCC tissues using Spearman's correlation analysis. (d) Wild‐type (WT) or mutant (MUT) miR‐26a target sequences of the ST3GAL6 mRNA 3′‐UTR. (e) Relative luciferase activity of HEK 293T cells after cotransfection with WT or MUT ST3GAL6 3′‐UTR reporter genes or pGL3‐control vector and miR‐26a mimics or miR‐control. (f) Western blot analysis of ST3GAL6 protein expression in MHCC97‐H and MHCC97‐L cells. (g) Relative expression of ST3GAL6, detected by quantitative real‐time PCR after transfection in MHCC97‐H and MHCC97‐L cells. Each experiment was independently performed at least three times. Data are presented as mean ± SD. *P < 0.05.
MicroRNA‐26a suppresses proliferation, migration, and invasion of HCC cells by negatively regulating ST3GAL6
Next, to verify whether miR‐26a could repress the migration, invasion, and proliferation of HCC cells by regulating ST3GAL6, we utilized gain‐of‐function and loss‐of‐function analyses (Fig. 4). MHCC97‐H cells were cotransfected with miRNA mimics (miR‐26a mimics or miR‐NC) and either the empty pcDNA3.1 vector or the pcDNA3.1/ST3GAL6 plasmid (ST3GAL6). After transfection with miR‐26a mimics, MHCC97‐H cells showed reduced proliferation (Fig. 4b,d), migration (Fig. 4h), and invasion (Fig. 4f) ability compared to the negative control. Interestingly, the overexpression of ST3GAL6 clearly abrogated the effect of miR‐26a. The experiments using HCCLM3 cells also generated results similar to those in MHCC97‐H cells (Fig. S2).
Figure 4.
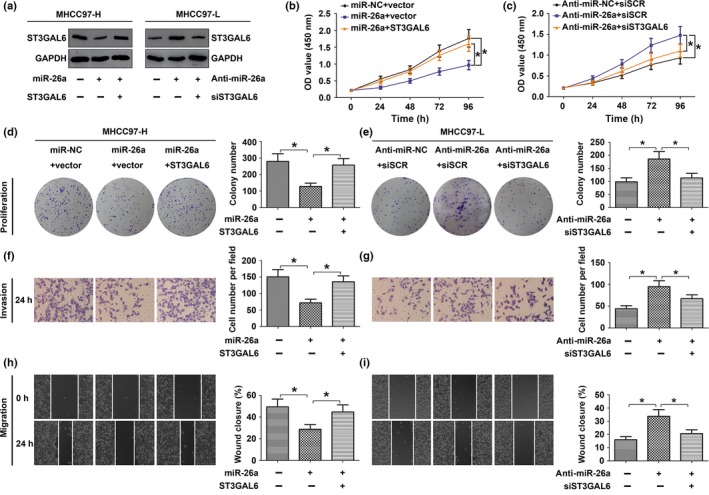
MicroRNA‐26a (miR‐26a) suppressed the proliferation, migration, and invasion of hepatocellular carcinoma (HCC) cells by negatively regulating sialyltransferase ST3GAL6. (a–f) MHCC97‐H cells were cotransfected with target mRNA (ST3GAL6 or control vector) and miRNA mimics (miR‐26a or miR‐NC); MHCC97‐L cells were cotransfected with siRNA (siST3GAL6 or scrambled siRNA [siSCR]) and miRNA inhibitor (anti‐miR‐26a or anti‐miR‐NC). (a) Western blot analysis of ST3GAL6 protein expression in MHCC97‐H and MHCC97‐L cells. (b–f) Upregulation of ST3GAL6 reversed the inhibitory effect of miR‐26a in MHCC97‐H cells and knockdown of ST3GAL6 abrogated the promoting effect of anti‐miR‐26a in MHCC97‐L cells. Proliferation curves according to results of the CCK‐8 assay in MHCC97‐H (b) and MHCC97‐L (c). Representative results of the colony formation assay in MHCC97‐H (d) and MHCC97‐L (e) cells. Representative results of wound‐healing assay in MHCC97‐H (h) and MHCC97‐L (i) cells and Transwell invasion assay in MHCC97‐H (f) and MHCC97‐L (g) cells. Each experiment was independently repeated at least three times. Data are presented as mean ± SD. *P < 0.05. OD, optical density.
To further confirm our hypothesis, MHCC97‐L cells were cotransfected with miR‐26a inhibitor (anti‐miR‐26a or anti‐miR‐NC) and siRNA (siST3GAL6 or siSCR). Downregulation of ST3GAL6 by siST3GAL6 reversed the stimulatory effect of the miR‐26a inhibitor on the proliferation (Fig. 4c,e), migration (Fig. 4i), and invasion (Fig. 4g) ability of MHCC97‐L cells. Together, the data above suggested that miR‐26a suppressed the proliferation, migration, and invasion of HCC cells at least partly by targeting ST3GAL6.
ST3GAL6 mediates the regulation of miR‐26a through the Akt/mTOR signaling pathway
As a classic signaling pathway, the Akt/mTOR pathway has been implicated in most cancers, especially HCC. Previous studies have found that STs can stimulate the phosphatidylinositol 3‐kinase (PI3K)/Akt pathway in HCC. This prompted us to investigate the effect of miR‐26a on the ST3GAL6/Akt/mTOR pathway. Activation of Akt and mTOR was detected by Western blot analysis. As expected, after increasing the expression level of miR‐26a by transfecting cells with miR‐26a mimics, the phosphorylation levels of Akt (p‐Akt) and mTOR (p‐mTOR) in MHCC97‐H cells decreased dramatically, whereas the total Akt and mTOR levels remained unchanged. However, these effects were attenuated with the cotransfection of ST3GAL6 (Fig. 5a). Furthermore, the effects of siST3GAL6 on p‐Akt and p‐mTOR were similar to the effects induced by miR‐26a (Fig. 5b). An experiment in HCCLM3 cells revealed the same effect as that observed in MHCC97‐H cells (Fig. S3). To further support our hypothesis, we knocked down the expression of miR‐26a in MHCC97‐L cells by transfecting these cells with the miR‐26a inhibitor. As expected, the downregulation of miR‐26a significantly increased the phosphorylation of Akt and mTOR with no change in the total Akt and mTOR level (Fig. 5c). Reducing the expression of ST3GAL6 with siST3GAL6 clearly reversed the effect of decreased miR‐26a levels. Moreover, the effects of ST3GAL6 on Akt and mTOR were similar to the effect induced by the miR‐26a inhibitor, which was consistent with our previous results (Fig. 5d). Taken together, these results suggest that ST3GAL6 mediates the regulation of miR‐26a through the Akt/mTOR signaling pathway.
Figure 5.
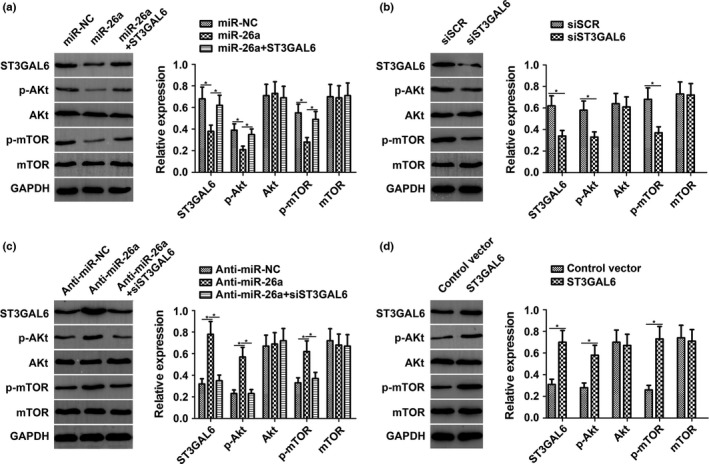
Sialyltransferase ST3GAL6 mediated the regulation of microRNA‐26a (miR‐26a) to the protein kinase B/mammalian target of rapamycin (Akt/mTOR) signaling pathway. (a) In MHCC97‐H hepatocellular carcinoma cells with miR‐26a overexpression, the expression levels of ST3GAL6, phosphorylated (p‐)Akt, and p‐mTOR were significantly decreased compared with the control. ST3GAL6 treatment abrogated the decreased expression of these genes induced by miR‐26a. (b) Knockdown of ST3GAL6 by siST3GAL6 inhibited expression of p‐Akt and p‐mTOR, similar to miR‐26a. (c) In MHCC97‐L cells with miR‐26a downregulation, the expression levels of ST3GAL6, p‐Akt, and p‐mTOR were dramatically increased compared with the control. siST3GAL6 treatment reversed the promoting effect of decreased miR‐26a expression. (d) Overexpression of ST3GAL6 promoted the expression of p‐Akt and p‐mTOR, similar to anti‐miR‐26a. Each experiment was independently repeated at least three times. Data are presented as mean ± SD. *P < 0.05.
MicroRNA‐26a suppresses tumor growth through ST3GAL6/Akt/mTOR pathway in vivo
To explore whether miR‐26a was involved in tumorigenesis in vivo, a xenograft tumor model was used in nude mice. MHCC97‐H cells were s.c. implanted into the right flank of each nude mouse to form tumors. As miR‐26a is downregulated in HCC cells, we constructed a lentiviral vector named LV‐miR‐26a (and LV‐miR‐NC as a control) to inject into the tumor to increase miR‐26a expression. Nude mice were divided into two groups (the LV‐miR‐26a group and the LV‐miR‐NC group) based on which vector was injected. Compared with the LV‐miR‐NC group, the LV‐miR‐26a group showed dramatically reduced tumor volume and weight during the 5 weeks after the tumors became palpable (Fig. 6a,b). Moreover, the expression of Ki67, which is a marker of proliferation, was detected in the resected tumor tissues from these s.c. xenograft tumors. Consistent with this, decreased expression of Ki67 was found in the LV‐miR‐26a group, indicating attenuated cell proliferation (Fig. 6c,e). To further clarify the cellular mechanisms underlying the miR‐26a‐mediated modulation of tumor characteristics, these resected tissues were evaluated to test ST3GAL6 expression and the activity of the downstream Akt/mTOR pathway. As shown in Figure 6(d,f), the LV‐miR‐26a group showed decreased ST3GAL6 expression and reduced activity of the Akt/mTOR pathway. These data provide strong evidence that miR‐26a inhibits HCC proliferation through the ST3GAL6/Akt/mTOR axis in vivo.
Figure 6.
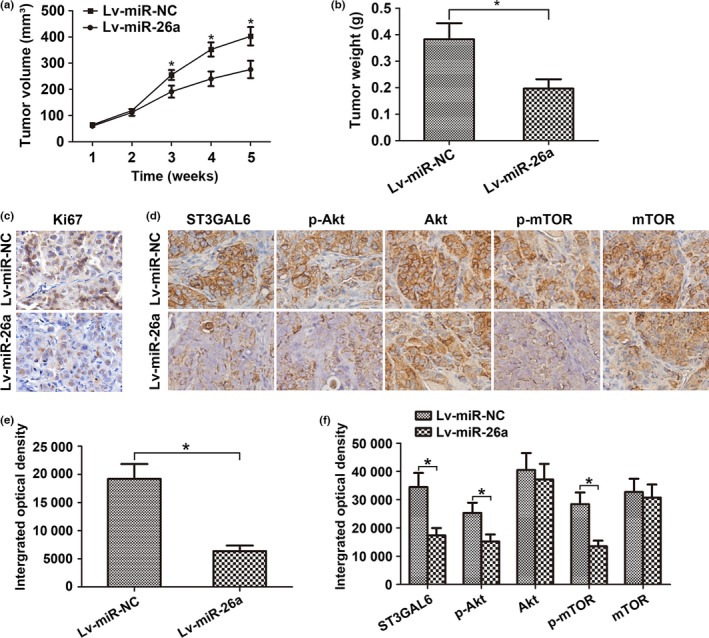
MicroRNA‐26a (miR‐26a) suppressed tumor growth through the sialyltransferase ST3GAL6/protein kinase B (Akt)/mammalian target of rapamycin (mTOR) pathway in vivo. (a–f) MHCC97‐H hepatocellular carcinoma cells were s.c. implanted into the right flank of each nude mouse to form tumors. miR‐26a lentivirus (Lv‐miR‐26a) or control lentivirus (Lv‐miR‐NC) was injected into each tumor. (a) Growth curve of tumor volumes. (b) Weight of xenograft tumors. Data represent mean ± SD from six nude mice. The expression of Ki67 (c, e) and activity of the ST3GAL6/Akt/mTOR pathway (d, f) in tumor tissues was examined by immunohistochemical staining. *P < 0.05.
Discussion
The ST family is frequently found to be deregulated in diverse types of cancers, resulting in a high mortality rate. In this study, we observed that: (i) ST3GAL6 was upregulated both in HCC cell lines and in sample specimens, promoting the proliferation, migration, and invasion capacity of HCC cells; (ii) miR‐26a functioned as a negative regulator of ST3GAL6 and was inversely correlated with ST3GAL6 expression; (iii) overexpression of miR‐26a suppressed cell proliferation, migration, and invasion in vitro and inhibited tumor growth in vivo; and (iv) ST3GAL6 overexpression dramatically attenuated the effect of miR‐26a and vice versa. Taken together, our work suggests that miR‐26a has a suppressive effect on cell growth, migration, and invasion in HCC by regulating ST3GAL6 expression.
The deregulation of cell proliferation and frequent metastasis from the primary tumor mass represent the two predominant processes that affect the prognosis of most cancers, including HCC. The elevated expression of sialic acids caused by the overexpression of sialyltransferases can protect cancer cells from apoptosis and prompt metastasis, and has been indicated to affect therapeutic efficacy.6, 19, 20 Certain reports have shown that ST3GAL6 promotes cell metastasis in breast cancer and multiple myeloma, but suppresses cell metastasis in gastrointestinal cancer.21, 22, 23A study has shown that ST3GAL6 expression was significantly reduced in HCC.24 However, in our previous study, we found that ST3GAL6 was upregulated in MHCC97‐H cells relative to MHCC97‐L cells.25 To further investigate the effect of ST3GAL6 in HCC, we measured the ST3GAL6 expression level in HCC cells and tissues relative to non‐cancerous cells and tissues. Consistent with our previous findings, a significant increase in ST3GAL6 was observed in HCC cells and tumor tissues. Moreover, we found that downregulation of ST3GAL6 dramatically attenuated the growth, migration, and invasion properties of HCC cells and vice versa. These data indicate that ST3GAL6 might function as a tumor promoter in HCC.
We then investigated the mechanism underlying ST3GAL6 upregulation in HCC. Recently, much attention has been paid to detecting miRNAs and their target mRNAs in several cancers, including HCC. For example, miR‐486‐5p suppressed the progression and metastasis of non‐small‐cell lung cancer by targeting ARHGAP5.26 Moreover, elevated oncofetal miR‐17‐5p expression regulated colorectal cancer progression by repressing the target gene P130.27 To further screen for miRNAs targeting ST3GAL6, we selected 20 miRNAs using bioinformatic analysis and finally identified miR‐26a as the most significantly decreased miRNA in highly invasive HCC cells. Furthermore, a dual‐luciferase assay indicated that miR‐26a can directly target the ST3GAL6 3′‐UTR. MicroRNA‐26a has been proven to be responsible for regulating the expression of many tumor‐associated factors, such as nuclear factor‐κ‐gene binding (NF‐κB),28 metadherin,29 and high mobility group A1,30 among others. In the present study, upregulation of miR‐26a, which clearly suppressed the expression of ST3GAL6, had little effect on other targets (Fig. S4). Moreover, downregulation of ST3GAL6 by miR‐26a overexpression caused subsequent reductions in cell proliferation, migration, and invasion. Inhibiting miR‐26a in MHCC97‐L cells showed the opposite effects. However, altering the expression of ST3GAL6 in turn had little effect on miR‐26a. More importantly, altering the expression of ST3GAL6 dramatically reversed the effect of miR‐26a. Therefore, these results suggest that the inhibitory effects of miR‐26a on HCC progression might correlate with ST3GAL6 to some extent. Previous studies have verified that c‐Myc and lncRNA taurine‐upregulated gene 1 can regulate the expression of miR‐26a.31, 32 This provides us with valuable evidence to explore the possible mechanisms underlying miR‐26a suppression in HCC in future research.
To further explore the ST3GAL6‐mediated regulatory mechanism underlying the progression of HCC, we searched for the downstream targets of ST3GAL6. A study has revealed a direct correlation between ST6GalNAcII and the PI3K/Akt/NF‐κB signaling pathway in breast cancer.33 ST3GAL5, also known as GM3S, was found to mediate migration and invasion in murine breast cancer cells by inhibiting the PI3K/Akt pathway.34 ST3GAL6 was reported to participate in the synthesis of sialyl‐paragloboside, a precursor of the sialyl‐Lewis X (sLe X) determinant.35 As one type of Lewis ligand, sLe X on the cell surface plays important roles in signal transduction and is correlated with PI3K/Akt signaling pathway activity.36 In HCC, the AKT/mTOR pathway is frequently overactivated, and a clinical study has indicated that activation of the AKT/mTOR pathway is correlated with tumor progression and reduced patient survival.37, 38 Moreover, extensive evidence indicates that the regulation of miRNAs in the Akt/mTOR signaling pathway is mediated by several molecules, such as c‐MET,39 phosphatase and tensin homolog,40 and methyl‐CpG binding protein 2.41 However, none of these reports has discussed a correlation between the Akt/mTOR pathway and the ST family. To test the effects of miR‐26a and ST3GAL6 on the Akt/mTOR signaling pathway, we used Western blot analysis. Consistent with other results, upregulation of ST3GAL6 attenuated the inhibitory effect of overexpressed miR‐26a on the ST3GAL6/Akt/mTOR pathway. Further investigation revealed that inhibition of ST3GAL6 could reverse the promoting effect induced by repression of miR‐26a in MHCC97‐L cells. Moreover, siST3GAL6 had the same effect as miR‐26a, and the ST3GAL6 expression vector had an effect similar to that of anti‐miR‐26a. From these data, we can draw the conclusion that miR‐26a regulates the Akt/mTOR pathway, at least in part, by targeting ST3GAL6.
In summary, our research revealed that ST3GAL6 mediated the effect of miR‐26a on the proliferation, migration, and invasion of HCC in vitro and tumor growth in vivo through the Akt/mTOR signaling pathway. This highlights the therapeutic potential of ST3GAL6 in HCC and provides a novel candidate target for the prevention and treatment of HCC. However, further investigations are still needed to elucidate the specific mechanism underlying the ST3GAL6‐mediated regulation of the Akt/mTOR signaling pathway.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Fig. S1. ST3GAL6 regulated invasion, migration, and proliferation of HCCLM3 hepatocellular carcinoma cells.
Fig. S2. MicroRNA‐26a (miR‐26a) suppressed the proliferation, migration, and invasion of HCCLM3 hepatocellular carcinoma cells by negatively regulating ST3GAL6.
Fig. S3. ST3GAL6 mediated the regulation of microRNA‐26a (miR‐26a) to the protein kinase B/mammalian target of rapamycin (Akt/mTOR) signaling pathway in HCCLM3 hepatocellular carcinoma cells.
Fig. S4. Effect of microRNA‐26a (miR‐26a) on ST3GAL6 and two other targets.
Acknowledgments
This work was supported by grants from the Natural Science Foundation of Liaoning Province, China (2015020252) and the Science and Technology Project of Dalian (2015E12SF148).
Cancer Sci 108 (2017) 267–276
Funding Information
Natural Science Foundation of Liaoning Province, China (2015020252), Science and Technology Project of Dalian (2015E12SF148)
References
- 1. Schlachterman A, Craft WW Jr, Hilgenfeldt E, Mitra A, Cabrera R. Current and future treatments for hepatocellular carcinoma. World J Gastroenterol 2015; 21: 8478–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou XD, Tang ZY, Yang BH et al Experience of 1000 patients who underwent hepatectomy for small hepatocellular carcinoma. Cancer 2001; 91: 1479–86. [DOI] [PubMed] [Google Scholar]
- 3. Waly Raphael S, Yangde Z, Yuxiang C. Hepatocellular carcinoma: focus on different aspects of management. ISRN Oncol 2012; 2012: 421673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Varki A, Schauer R. Sialic acids In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, eds. Essentials of Glycobiology, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2009. [PubMed] [Google Scholar]
- 5. Schneider F, Kemmner W, Haensch W et al Overexpression of sialyltransferase CMP‐sialic acid: Galbeta 1, 3GalNAc‐R alpha6‐Sialyltransferase is related to poor patient survival in human colorectal carcinomas. Cancer Res 2001; 61: 4605–11. [PubMed] [Google Scholar]
- 6. Cui H, Lin Y, Yue L, Zhao X, Liu J. Differential expression of the alpha2,3‐sialic acid residues in breast cancer is associated with metastatic potential. Oncol Rep 2011; 25: 1365–71. [DOI] [PubMed] [Google Scholar]
- 7. Schultz MJ, Holdbrooks AT, Chakraborty A et al The tumor‐associated glycosyltransferase ST6Gal‐I regulates stem cell transcription factors and confers a cancer stem cell phenotype. Cancer Res 2016; 76: 3978–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bai Q, Liu L, Xia Y et al Prognostic significance of ST3GAL‐1 expression in patients with clear cell renal cell carcinoma. BMC Cancer 2015; 15: 880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chong YK, Sandanaraj E, Koh LW et al ST3GAL1‐associated transcriptomic program in glioblastoma tumor growth, invasion, and prognosis. J Natl Cancer Inst 2015; 108: pii: djv326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu H, Shi XL, Zhang HJ et al Overexpression of ST3Gal‐I promotes migration and invasion of HCCLM3 in vitro and poor prognosis in human hepatocellular carcinoma. Onco Targets Ther 2016; 9: 2227–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pérez‐Garay M, Arteta B, Pagès L et al alpha2,3‐sialyltransferase ST3Gal III modulates pancreatic cancer cell motility and adhesion in vitro and enhances its metastatic potential in vivo. PLoS One 2010; 5: pii: e12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–97. [DOI] [PubMed] [Google Scholar]
- 13. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post‐transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 2008; 9: 102–14. [DOI] [PubMed] [Google Scholar]
- 14. Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet 2012; 13: 358–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hale BJ, Yang CX, Ross JW. Small RNA regulation of reproductive function. Mol Reprod Dev 2014; 81: 148–59. [DOI] [PubMed] [Google Scholar]
- 16. Wang H, Zhang X, Liu Y et al Downregulated miR‐31 level associates with poor prognosis of gastric cancer and its restoration suppresses tumor cell malignant phenotypes by inhibiting E2F2. Oncotarget 2016; 7: 36577–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ling H, Pickard K, Ivan C et al The clinical and biological significance of MIR‐224 expression in colorectal cancer metastasis. Gut 2016; 65: 977–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu Q, Xu H, Xu Q, Yan W, Tian D. Expression of Twist gene in human hepatocellular carcinoma cell strains of different metastatic potential. J Huazhong Univ Sci Technolog Med Sci 2008; 28: 144–6. [DOI] [PubMed] [Google Scholar]
- 19. Swindall AF, Bellis SL. Sialylation of the Fas death receptor by ST6Gal‐I provides protection against Fas‐mediated apoptosis in colon carcinoma cells. J Biol Chem 2011; 286: 22982–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Amano M, Eriksson H, Manning JC et al Tumour suppressor p16(INK4a) – anoikis‐favouring decrease in N/O‐glycan/cell surface sialylation by down‐regulation of enzymes in sialic acid biosynthesis in tandem in a pancreatic carcinoma model. FEBS J 2012; 279: 4062–80. [DOI] [PubMed] [Google Scholar]
- 21. Julien S, Ivetic A, Grigoriadis A et al Selectin ligand sialyl‐Lewis x antigen drives metastasis of hormone‐dependent breast cancers. Cancer Res 2011; 71: 7683–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Glavey SV, Manier S, Natoni A et al The sialyltransferase ST3GAL6 influences homing and survival in multiple myeloma. Blood 2014; 124: 1765–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kawamura YI, Toyota M, Kawashima R et al DNA hypermethylation contributes to incomplete synthesis of carbohydrate determinants in gastrointestinal cancer. Gastroenterology 2008. July; 135(1): 142–51.e3. [DOI] [PubMed] [Google Scholar]
- 24. Souady J, Hülsewig M, Distler U et al Differences in CD75s‐ and iso‐CD75s‐ganglioside content and altered mRNA expression of sialyltransferases ST6GAL1 and ST3GAL6 in human hepatocellular carcinomas and nontumoral liver tissues. Glycobiology 2011; 21: 584–94. [DOI] [PubMed] [Google Scholar]
- 25. Zhao Y, Li Y, Ma H et al Modification of sialylation mediates the invasive properties and chemosensitivity of human hepatocellular carcinoma. Mol Cell Proteomics 2014; 13: 520–36. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26. Wang J, Tian X, Han R et al Downregulation of miR‐486‐5p contributes to tumor progression and metastasis by targeting protumorigenic ARHGAP5 in lung cancer. Oncogene 2014; 33: 1181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma Y, Zhang P, Wang F et al Elevated oncofoetal miR‐17‐5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat Commun 2012; 3: 1291. [DOI] [PubMed] [Google Scholar]
- 28. Rasheed Z, Al‐Shobaili HA, Rasheed N, Mahmood A, Khan MI. MicroRNA‐26a‐5p regulates the expression of inducible nitric oxide synthase via activation of NF‐κB pathway in human osteoarthritis chondrocytes. Arch Biochem Biophys 2016; 594: 61–7. [DOI] [PubMed] [Google Scholar]
- 29. Zhang B, Liu XX, He JR et al Pathologically decreased miR‐26a antagonizes apoptosis and facilitates carcinogenesis by targeting MTDH and EZH2 in breast cancer. Carcinogenesis 2011. Jan; 32(1): 2–9. [DOI] [PubMed] [Google Scholar]
- 30. Lin Y, Chen H, Hu Z et al miR‐26a inhibits proliferation and motility in bladder cancer by targeting HMGA1. FEBS Lett 2013; 587: 2467–73. [DOI] [PubMed] [Google Scholar]
- 31. Sander S, Bullinger L, Klapproth K et al MYC stimulates EZH2 expression by repression of its negative regulator miR‐26a. Blood 2008; 112: 4202–12. [DOI] [PubMed] [Google Scholar]
- 32. Li J, An G, Zhang M, Ma Q. Long non‐coding RNA TUG1 acts as a miR‐26a sponge in human glioma cells. Biochem Biophys Res Commun 2016; 477: 743–8. [DOI] [PubMed] [Google Scholar]
- 33. Ren D, Jia L, Li Y et al ST6GalNAcII mediates the invasive properties of breast carcinoma through PI3K/Akt/NF‐κB signaling pathway. IUBMB Life 2014; 66: 300–8. [DOI] [PubMed] [Google Scholar]
- 34. Gu Y, Zhang J, Mi W et al Silencing of GM3 synthase suppresses lung metastasis of murine breast cancer cells. Breast Cancer Res 2008; 10(1): R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Okajima T, Fukumoto S, Miyazaki H et al Molecular cloning of a novel alpha2,3‐sialyltransferase (ST3Gal VI) that sialylates type II lactosamine structures on glycoproteins and glycolipids. J Biol Chem 1999; 274: 11479–86. [DOI] [PubMed] [Google Scholar]
- 36. Guo Q, Guo B, Wang Y et al Functional analysis of α1,3/4‐fucosyltransferase VI in human hepatocellular carcinoma cells. Biochem Biophys Res Commun 2012. Jan 6; 417(1): 311–7. [DOI] [PubMed] [Google Scholar]
- 37. Tommasi S, Pinto R, Pilato B, Paradiso A. Molecular pathways and related target therapies in liver carcinoma. Curr Pharm Des 2007; 13: 3279–87. [DOI] [PubMed] [Google Scholar]
- 38. Sun CH, Chang YH, Pan CC. Activation of the PI3K/Akt/mTOR pathway correlates with tumour progression and reduced survival in patients with urothelial carcinoma of the urinary bladder. Histopathology 2011; 58: 1054–63. [DOI] [PubMed] [Google Scholar]
- 39. Chen QY, Jiao DM, Wu YQ et al MiR‐206 inhibits HGF‐induced epithelial‐mesenchymal transition and angiogenesis in non‐small cell lung cancer via c‐Met /PI3k/Akt/mTOR pathway. Oncotarget 2016; 7: 18247–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xue M, Yao S, Hu M et al HIV‐1 Nef and KSHV oncogene K1 synergistically promote angiogenesis by inducing cellular miR‐718 to regulate the PTEN/AKT/mTOR signaling pathway. Nucleic Acids Res 2014; 42: 9862–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsujimura K, Irie K, Nakashima H et al miR‐199a links MeCP2 with mTOR signaling and its dysregulation leads to Rett syndrome phenotypes. Cell Rep 2015; 12: 1887–901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. ST3GAL6 regulated invasion, migration, and proliferation of HCCLM3 hepatocellular carcinoma cells.
Fig. S2. MicroRNA‐26a (miR‐26a) suppressed the proliferation, migration, and invasion of HCCLM3 hepatocellular carcinoma cells by negatively regulating ST3GAL6.
Fig. S3. ST3GAL6 mediated the regulation of microRNA‐26a (miR‐26a) to the protein kinase B/mammalian target of rapamycin (Akt/mTOR) signaling pathway in HCCLM3 hepatocellular carcinoma cells.
Fig. S4. Effect of microRNA‐26a (miR‐26a) on ST3GAL6 and two other targets.


