Abstract
Genome‐wide association studies have linked genetic variants at 9p21.3 to the risk of multiple cancers. However, the roles of genetic variants at 9p21.3 in esophageal squamous cell carcinoma (ESCC) development are largely unknown. We evaluated the genetic variants at 9p21.3 reported in cancer genome‐wide association studies with a case–control study including 2139 ESCC cases and 2273 controls in a Chinese population, and measured the mRNA expression levels of MTAP,CDKN2A,CDKN2B, and CDKN2B‐AS1 in paired ESCC tumor and adjacent normal tissues. We found that the G allele of rs7023329 was significantly associated with a decreased risk of ESCC with a per‐allele odds ratio of 0.84 (95% confidence interval, 0.77–0.91; P = 2.95 × 10−5). The rs7023329‐G allele was related to a high expression of MTAP (P = 0.020). The rs1679013‐C allele was independently associated with an increased risk of ESCC with a per‐allele odds ratio of 1.12 (95% confidence interval, 1.01–1.24; P = 0.039). We also found that the carriers of the risk allele rs1679013‐C had lower expression of CDKN2B than non‐carriers (P = 0.035). CDKN2B was also significantly downregulated in ESCC tumor tissues compared with adjacent normal tissues (P = 3.50×10−5). Therefore, our findings indicate that genetic variants at 9p21.3 may modulate the expression of MTAP and CDKN2B and contribute to ESCC susceptibility. This may further advance our understanding of the 9p21.3 locus in cancer development.
Keywords: 9p21.3, esophageal squamous cell carcinoma, gene expression, genetic variants, susceptibility
Esophageal cancer is the third most diagnosed cancer and the fourth leading cause of cancer death in China.1 Esophageal squamous cell carcinoma is the predominant histologic type (90–95%); the incidence of esophageal adenocarcinoma remains extremely low in China.2 Epidemiological and etiological studies have shown that environmental and genetic factors have important roles in esophageal carcinogenesis.3, 4 A series of studies have shown that tobacco smoking, heavy alcohol drinking, nutritional deficiencies, and dietary carcinogen exposure may cause this malignancy.5 There is a strong tendency toward familial aggregation of ESCC in the high incident rates of ESCC, suggesting that genetic susceptibility, in combination with exposure to environmental risk factors, contributes to ESCC.6 However, the exact mechanism for the carcinogenesis of ESCC is still unclear.
Up to now, three ESCC GWAS6, 7, 8 and relevant pooling analyses9, 10, 11 have identified a number of susceptibility loci associated with ESCC risk in Chinese populations. However, these loci only explain a small fraction of the heritability of ESCC and the other genetic variants associated with ESCC risk need to be further explored. Of interest, the GWAS have shown that genetic variants located at chromosome 9p21.3, consisting of MTAP, CDKN2A, CDKN2B, and CDKN2B‐AS1 genes, are related to multiple cancer risk, including glioma,12, 13, 14 melanoma,15, 16 basal cell carcinoma,17 nasopharyngeal carcinoma,18 breast cancer,19, 20 chronic lymphocytic leukemia,21 and childhood acute lymphoblastic leukemia.22 This evidence collectively suggests a pleiotropic effect of the 9p21.3 locus on cancer development.
Recently, Song et al.23 identified eight significantly mutated genes in ESCC, including CDKN2A located at the 9p21.3 region, by whole‐genome sequencing in 17 ESCC cases and whole‐exome sequencing in 71 ESCC cases. Subsequently, Gao et al.24 confirmed that genes involved in cell cycle and apoptosis regulation, including CDKN2A, were mutated in 99% of ESCC cases by sequencing on 113 tumor–normal pairs. These findings highlight the important role of genetic alterations at the 9p21.3 region in the development of ESCC and suggest that 9p21.3 may be a crucial genomic region for the regulation of ESCC development.
In the current study, we undertook a case–control study including 2139 ESCC cases and 2273 controls in a Chinese population, to investigate the role of genetic variants at 9p21.3 in ESCC susceptibility. We also evaluated the mRNA expression levels of MTAP, CDKN2A, CDKN2B, and CDKN2B‐AS1 in paired ESCC tumor and adjacent normal tissues to further explore the genotype–phenotype correlation.
Materials and Methods
Study subjects
Esophageal squamous cell carcinoma cases were consecutively recruited from the Cancer Hospital of Jiangsu Province (Nanjing, China) and The Affiliated Huaian First People's Hospital of Nanjing Medical University (Huaian, China). All of the cases were histopathologically confirmed and the cases were excluded if they had a cancer history. A total of 2139 incident ESCC cases were included in this study. After being frequency‐matched to the cases on age (5‐year interval) and gender, 2273 controls were selected from cancer‐free participants in a community‐based screening program for non‐infectious diseases in Jiangsu province. All of the subjects were self‐reported ethnic Han Chinese. After signing informed consent, each subject was interviewed face‐to‐face with a standard questionnaire including information about demographic information and relevant risk factors, such as tobacco smoking and alcohol consumption. Each participant donated a ~5‐mL venous blood sample. The characteristics of cases and controls are shown in Table S1. The cases had higher proportions of smokers and drinkers as compared with controls. This study was approved by the institutional review board of Nanjing Medical University (Nanjing, China).
Polymorphism selection strategy and genotyping assays
Genetic variants were selected mainly from the published GWAS that reported significant association between genetic variants located at chromosome 9p21.3 and the risk of various types of malignant tumor, including glioma (rs4977756; rs1412829),12, 13, 14 melanoma (rs7023329),15, 16 basal cell carcinoma (rs2151280),17 nasopharyngeal carcinoma (rs1412829),18 breast cancer (rs1011970),19, 20 chronic lymphocytic leukemia (rs1679013),21 and childhood acute lymphoblastic leukemia (rs3731217).22 In addition, a recent study explored the associations between common genetic variants in this region and multiple tumors, and confirmed that rs3731239 was significantly associated with ESCC in Chinese patients.25 We filtered the above reported variants according to the following items: (i) only one variant was selected as multiple one were in LD at r 2 > 0.2 (1000 Genomes Project data of Chinese population, http://browser.1000genomes.org/index.html); (ii) those variants with a minor allele frequency <5% in the Chinese population were excluded. Finally, we included rs7023329, rs3731217, rs3731239, rs1011970, and rs1679013 as tag SNPs to be genotyped in this study.
Genotyping was carried out using the TaqMan allelic discrimination assay on the ABI 7900 system (Applied Biosystems, Foster City, CA, USA). Two negative controls were included in each 384‐well plate for quality control. Detailed information regarding the primers and probes is shown in Table S2. Technicians were blinded to the status of subjects (case or control) when carrying out the genotyping. The genotypes were called using the SDS 2.3 Allelic Discrimination Software (Applied Biosystems, Foster City, CA, USA).
RNA extraction, reverse transcription, and quantitative real‐time PCR assay
Total RNA was extracted from ESCC tumor and adjacent normal tissues using TRIzol LS Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. The concentration of the RNA was measured by NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA) and the absorbance values at 260 and 280 nm were acquired. All of the samples were of good quality, and 500 ng RNA was reverse transcribed to cDNA by using the PrimeScript RT Master Mix (TaKaRa, Kyoto, Japan) in accordance with the manufacturer's instructions. MTAP, CDKN2A, and CDKN2B mRNA expression levels were tested using the Hot Taq PCR Reaction Mix (BioSteed BioTechnologies, Nanjing, China), while quantitation of CDKN2B‐AS1 transcripts was completed using SYBR Green labeling (TaKaRa, Kyoto, Japan) as implemented in the ABI 7900 real‐time PCR System (Applied Biosystems, Foster City, CA, USA). Experiments were carried out in duplicate in three independent assays. The results of mRNA expression were normalized using the threshold cycle of β‐actin (ACTB).
Statistical analysis
The chi‐square‐test was used to analyze distribution differences of the demographic characteristics, selected variables, and genotypes between cases and controls. The HWE was tested using a goodness‐of‐fit chi‐square‐test among the control subjects. Logistic regression analysis was used to test the association between SNPs and ESCC risk. The ORs and 95% CIs were calculated with adjustment for age, gender, smoking, and drinking status. The chi‐square‐based Q‐test was used to test the heterogeneity of effect sizes (ORs and 95% CIs) between subgroups.26 Linear regressing analysis was used to test the genotype–phenotype correlation. Paired two‐sample Student's t‐test was used to compare the MTAP, CDKN2A, CDKN2B, and CDKN2B‐AS1 expression in paired tumor and non‐tumor tissues. All the statistical analyses were carried out using STATA version 12.0 software (STATA, College Station, TX, USA).
Results
Genotyping call rates were >95% for all five SNPs and the observed genotype frequencies were all in agreement with HWE in the controls (Table S3). The genotype distributions of the five SNPs between the cases and the controls are summarized in Table 1. In the additive model, the G allele of rs7023329 was significantly associated with a decreased risk of ESCC with a per‐allele OR of 0.84 (95% CI, 0.77–0.91; P = 2.95 × 10−5); in the codominant model, individuals carrying either AG or GG genotype showed a significantly reduced ESCC risk (OR = 0.68 and 0.72, respectively) compared with those with AA genotype. The C allele of rs1679013 was associated with an increased risk of ESCC with a per‐allele OR of 1.12 (95% CI, 1.01–1.24; P = 0.039) in the additive model, and in the codominant model, individuals carrying CC genotype showed an increased ESCC risk (OR = 1.44, 95% CI, 1.06–1.96; P = 0.020) compared with those with TT genotype. After conditioned on each other for these two SNPs, the association results for both variants changed little; rs1679013 and rs7023329 might be independent loci for ESCC susceptibility (Table S4). Furthermore, subgroup analyses stratified by age, gender, and smoking and drinking status, were carried out for the association of two SNPs (rs7023329 and rs1679013) and ESCC risk. As shown in Table 2, there were no significant differences between subgroups (P > 0.05 for heterogeneity tests) for associations of rs7023329 and rs1679013 with ESCC risk. However, no significant associations were observed for the other three SNPs with ESCC risk (additive P = 0.581, 0.909, and 0.183 for rs3731239, rs3731217, and rs1011970, respectively) (Table 1).
Table 1.
Summary of associations between genetic variants at 9p21.3 and esophageal squamous cell carcinoma risk in 2139 cases and 2273 controls in a Chinese population
| SNP | Cases, n (%) | Controls, n (%) | OR (95% CI)a | P‐valuea |
|---|---|---|---|---|
| rs7023329 | n = 2121 | n = 2239 | ||
| AA | 706 (33.29) | 576 (25.73) | 1.00 (reference) | |
| AG | 950 (44.79) | 1129 (50.42) | 0.68 (0.59–0.79) | 1.47 × 10−7 |
| GG | 465 (21.92) | 534 (23.85) | 0.72 (0.61–0.85) | 1.12 × 10−4 |
| G allele | 0.84 (0.77–0.91) | 2.95 × 10−5 | ||
| rs3731239 | n = 2095 | n = 2235 | ||
| TT | 1572 (75.04) | 1697 (75.93) | 1.00 (reference) | |
| TC | 498 (23.77) | 503 (22.51) | 1.12 (0.97–1.29) | 0.118 |
| CC | 25 (1.19) | 35 (1.75) | 0.76 (0.45–1.18) | 0.312 |
| C allele | 0.96 (0.84–1.10) | 0.581 | ||
| rs3731217 | n = 2058 | n = 2224 | ||
| TT | 1325 (64.38) | 1421 (63.89) | 1.00 (reference) | |
| TG | 641 (31.15) | 715 (32.15) | 0.95 (0.83–1.08) | 0.423 |
| GG | 92 (4.47) | 88 (3.96) | 1.13 (0.83–1.53) | 0.432 |
| G allele | 0.99 (0.89–1.11) | 0.909 | ||
| rs1011970 | n = 2088 | n = 2227 | ||
| GG | 1745 (83.57) | 1835 (82.40) | 1.00 (reference) | |
| GT | 331 (15.85) | 381 (17.11) | 0.88 (0.75–1.03) | 0.112 |
| TT | 12 (0.57) | 11 (0.49) | 1.20 (0.52–2.75) | 0.667 |
| T allele | 0.90 (0.77–1.05) | 0.183 | ||
| rs1679013 | n = 2130 | n = 2182 | ||
| TT | 1247 (58.54) | 1366 (62.60) | 1.00 (reference) | |
| TC | 778 (36.53) | 739 (33.87) | 1.07 (0.94–1.21) | 0.322 |
| CC | 105 (4.93) | 77 (3.53) | 1.44 (1.06–1.96) | 0.020 |
| C allele | 1.12 (1.01–1.24) | 0.039 |
Derived from logistic regression with an adjustment for age, sex, and smoking and drinking status. CI, confidence interval; OR, odds ratio.
Table 2.
Stratified analyses of association between rs7023329 and rs1679013 and esophageal squamous cell carcinoma risk in 2139 cases and 2273 controls in a Chinese population
| Variables | rs7023329 (AA/AG/GG) | rs1679013 (TT/TC/CC) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | OR (95% CI) | P‐value† | P‐value‡ | Cases | Controls | OR (95% CI) | P‐value† | P‐value‡ | |
| Age, years | ||||||||||
| <60 | 302/426/193 | 256/501/240 | 0.83 (0.73–0.94) | 3.61 × 10−3 | 0.782 | 560/324/43 | 638/313/20 | 1.29 (1.09–1.52) | 2.88 × 10−3 | 0.146 |
| ≥60 | 404/524/272 | 320/628/294 | 0.85 (0.76–0.95) | 4.10 × 10−3 | 687/454/62 | 728/426/57 | 1.10 (0.96–1.26) | 0.173 | ||
| Gender | ||||||||||
| Female | 199/243/123 | 147/299/127 | 0.82 (0.70–0.97) | 0.019 | 0.806 | 335/206/26 | 333/201/22 | 1.03 (0.84–1.26) | 0.772 | 0.161 |
| Male | 507/707/342 | 429/830/407 | 0.84 (0.76–0.93) | 4.83 × 10−4 | 912/572/79 | 1,033/538/55 | 1.22 (1.08–1.38) | 1.81 × 10−3 | ||
| Smoking | ||||||||||
| Never | 325/440/211 | 283/560/262 | 0.83 (0.73–0.94) | 2.28 × 10−3 | 0.892 | 581/353/43 | 666/373/39 | 1.09 (0.94–1.27) | 0.248 | 0.232 |
| Ever | 381/510/254 | 293/569/272 | 0.84 (0.75–0.95) | 4.10 × 10−3 | 666/425/62 | 700/366/38 | 1.24 (1.07–1.44) | 4.10 × 10−3 | ||
| Drinking | ||||||||||
| Never | 366/497/237 | 372/706/356 | 0.82 (0.73–0.91) | 3.85 × 10−4 | 0.590 | 644/413/46 | 873/478/52 | 1.12 (0.98–1.29) | 0.112 | 0.431 |
| Ever | 340/452/228 | 204/423/178 | 0.86 (0.75–0.98) | 0.020 | 602/365/59 | 493/261/25 | 1.22 (1.04–1.44) | 0.016 | ||
†Derived from additive model using logistic regression analyses with an adjustment for age, sex, smoking, and drinking status. ‡P‐values were from heterogeneity test based on chi‐square‐based Q test. CI, confidence interval; OR, odds ratio.
Around the selected variants at 9p21.3 region, there are four important genes: MTAP, CDKN2A, CDKN2B, and CDKN2B‐AS1 (Fig. 1). We next sought to determine whether rs7023329 and rs1679013 genotypes were associated with expression of these genes by quantitative RT‐PCR analysis of ESCC tumors and adjacent normal tissues (Figs 2,S1). We observed that the protective allele rs7023329‐G was related to a high expression of MTAP (P = 0.020), and the carriers of the risk allele rs1679013‐C had significantly lower expressions of CDKN2B than did non‐carriers (P = 0.035) among adjacent normal tissues (Fig. 2a,b). We found that CDKN2B was also significantly downregulated in ESCC tumor tissues compared with adjacent normal tissues (P = 3.50 × 10−5; Fig. 2c) while CDKN2B‐AS1 was significantly upregulated (P = 3.34 × 10−3; Fig. 2d). These findings indicate that genetic variants at 9p21.3 may modulate the expression of MTAP and CDKN2B and contribute to ESCC susceptibility.
Figure 1.
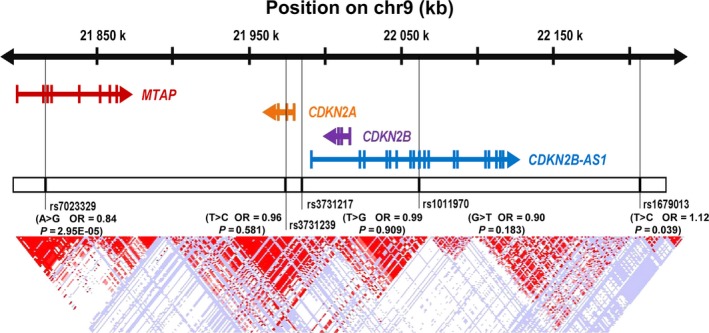
Diagram showing the 9p21.3 locus containing the MTAP,CDKN2A,CDKN2B, and CDKN2B‐AS1 genes and the five selected genetic variants. chr, chromosome; OR, odds ratio.
Figure 2.
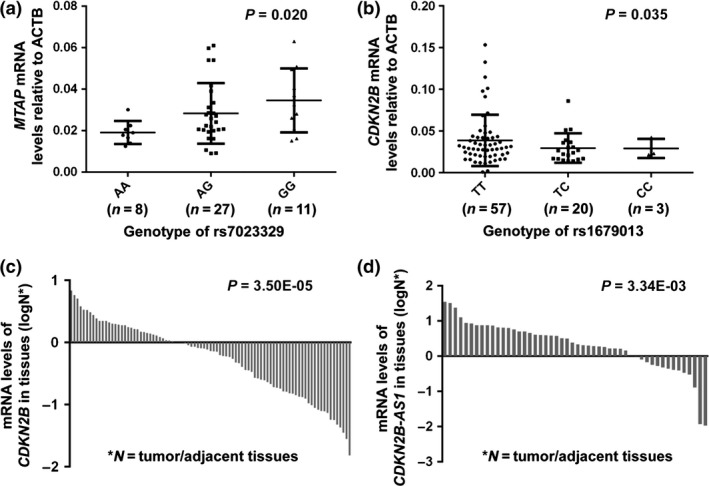
Genotype‐phenotype correlation and differential expression analysis between esophageal squamous cell carcinoma (ESCC) tumor and adjacent normal tissues. (a, b) The relationship between genotype and expression for rs7023329 and MTAP (a), and rs1679013 and CDKN2B (b) in ESCC adjacent normal tissues. (c, d) Differential expressions of CDKN2B (c) and CDKN2B‐AS1 (d) in paired ESCC tumor and adjacent normal tissues. ACTB, β‐actin.
Discussion
In this study, we investigated the associations of genetic variants at 9p21.3 with the risk of ESCC in a Chinese case–control study with 2139 cases and 2273 controls. We found that the G allele of rs7023329 was significantly associated with a reduced risk of ESCC, whereas the C allele of rs1679013 was associated with an increased risk of ESCC. Further genotype–phenotype analysis confirmed that the protective allele rs7023329‐G was related to a high expression of MTAP, which might increase the role of tumor suppressor MTAP. We also found that the carriers of the risk allele rs1679013‐C had lower expression of CDKN2B than did non‐carriers. CDKN2B, known as a tumor suppressor, was significantly downregulated in ESCC tumor tissues compared with adjacent normal tissues, which indicated that genetic variants at 9p21.3 may also be implicated with ESCC susceptibility. These findings may further improve our understanding of 9p21.3 locus in ESCC development.
The chromosome 9p21.3 region has been identified as a genetic susceptibility locus for a number of disease phenotypes, including heart disease, stroke, diabetes, and especially cancers.27, 28, 29 Recently, several GWAS have confirmed that variants in this region are related to multiple cancer risks, including glioma,12, 13, 14 melanoma,15, 16 basal cell carcinoma,17 nasopharyngeal carcinoma,18 breast cancer,19, 20 chronic lymphocytic leukemia,21 and childhood acute lymphoblastic leukemia.22 These associations suggest that multiple functional genetic variants may exist in this region and the genetic variants at 9p21.3 may also be pleiotropic.
Two studies have reported that genetic variants in 9p21.3 (including rs3731239, rs1063192, rs2157719, rs615552, rs573687, rs4977756, and rs564398) were associated with ESCC in a Chinese population.25, 29 These variants mainly represent three independent signals according to the LD pattern in the 1000 Genomes Project data of Chinese population: signal 1, rs1063192 and rs4977756 (r 2 = 0.91); signal 2, rs2157719, rs615552, rs573687, and rs564398 (r 2 = 1.00); and signal 3, rs3731239. However, our findings with a larger sample size did not find a significant association between rs3731239 (signal 3) and ESCC risk. Instead, we found that rs7023329 and rs1679013 were independently associated with ESCC risk. These two variants were also not in LD with previous reported variants (r 2 < 0.1). However, large well‐designed studies are warranted to further confirm our findings.
Recent studies showed that the 9p21.3 region was enriched in regulatory sequences such as enhancers that regulate the expression of genes in this region and downstream, thereby establishing a functional link between 9p21 genetic variation and immune signaling pathways.30 Our results confirmed that the carriers of the protective allele rs7023329‐G had significantly higher expressions of MTAP than did non‐carriers among adjacent normal tissues. It has been reported that loss of MTAP expression can exert a tumor‐promoting effect in multiple cancers and MTAP may function as a tumor suppressor gene.31, 32, 33 Therefore, the rs7023329‐G allele was related to high expression of MTAP, which might increase the role of tumor suppressor MTAP and ultimately result in a reduced ESCC risk. Moreover, the ESCC‐risk allele rs1679013‐C was associated with reduced CDKN2B expression in our study. Analysis of the Encyclopedia of DNA Elements data, as implemented in the online tool HaploReg (http://broadinstitute.org/mammals/haploreg/haploreg.php), indicated that rs1679013 was located in a transcription regulatory region and may influence the chromatin structure and histone modifications by altering the transcription factor binding sites of OTX2, which may serve as a regulatory variant in the expression of CDKN2B, suggesting that modulation of CDKN2B expression may mediate ESCC susceptibility. CDKN2B encodes the p15INK4B protein, which was subject to frequent inactivation in human primary ESCC.34 Moreover, CDKN2B has previously been linked to cellular senescence and shown to act as a tumor suppressor protein.35, 36 Our data showed that CDKN2B was significantly downregulated in ESCC tumor tissues compared with adjacent normal tissues, suggesting that CDKN2B underexpression may also mediate susceptibility to ESCC in Chinese patients. These data suggest that genetic variants in the MTAP/CDKN2A/2B/2B‐AS1 cluster may modulate ESCC susceptibility through regulating expression levels of genes in the cluster. Although we evaluated the possible mechanism by genotype–phenotype analysis and bioinformatics annotations, it is not enough to illustrate the exact biological function between these variants and ESCC. Further functional evaluations are warranted to further clarify these findings.
In summary, in a relatively large case–control study in a Chinese population, we found two independent variants in 9p21.3 (rs7023329 and rs1679013) associated with ESCC risk and provided additional evidence for the role of 9p21.3 locus in ESCC development. Further work is needed to validate our findings in diverse ethnic populations and to clarify the functional variants at 9p21.3 contributing to the development of ESCC.
Disclosure Statement
The authors have no conflict of interest.
Abbreviations
- CI
confidence interval
- ESCC
esophageal squamous cell carcinoma
- GWAS
genome‐wide association studies
- HWE
Hardy–Weinberg equilibrium
- LD
linkage disequilibrium
- MTAP
methylthioadenosine phosphorylase
- OR
odds ratio
Supporting information
Fig. S1. Genotype–phenotype correlation and differential expression analysis between esophageal squamous cell carcinoma tumor and adjacent normal tissues.
Table S1. Comparison of characteristics between esophageal squamous cell carcinoma cases and controls.
Table S2. Primers and probes used for genotyping.
Table S3. Selected SNP information and genotyping results.
Table S4. Multivariate regression analysis of rs7023329 and rs1679013 with esophageal squamous cell carcinoma risk.
Acknowledgments
This work was supported by grants from: the National Basic Research Program (973) (2013CB910304); the National Key Research and Development Program of China (2016YFC1302703); the National Natural Science Foundation of China (81422042, 81373090, 81230067, 81572421); the Science Foundation for Distinguished Young Scholars in Jiangsu (BK20130042); the Key Grant of Natural Science Foundation of Jiangsu Higher Education Institutions (15KJA330002); the Key Program of Science and Technology Development Foundation of Nanjing Medical University (2015NJMUZD019); the Top‐notch Academic Programs Project of Jiangsu Higher Education Institution (PPZY2015A067); and the Priority Academic Program for the Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine).
Cancer Sci 108 (2017) 250–255
Funding Information
National Basic Research Program (973) (2013CB910304); National Key Research and Development Program of China (2016YFC1302703); National Natural Science Foundation of China (81422042, 81373090, 81230067, 81572421); Science Foundation for Distinguished Young Scholars in Jiangsu (BK20130042); Natural Science Foundation of Jiangsu Higher Education Institutions (15KJA330002); Key Program of Science and Technology Development Foundation of Nanjing Medical University (2015NJMUZD019); Top‐notch Academic Programs Project of Jiangsu Higher Education Institutions (PPZY2015A067); Priority Academic Program for the Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine).
Contributor Information
Guangfu Jin, Email: guangfujin@njmu.edu.cn.
Lin Xu, Email: xulin83@vip.sina.com.
Hongbing Shen, Email: hbshen@njmu.edu.cn.
References
- 1. Chen W, Zheng R, Baade PD et al Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66: 115–32. [DOI] [PubMed] [Google Scholar]
- 2. Wang AH, Liu Y, Wang B, He YX, Fang YX, Yan YP. Epidemiological studies of esophageal cancer in the era of genome‐wide association studies. World J Gastrointest Pathophysiol 2014; 5: 335–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hongo M, Nagasaki Y, Shoji T. Epidemiology of esophageal cancer: orient to occident. Effects of chronology, geography and ethnicity. J Gastroenterol Hepatol 2009; 24: 729–35. [DOI] [PubMed] [Google Scholar]
- 4. Cheung WY, Liu G. Genetic variations in esophageal cancer risk and prognosis. Gastroenterol Clin North Am 2009; 38: 75–91, viii. [DOI] [PubMed] [Google Scholar]
- 5. Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut 2015; 64: 381–7. [DOI] [PubMed] [Google Scholar]
- 6. Wang LD, Zhou FY, Li XM et al Genome‐wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet 2010; 42: 759–63. [DOI] [PubMed] [Google Scholar]
- 7. Abnet CC, Freedman ND, Hu N et al A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet 2010; 42: 764–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu C, Hu Z, He Z et al Genome‐wide association study identifies three new susceptibility loci for esophageal squamous‐cell carcinoma in Chinese populations. Nat Genet 2011; 43: 679–84. [DOI] [PubMed] [Google Scholar]
- 9. Abnet CC, Wang Z, Song X et al Genotypic variants at 2q33 and risk of esophageal squamous cell carcinoma in China: a meta‐analysis of genome‐wide association studies. Hum Mol Genet 2012; 21: 2132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu C, Kraft P, Zhai K et al Genome‐wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene‐environment interactions. Nat Genet 2012; 44: 1090–7. [DOI] [PubMed] [Google Scholar]
- 11. Wu C, Wang Z, Song X et al Joint analysis of three genome‐wide association studies of esophageal squamous cell carcinoma in Chinese populations. Nat Genet 2014; 46: 1001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shete S, Hosking FJ, Robertson LB et al Genome‐wide association study identifies five susceptibility loci for glioma. Nat Genet 2009; 41: 899–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wrensch M, Jenkins RB, Chang JS et al Variants in the CDKN2B and RTEL1 regions are associated with high‐grade glioma susceptibility. Nat Genet 2009; 41: 905–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rajaraman P, Melin BS, Wang Z et al Genome‐wide association study of glioma and meta‐analysis. Hum Genet 2012; 131: 1877–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bishop DT, Demenais F, Iles MM et al Genome‐wide association study identifies three loci associated with melanoma risk. Nat Genet 2009; 41: 920–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barrett JH, Iles MM, Harland M et al Genome‐wide association study identifies three new melanoma susceptibility loci. Nat Genet 2011; 43: 1108–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stacey SN, Sulem P, Masson G et al New common variants affecting susceptibility to basal cell carcinoma. Nat Genet 2009; 41: 909–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bei JX, Li Y, Jia WH et al A genome‐wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat Genet 2010; 42: 599–603. [DOI] [PubMed] [Google Scholar]
- 19. Turnbull C, Ahmed S, Morrison J et al Genome‐wide association study identifies five new breast cancer susceptibility loci. Nat Genet 2010; 42: 504–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Michailidou K, Hall P, Gonzalez‐Neira A et al Large‐scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet 2013; 45: 353–61, 61e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Berndt SI, Skibola CF, Joseph V et al Genome‐wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat Genet 2013; 45: 868–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sherborne AL, Hosking FJ, Prasad RB et al Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat Genet 2010; 42: 492–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Song Y, Li L, Ou Y et al Identification of genomic alterations in oesophageal squamous cell cancer. Nature 2014; 509: 91–5. [DOI] [PubMed] [Google Scholar]
- 24. Gao YB, Chen ZL, Li JG et al Genetic landscape of esophageal squamous cell carcinoma. Nat Genet 2014; 46: 1097–102. [DOI] [PubMed] [Google Scholar]
- 25. Gu F, Pfeiffer RM, Bhattacharjee S et al Common genetic variants in the 9p21 region and their associations with multiple tumours. Br J Cancer 2013; 108: 1378–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta‐analysis. Stat Med 2002; 21: 1539–58. [DOI] [PubMed] [Google Scholar]
- 27. Wellcome Trust Case Control C . Genome‐wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007; 447: 661–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Helgadottir A, Thorleifsson G, Manolescu A et al A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 2007; 316: 1491–3. [DOI] [PubMed] [Google Scholar]
- 29. Li WQ, Pfeiffer RM, Hyland PL et al Genetic polymorphisms in the 9p21 region associated with risk of multiple cancers. Carcinogenesis 2014; 35: 2698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Harismendy O, Notani D, Song X et al 9p21 DNA variants associated with coronary artery disease impair interferon‐gamma signalling response. Nature 2011; 470: 264–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kryukov GV, Wilson FH, Ruth JR et al MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science 2016; 351: 1214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang HY, Li SH, Yu SC et al Homozygous deletion of MTAP gene as a poor prognosticator in gastrointestinal stromal tumors. Clin Cancer Res 2009; 15: 6963–72. [DOI] [PubMed] [Google Scholar]
- 33. Christopher SA, Diegelman P, Porter CW, Kruger WD. Methylthioadenosine phosphorylase, a gene frequently codeleted with p16(cdkN2a/ARF), acts as a tumor suppressor in a breast cancer cell line. Cancer Res 2002; 62: 6639–44. [PubMed] [Google Scholar]
- 34. Xing EP, Nie Y, Wang LD, Yang GY, Yang CS. Aberrant methylation of p16INK4a and deletion of p15INK4b are frequent events in human esophageal cancer in Linxian, China. Carcinogenesis 1999; 20: 77–84. [DOI] [PubMed] [Google Scholar]
- 35. Erickson S, Sangfelt O, Heyman M, Castro J, Einhorn S, Grander D. Involvement of the Ink4 proteins p16 and p15 in T‐lymphocyte senescence. Oncogene 1998; 17: 595–602. [DOI] [PubMed] [Google Scholar]
- 36. Fuxe J, Akusjarvi G, Goike HM, Roos G, Collins VP, Pettersson RF. Adenovirus‐mediated overexpression of p15INK4B inhibits human glioma cell growth, induces replicative senescence, and inhibits telomerase activity similarly to p16INK4A. Cell Growth Differ 2000; 11: 373–84. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Genotype–phenotype correlation and differential expression analysis between esophageal squamous cell carcinoma tumor and adjacent normal tissues.
Table S1. Comparison of characteristics between esophageal squamous cell carcinoma cases and controls.
Table S2. Primers and probes used for genotyping.
Table S3. Selected SNP information and genotyping results.
Table S4. Multivariate regression analysis of rs7023329 and rs1679013 with esophageal squamous cell carcinoma risk.


