Abstract
MicroRNAs (miRNAs) are pervasively expressed and regulate most biological functions. They function by modulating transcriptional and translational programs and therefore they orchestrate both physiological and pathological processes, such as development, cell differentiation, proliferation, apoptosis and tumor growth. miRNAs work as small guide molecules in RNA silencing, by negatively regulating the expression of several genes both at mRNA and protein level, by degrading their mRNA target and/or by silencing translation. One of the most recent advances in the field is the comprehension of their role in oncogenesis. The number of miRNA genes is increasing and an alteration in the level of miRNAs is involved in the initiation, progression and metastases formation of several tumors. Some tumor types show a distinct miRNA signature that distinguishes them from normal tissues and from other cancer types. Genetic and biochemical evidence supports the essential role of miRNAs in tumor development. Although the abnormal expression of miRNAs in cancer cells is a widely accepted phenomenon, the cause of this dysregulation is still unknown. Here, we discuss the biogenesis of miRNAs, focusing on the mechanisms by which they regulate protein synthesis. In addition we debate on their role in cancer, highlighting their potential to become therapeutic targets.
Keywords: MicroRNA, Translation, Cancer, OncomiR, Tumor suppressor
Core tip: MicroRNAs (miRNAs) are short non-coding RNAs (19-25 bp in length) which negatively regulate gene expression at the mRNA and protein level. By binding coding transcripts, miRNAs cause degradation or translation inhibition of their target genes and affect a multitude of biological processes, such as proliferation and tumor growth. In this review we critically analyze the mechanism of action of miRNAs and their potential role in cancer, opening a window on future perspectives for their use as novel therapeutic targets.
INTRODUCTION
Recent advances in transcriptome analysis and high-throughput technologies highlighted an impressive complexity in the RNA world. The most studied RNA regions are protein-coding genes, mRNAs, accounting for around 1.5% of the human genome[1]. The importance of coding mRNAs is undisputable as they have been for years the building brick of experimental biology, culminating in the systematic deletion of coding genes in several species. Not less important are retrotransposons, specific genetic elements which are known to regulate gene expression[2,3]. Since RNA was identified as the crux of genetic regulation, the idea that it carries fundamental information has been extended to novel classes of RNA. More recently, the non-protein coding portion of the genome gained attention due to its unexpected role in regulating development and disease[4]. Nowadays, most scientists agree in stating that transcription of the human genome is pervasive, therefore raising questions on the function of many uncharacterized RNAs.
The discovery of non-coding RNAs (ncRNAs) has changed the way we look at the human genome and led the scientific world to characterize the different types of ncRNAs transcribed in human cells. Although there is not a clear delineation of ncRNA classes, they are usually classified, according to their nucleotides length, in three main groups: Short ncRNAs, mid-size ncRNAs and long ncRNAs[5]. Among short ncRNAs we can distinguish between microRNAs (miRNA) and piwi-interacting RNAs (piRNAs), respectively 19-25 base pairs (bp) and 26-31 bp long. miRNAs are involved in the regulation of gene expression at the translational and stability level[6-8], while piRNAs are involved in DNA methylation and transposon repression[9-11]. Small nucleolar RNAs (60-300 bp) are part of mid-size RNAs and act as guides for rRNA modifications[12], Promoter Associated RNAs (22-200 bp) belong to the same group but their function is obscure[13]. Last but not least, long non-coding RNAs (lncRNAs) comprise all ncRNAs longer than 200 nucleotides and include the largest portion of the non-coding transcriptome[4]. lncRNAs are involved in several biological and pathological processes, such as genomic imprinting, telomere regulation, X-chromosome inactivation, development, stem cell pluripotency, immune regulation, cancer progression and in metastatic potential[14,15]. In particular, a subset of lncRNAs, the T-UCR, themselves target specific miRNAs. The binding between these lncRNAs and miRNAs prevents target transcription degradation determining an intricate co-regulation between lncRNAs and miRNAs[16-18] and strictly linking these two different types of ncRNAs. It should be however stressed out that the definition of ncRNA relies mainly on bioinformatic tools that are likely to be challenged in the next future. In particular, open reading frames (ORF) shorter than 100 nucleotides and/or lacking a strong ATG consensus sequence for translational start are considered noncoding. In view of the emergence of alternative translational start sites[19], we may discover that at least some ncRNAs are indeed “coding” for small peptides.
The relevance of the non-coding transcriptome in the comprehension of human diseases is highlighted by the impressive number of ncRNAs that are abnormally expressed in cancer, in neurological and heart diseases or in immune disorders. In this context, short RNAs have attracted the attention of most researchers. Here, we focus on miRNA and on their role in translation regulation and cancer. In particular we zoom in the known mechanisms of miRNA-regulated translation, after a brief elucidation of their discovery and biogenesis. Finally, we account for the aberrant expression of miRNAs in cancer and for their therapeutical potential as new drugs.
miRNA: DISCOVERY AND BIOGENESIS
A brief history
miRNAs are endogenous, non-coding single stranded RNAs of approximately 19-25 nucleotides in length, found both in animals and plants and involved in post transcriptional regulation[7,20]. Two decades ago the existence of miRNAs was obscure and the scientific community was focused largely on protein-coding genes.
However in 1993 the discovery of the first small ncRNA lin-4, in C. elegans, has totally changed the scientists’ point of view[21]. At the time of the first discoveries, two main questions were raised: (1) what is the role of lin-4; and (2) what is its mechanism of action? Genetic studies showed that lin-4 is one of the most relevant genes involved in the control of temporal development of larval stages[22,23]. Almost simultaneously, Lee and collaborators discovered that null mutations of the lin-14 gene were able to cause an opposite phenotype to null lin-4 mutations, suggesting that lin-4 could regulate lin-14[23,24]. How was this regulation taking place? Several groups unequivocally demonstrated that the introduction of mutations in the putative ORF of the lin-4 gene, did not affect its function, concluding that lin-4 did not encode for a protein. Mature lin-4 was found to be present in two small transcripts with different lengths, 22 and 61 nucleotides[24]. Furthermore, mutations in the 3’UTR of lin-14 mRNA and gene fusion experiments showed that lin-14 was downregulated posttranscriptionally by lin-4, delineating the 3’UTR of lin-14 as necessary for the regulation of LIN-4 protein levels[25,26]. These data led to a unified conclusion: lin-4 transcripts were complementary to the 3’UTR of the lin-14 gene and regulated its expression by annealing to its 3’UTR. With a similar approach, seven years later another miRNA was discovered, let-7, which was able to regulate lin-41 expression by binding to its 3’UTR[27,28]. Further, the sequence of let-7 was found conserved among species, from flies to humans. A new era in transcriptomics was now open for study by the entire scientific world!
miRNA biogenesis and function
miRNA biogenesis occurs in two main steps that take place in the nucleus and in the cytoplasm. miRNA genes are transcribed by RNA polymerase II and processed through both a canonical and a non-canonical biogenesis pathway. During canonical biogenesis primary miRNAs (pri-miRNAs) are processed into the nucleus by the RNase III Drosha generating an approximately 70 nucleotide-long precursor miRNA (pre-miRNA) product. In the non-canonical pathway pre-miRNAs are instead generated by the mRNA splicing machinery, avoiding Drosha digestion[29]. The subsequent steps are identical in both the canonical and non-canonical pathways. Pre-miRNAs are recognized by the Ran-GTP dependent transporter Exportin 5, which mediates their translocation to the cytoplasm. Here, Dicer, an other RNase type III enzyme, cleaves the pre-miRNA hairpins and the mature miRNAs generated by this mechanism are loaded into miRISC (miRNA associated RNA induced silencing complex), where, with the help of Argonaute proteins, they act as post-transcriptional regulators[30]. It is clear that, due to its complexity, the system of miRNA biogenesis requires a tight control. Transcriptional regulation remains the preferential process of miRNA expression control[31]. Knockout of Drosha causes the entire ablation of canonical miRNA production, suggesting its essential role in miRNA biogenesis[32]. DGCR8 is able to stabilize the Drosha complex by binding to Drosha itself. Drosha reduces DGCR8 expression[33,34]. It has also been shown that high levels of DGCR8 compromise Drosha activity[35]. Thus, complex networks may regulate Drosha complex activity. Dicer-deleted cells, instead, show some detectable canonical miRNAs, even if at reduced levels. What is more, Dicer is destabilized by low expression of TRBP. These data reveal the important, but not essential contribution of Dicer in the miRNA biogenesis pathway[32,36,37].
miRNA biogenesis is characterized by a physical separation between Drosha (nucleus) and Dicer (cytoplasm). Nervertheless, several mature miRNAs are located in the nucleus, like miR-29[38,39], or in the mithocondria, such as miR-1 and miR-181[40-42], or in small vesicles, suggesting non-canonical roles for miRNAs. Particularly, several studies reveal that miRNAs are transported into the nucleus, where they regulate the maturation of other miRNAs, by targeting their primary transcript, or control their own expression. Here, they can also bind long ncRNAs and thus regulate their expression and maturation[43]. In conclusion, it would be important for miRNA characterization to explore potential roles in non-canonical functions.
The function of miRNAs was first defined 20 years ago. A mature miRNA loaded into the RISC, is capable to bind and regulate the expression of target mRNA via base-pairing. In particular, miRNAs bind the 3’UTR of target mRNAs through a sequence of 2-8 nucleotides in their 5’end, termed seed region (Figure 1A). The partial or perfect complementarity between miRNAs and target mRNAs causes repression of translation or mRNA degradation, respectively[44] (Figure 1B). Owing to the short base pairing between a miRNA and the 3’UTR of its target mRNA, the interaction is dynamic: One miRNA can bind sequentially to hundreds of target mRNAs and a single mRNA can be targeted by several miRNAs[20]. miRNAs are able to select and interact with their targets based on I) their expression levels or II) expression levels of their mRNA targets. Since the expression and function of some miRNAs are species- and/or tissue-specific, the co-localization of a miRNA with its mRNA target is clearly necessary for its functionality[45]. Moreover, tissue-specific miRNAs can localize both in intragenic and intergenic regions, and consequently they could be under the control of host gene promoters or, alternatively, they could hold their own promoter. Hence, for the intragenic miRNAs, expression could also be dependent on the transcription of host genes[46], suggesting the latter to be able to influence miRNA function.
Figure 1.
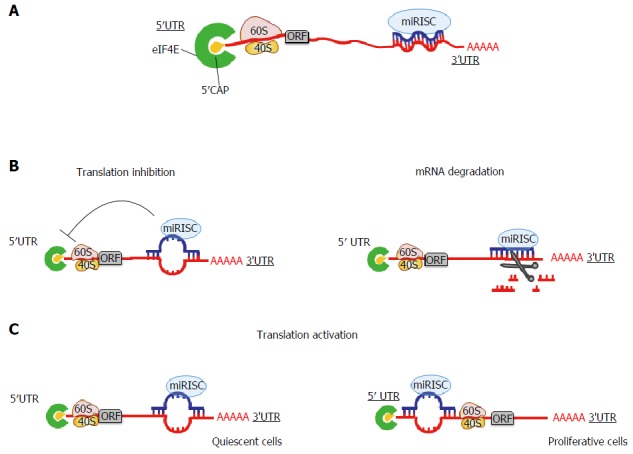
Mechanisms of action of microRNAs. A: miRNAs bind the 3’UTR of a target gene by base pairing. The binding between the miRNA seed sequence (nucleotides 2-8 at the 5’end of the miRNA sequence) and the miRNA regulatory element (MRE) at the 3’UTR of a target gene determines the specific type of regulation; B: miRNAs act as inhibitors of translation when the binding at the 3’UTR of target genes is only partially complementary. Instead, when the binding complementarity is perfect, miRNAs induce mRNA degradation; C: miRNAs can also function in an unconventional manner: Under specific conditions, particularly during quiescence, they can activate translation by binding non canonical sites in the 5’UTR of target genes. ORF: Open reading frames.
Summarizing, by selecting their targets in a dose-dependent manner, miRNAs could control the balance of specific cellular processes.
Recent reports have suggested that in addition to the classical binding of miRNAs to the 3’UTR of mRNAs, they are able to bind also the 5’UTR region and ORF[47,48]. Sites located in coding regions and in 5’UTRs appear to be less robust than those in 3’UTR and, surprisingly, determine translational activation, and not repression, of miRNA-targeted mRNAs. This situation has been described upon growth arrest conditions[49,50] (Figure 1C). However, these models are not universally accepted because ribosomes that scan the 5’UTR and the ORF are expected to remove annealed miRNAs.
Nowadays, the miRNA landscape is very tangled as the number of miRNA genes is exponentially growing[51], rendering it much more difficult to clearly define their function. miRNA genes have been clustered into different groups, known as miRNA families, based on the sequence of mature miRNAs or on the structure of pre-miRNAs. This clusterization is really relevant for studying miRNA functions, since miRNA genes belonging to the same family co-localize and take place in the same specific mechanism, e.g., immune system regulation, development or cancer[52]. Moreover, the increasing number of novel miRNAs reveals that some of them are evolutionarily conserved whilst others are species- and/or tissues-specific. The expression levels of both the newly discovered and the long-known miRNAs are different from tissue to tissue, unraveling a differential tissue- and cell specific-functional impact of miRNAs[53].
miRNAs AND TRANSLATION REGULATION
mRNA translation is a cellular process finely regulated during growth and development, and its control is essential to maintain physiological processes in the cell. Translational control plays the major role in regulation of gene expression[54] and miRNAs take part in the regulation of mRNA translation.
Since a miRNA binds the 3’UTR of a target mRNA, how can it inhibit its translation? To date it is very clear that miRNAs contribute to the regulation of protein synthesis in two ways, mRNA destabilization or translational repression. Unfortunately, to date, a general mechanism for the translational inhibition by miRNAs has not been widely accepted; we rely on several different models that will be critically presented[55]. mRNA translation is divided in four phases: Initiation, elongation, termination and recycling. Here we will review in detail how miRNAs can repress translation at the initiation, post-initiation and elongation steps (Figure 2).
Figure 2.
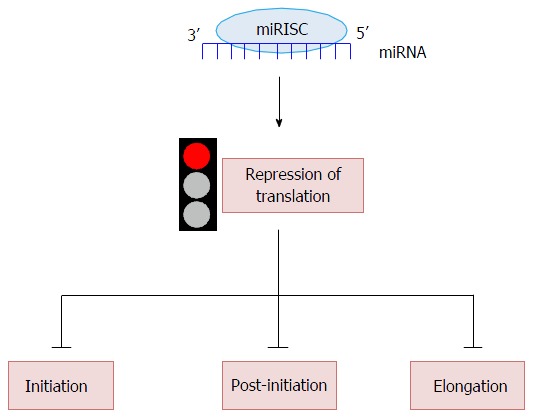
Schematic representation of microRNAs role in translation. miRNAs can inhibit protein synthesis at three different stages of translation: Initiation, the rate limiting step, post initiation and elongation. For details refer to text.
Initiation of translation
Initiation is the rate-limiting step in translation of a given mRNA and leads to the formation of the ribosome-tRNA-mRNA complex. The golden method to analyze the step at which the translation of a specific mRNA is blocked is measuring the localization of the same mRNA in a sucrose polysome gradient. The general assumption is that a translated mRNA associates with multiple ribosomes (polysomes) and co-sediments to the heavy part of the gradient. Several in vitro studies reveal that repressed mRNAs shift to the lighter region of the sedimentation gradient, indicating reduced ribosome loading of the repressed mRNAs[56]. The mechanistic effect of miRNAs at initiation is confirmed by studies in which Ago was found bound to the translational machinery. Briefly, mammalian Ago2 is able to bind the m7G-cap of mRNA directly, suggesting that Ago2 and the cap binding protein eIF4E compete for association with the cap structure[57]. In this model miRNAs prevent translation of capped but not Internal Ribosome Entry Site (IRES) containing mRNAs. The discovery of a specific Ago2 domain responsible for the interaction with the cap structure supports the above hypothesis. Mutation of two key amino acids in Ago2 disrupts cap-Ago2 binding, and abolishes also the association between Ago2 and GW182, the latter being an important factor in miRNA mediated repression[58,59].
In vitro studies suggest other mechanisms of miRNA repression at the initiation step. For some studies, the presence of the m7G-cap is necessary for translational inhibition. Other studies demonstrated that miRNA-mediated repression impairs also cap-independent, IRES-initiated translation, and exclude eIF4E-cap recognition as a target for miRNA function[60,61]. This consideration is complicated by kinetic issues, as IRES-containing mRNAs are in general less efficiently translated, but also by the fact that some mRNAs have both a m7G-cap and an IRES.
Studies performed on D. Melanogaster and mouse cells indicate that miRNAs impair the association of mRNAs to 40S or 80S ribosomes, probably disrupting the mRNA-40S complex[62]. In this study an excess of eIF4F ameliorates the miRNA mediated inhibition of a specific mRNA. Other in vitro studies confirmed that translational repression exists only in the presence of both the m7G cap and poly-A tail, indicating that polyadenylation itself may have a role in miRNA mediated repression[63-66]. Several hypotheses were proposed: (1) the CCR4-NOT deadenylation complex is sufficient to mediate silencing and could inhibit mRNA translation independently of its deadenylation activity[67,68]; (2) miRISC is able to inhibit 43S scanning by impairing eIF4F function[69,70], in particular NOT1 interaction with eIF4A2 could block eIF4A2 function and consequently 43S scanning; and (3) in contrast to hypothesis 2, a very recent study suggests that eIF4A activity and 43S ribosomal scanning are not required for miRNA silencing. In this context AGOs, GW182, CCR4-NOT and DDX6 complexes are able to repress and degrade mRNAs in a 43S scanning-independent manner[65]. It is evident that conflicting reports may be due to difficulties in the analysis of the fast translation inhibition driven by miRNAs.
In addition, in the intertwined scenario of mRNA translational inhibition and mRNA degradation driven by miRNAs, the recurrent question is which step precedes the other. Recent studies performed in human cells, zebrafish, and D. Melanogaster, show that miRNAs reduce translation just before mRNA deadenylation and decay. Kinetic analysis monitoring in parallel the level of mRNAs, proteins and poly-A tail lengths coupled with ribosome profiling data, revealed that protein levels are affected prior to mRNA stability and poly-A tail lenght[63,64,71]. A recent work presents another interesting hypothesis: miRNAs destabilize mRNAs when they are in a ribosome free state, but at the same time mRNAs targeted by miRNAs are fully polysome associated. The authors demonstrate that while mRNAs are associated with polysomes, the decapping mechanism occurs proceeding in a 5’ to 3’ direction following the last translating ribosome[66]. According to this model miRNA mediated mRNA decay occurs cotranslationally, providing a solution to this complex mistery.
Finally, it has been also suggested that miRNAs act at the level of active 80S complex formation, by affecting 60S joining. Eukaryotic Initiation Factor 6 (eIF6) associates with the 60S ribosomal subunit and is able to coimmunoprecipitate with the Ago2-Dicer-TRBP complex. Studies in human cells and C. Elegans led to the conclusion that miRISC, when associated with eIF6, abolishes polysome formation, disrupting 80S ribosomes assembly[72]. However, these results were not confirmed in D. Melanogaster cells, where depletion of eIF6 had no effect in miRNA-mediated inhibition[73]. Furthermore all published data converge on the idea that eIF6 acts as an anti-association factor that, by binding the 60S ribosomal subunit, prevents the formation of active 80S[74,75]. This mechanism is regulated by post-translational modifications. Indeed eIF6 is activated downstream of the RACK1-PKCβIIaxis[76,77] through phosphorylation on residue Ser235, an event which is found deregulated in several types of cancer[78-80].
Post-initiation and elongation of translation
At the end of the initiation step, the mRNA is positioned on the ribosome and amino acids are bound together to form a polypeptide chain, thus determining the intermediate step of translation. Since 1999, by sedimentation velocity ultracentrifugation in a sucrose gradient, it has been reported that some miRNAs fully associate with polysomes[81,82]. The copurification of miRNAs with polysomes, confirmed by many studies in the last years, not only proves that miRNAs are involved in translational repression, but suggests that miRNA targets are actively translated. Taken together, these data suggest that mRNAs could be silenced by miRNAs at the post initiation step. Some examples will clarify the situation. Most miRNAs are shown to be associated with polysomes and in particular let-7 is capable to cosediment with polyribosomes[82,83]. When translation is blocked, by either hypertonic stress or puromycin treatment, miRNAs are no longer associated with polysomes, differently from their mRNA targets. The latter in fact dissociate from polyribosomes only partially, suggesting a reduction of translational elongation or impairment of the post initiation step. The capability of miRNAs to repress translation could also depend on their strength in associating with polyribosomes, i.e., the amount of a specific miRNA in polysomes relative to its total quantity. Molotski and coworkers quantified the association of miRNAs with polysomes[83], and, in line with this study, we discovered that in a Mesothelioma cellular model, only 8% of the miRNAs analyzed is stable and enriched on polysomes (data not published), suggesting that the preference for a microRNA to bind polyribosomes might depend on: (1) the specific seed sequence of the microRNA; (2) the level of pairing energy between the miRNA and its target mRNA; and (3) the fact that target mRNAs are being translated or not. Overall what these data suggest is that the miRNA ability to repress translation could also depend on the rate of association with polysomes. However, how these small non-coding RNAs are capable to impair elongation or termination of translation is unclear. It has been also proposed that proteins synthesized from miRNA-targeted mRNAs are not able to accumulate because they are degraded by certain proteases employed by microRNA ribonucleoprotein complexes (miRNPs)[81], thus proposing another mechanism on how miRNAs might function.
miRNA “surprise” role: miRNA-dependent activation of translation
The dynamic interactions between miRNAs and mRNAs open new frontiers in the field of miRNome studies. Most miRNAs negatively regulate gene expression and led scientists to deeply characterize the binding mechanism of miRNAs seed sequence to mRNAs. It is clear that miRNAs bind 3’UTRs and repress translation of target mRNAs. The demonstration that miR-369 has the capacity to either activate or repress protein translation[84] raised the question on why and how miRNAs activate translation of their target mRNAs. When cells are grown in normal growth factor conditions, target mRNAs are translationally inhibited or decayed. Instead, in the absence of growth factors, i.e., in serum starved conditions, the same miRNAs are able to activate translation and increase the protein levels of their target mRNAs, as it happens with miR-369 and its target TNF alfa[84-86]. Nevertheless this is not true for all miRNAs. For example, when miR-16 targets TNF alfa in a different 3’UTR region from that targeted by miR-369, it inhibits translation also in quiescent conditions[86,87]. This suggests that, when a cell exits the cell cycle, activation of translation depends on miRNAs seed sequences and on miRNA-mRNA base pairing. It has also been demonstrated that the repression or activation of translation requires the FXR1 protein and Ago2, and that other miRNAs, among which let-7, respond to serum starvation upregulating translation of their target mRNAs[85,86].
New paradigms discovered more recently make the mechanism of miRNA regulation even more puzzling. In the recent years, non-canonical sites of binding have been reported. Such sites map to the 5’UTR and coding regions of mRNAs[88,89]. Several studies reported that miR-122 and miR-103a-3p have their target sites in 5’UTR[90,91], and that some miRNAs are even able to target both 3’ and 5’UTRs. Moreover, under cellular stress miR-10a activates translation by binding the 5’UTR of its target ribosomal protein coding mRNA[48]. All of these mechanisms, and probably several others yet unknown, render the landscape of miRNAs mode of action even more difficult to assess. In conclusion, it is essential to study the function of each miRNA singularly and in a specific cellular context in order to understand its precise function.
miRNAs AND CANCER
Cancer is a pathological condition in which gene expression is dramatically deregulated. miRNAs affect all steps of tumor progression including tumor growth, invasion, metastatic capability and angiogenesis. The relevance of miRNAs in cancer has been highlighted by alterations in their expression (Table 1) and consequently by the deregulation of the expression of their target mRNAs[92,93]. The first evidence of the involvement of miRNA in cancer derived from studies on chronic lymphocytic leukemia (CLL). Croce’s group discovered that two miRNAs, miR-15a and miR-16-1 derive from the same polycistronic RNA which is transcribed from a specific region of chromosome 13, frequently found deleted in CLL. Analyzing a set of CLL patients, they found that 69% of them presented the deletion of miR-15-a and miR-16-1[44]. Moreover, they realized that a significant percentage of miRNA genes localizes in fragile sites and/or in genome regions which often show chromosomal alterations, including amplifications or deletions. This last finding suggested that miRNAs are a new class of genes important in regulating cancer pathogenesis and development. These relevant and preliminary observations implemented the need for investigation with new advanced technologies. All known miRNAs are now mapped and the development of several new platforms is helpful to study the miRNome in both normal and pathological tissues and for the estabilishment of tumor classification, diagnosis and prognosis by miRNA profiling[94-96].
Table 1.
List of selected tumor suppressor microRNAs and oncomiRs in cancer
| Cancer type | |
| OncomiRs | |
| miR17-92[108] | B-cell lymphoma, small cell lung cancer, colon cancer, gastric cancer |
| miR-21[107] | Breast, colon and lung cancer, glioblastoma |
| miR-106[133] | Gastric cancer, colorectal cancer |
| miR-10b[113] | Breast cancer |
| miR-191[134,135] | Human colorectal and breast cancer |
| Tumor suppressor miRNAs | |
| let-7[105,106] | Lung cancer, Burkitt lymphoma |
| miR-15a, miR16-1[103,104] | CLL, prostate cancer, mesothelioma |
| miR-29[136] | Lung cancer, breast cancer |
| miR-34a[116] | Prostate cancer, mesothelioma, HCC |
| miR-126[114] | Lung and breast cancer |
| Both O and TS | |
| miR-24[137,138] | Breast cancer, glioma (O) |
| Laryngeal carcinoma (TS) | |
| miR-125[122,123] | Pancreatic and prostate cancer (O) |
| Melanoma, osteosarcoma, ovarian cancer (TS) | |
| miR-155[120,121] | Lymphoma, breast cancer (O) |
| Melanoma, ovarian and gastric cancer (TS) | |
| miR-221/222[139] | Glioblastoma, HCC, breast cancer (O) |
| Tongue squamous cell carcinoma (TS) |
CLL: Chronic lymphocytic leukemia; HCC: Hepatocellular carcinoma; O: OncomiR; TS: Tumor suppressor miRNA.
Just like classical protein-coding genes, also miRNA genes can be altered by promoter methylation, chromosomal amplifications, deletions and transcriptional activation. Genetic alterations may involve the miRNA machinery[97] or alter the target binding site[98], the processing of miRNAs and their post-transcriptional editing[99]. In cancer, dysregulated miRNAs can act as oncogenic miRNAs (oncomiRs) or tumor suppressor miRNAs, based on their capability to repress the expression of tumor suppressor genes or oncogenes, respectively (Figure 3). The inhibition or stimulation of oncomiRs or tumor suppressor miRNAs modulates cancer cell proliferation, tumor growth, metastasis formation and cell survival[100]. Generally, oncomiRs, which modulate tumor suppressor proteins, are overexpressed in cancer, whilst tumor suppressor miRNAs, which target oncoproteins, are downregulated or deleted. Tumor suppressors miR-15a and miR-16-1, whose target is Bcl-2[101,102], are downregulated in several cancers, such as mesothelioma[103], CLL and prostate carcinoma[104], tumor suppressor let-7 targets RAS and Myc[105,106], while oncomiR miR-21 is overexpressed in breast cancer, colon cancer and glioblastoma and targets PTEN in non-small cell lung cancer[107]. Furthermore, the most studied miRNA cluster, miR17-92 is able to induce lymphomagenesis in a B-cell specific transgenic mice[108], and miR-19, miR-20a and miR-92, which are part of this cluster, promote T cell ALL development in mouse models[109]. It has been established that knocking down the upregulated oncomiRs reduces cell proliferation and tumor growth both in vitro and in vivo tumor systems[110-112]. In addition to classical tumor suppressor or oncogene functions, miRNAs are also involved in cell migration and metastasis formation. In breast cancer miR-10b modulation increases cell invasion and migration by targeting HOXD10 and eliciting the expression of the pro-metastatic gene RHOC[113]. Other examples are miR-335 and miR-126, which act as negative regulators of metastasis and tumor invasion in lung and breast cancer[114,115]. miR-34a, instead, is lost in several tumors and is involved in the p53 pathway[116]. Moreover, miR-34a is able to inhibit migration and invasion downregulating MET expression in HCC cells[117].
Figure 3.
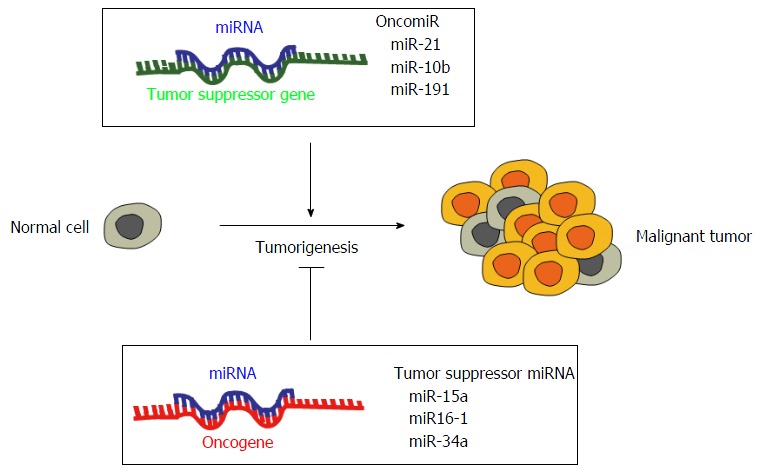
Roles of microRNAs in cancer. miRNAs suppress the expression of their target genes. An oncogenic miRNA, termed oncomiR, can repress the translation of a tumor suppressor gene, stimulating tumorigenesis and leading to tumor formation. Conversely, a tumor suppressor miRNA is able to inhibit the expression of oncogenes, blocking the tumorigenesis process and consequently the development of cancer.
Several miRNAs cannot be clearly and unequivocally categorized as tumor suppressors or oncomiRs given that the data in our hands are quite intricate and conflicting since they could act as tumor suppressors in one scenario or as oncomiRs in the other (Table 1). This is not surprising considering that the same miRNA may regulate from ten to hundreds of genes involved in completely different cellular pathways. If we consider miR-155, it works as an oncomiR in solid and hematological malignancies, such as lymphoma and breast cancer[118,119], but in melanoma, as well as in ovarian and gastric cancer, it shows a tumor suppressor role[120,121]. Another relevant example is represented by miR-125, which shows tumor suppressor properties in several cancers, like melanoma, osteosarcoma, ovarian and breast cancer, and tumor promoting functions in pancreatic and prostate cancers[122,123]. It is clear that the dual role of miRNAs could be due to the heterogeneity and variability of cancer, causing the same miRNA to carry out different effects in different tumors.
miRNAs have also an important role in the clinic where they are useful in terms of diagnosis, prognosis and prediction of therapy response. In this context miRNAs expression can be used as a tool to predict tumorigenesis and overall survival, but also to classify malignant and non-malignant tissues. To date, the clinical importance of miRNAs has been demonstrated for several types of cancers and by using also biopsies or surgery specimens[124].
To avoid the invasiveness of surgery techniques, several studies focused their attention on analyzing miRNA expression levels in human fluids, such as plasma/serum, saliva and urine, speculating the idea that circulating miRNAs could be stable and therefore useful clinical biomarkers. To support this idea, to date, miRNA deregulation in serum of cancer patients has been described for several types of tumors such as leukemia, lymphoma, gastric, lung, ovarian, prostate, pancreatic and breast cancer[125]. Most miRNAs found outside of cells, particularly in body fluids, are stable, and this is quite surprising since most RNA molecules in the extracellular environment are subjected to ribonucleases. These observations suggest that secreted miRNAs could be protected by degradation possibly by being packed in particular extracellular vesicles[126].
Among extracellular vesicles, exosomes turn out to be the most studied membrane bound vesicles released from cells into the extracellular space[127]. Exosomes play an important role in exchanging information between cancer cells, and such cell-to-cell communication is essential for tumor survival and progression and for metastases formation. Several studies identified exosomes as the key components of this process, and the idea that extracellular miRNA are among the mediators used by exosomes for this inter-cell communication makes this model even more attractive. In line with these data, several studies showed that exosomal miRNA expression is altered in cancer[128,129]. The function of exosomal miRNAs is poorly understood, but some reports showed that in this context they carry out their conventional role of negative regulators of gene expression. One example is miR-105 which, once released from breast cancer cell lines, reduces ZO-1 gene expression and promotes metastases formation in the lung and brain[130]. Recently, a novel and peculiar function of exosomal miR-21 and miR-29a was demonstrated: Such miRNAs are capable to activate immune cells, by acting as toll-like receptors ligands[131].
These observations and future progresses in the miRNA research field will be very helpful for the development of new therapeutical strategies to fight cancer. Indeed, when a cancer is characterized by the overexpression of specific miRNAs, the use of anti-miRs as drugs could help restoring the non-pathological condition. On the contrary, the same results could be obtained by the use of miRNA mimics in cancers in which specific miRNAs are downregulated. A similar approach was described by Kota et al[132]: The restoring of miR-26a in hepatocellular carcinoma is able to reduce cancer cell proliferation by triggering apoptosis. These data widely show that miRNAs have a precious potential to act as therapeutical targets.
CONCLUSION
In conclusions, the miRNA world is fascinating and mysterious. The number of miRNA genes that are being discovered is increasing and novel mechanisms of action might reveal possible new therapeutic strategies. The fact that miRNAs use non-canonical target sites to perform their function opens a puzzling scenario that could lead researchers to discover completely new miRNA functions and modes of action. Although great strides have been made in the recent years, the comprehension of the global miRNome and the establishment of functional therapeutic strategies in miRNA cancer research are yet far from being achieved. The discovery and development of miRNA inhibitors or miRNA mimics as novel drugs will offer new hopes in the fight against cancer.
Footnotes
Manuscript source: Invited manuscript
Specialty type: Biochemistry and molecular biology
Country of origin: Italy
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
Conflict-of-interest statement: Authors declare no conflict of interests for this article.
Peer-review started: August 24, 2016
First decision: October 8, 2016
Article in press: January 18, 2017
P- Reviewer: Bai G, Witzany G S- Editor: Kong JX L- Editor: A E- Editor: Li D
References
- 1.Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating non-coding regions of the genome. Nat Rev Genet. 2010;11:559–571. doi: 10.1038/nrg2814. [DOI] [PubMed] [Google Scholar]
- 2.Spadafora C. A LINE-1-encoded reverse transcriptase-dependent regulatory mechanism is active in embryogenesis and tumorigenesis. Ann N Y Acad Sci. 2015;1341:164–171. doi: 10.1111/nyas.12637. [DOI] [PubMed] [Google Scholar]
- 3.Elbarbary RA, Lucas BA, Maquat LE. Retrotransposons as regulators of gene expression. Science. 2016;351:aac7247. doi: 10.1126/science.aac7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 6.Mihailescu R. Gene expression regulation: lessons from noncoding RNAs. RNA. 2015;21:695–696. doi: 10.1261/rna.050815.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 11.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24:887–892. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deschamps-Francoeur G, Garneau D, Dupuis-Sandoval F, Roy A, Frappier M, Catala M, Couture S, Barbe-Marcoux M, Abou-Elela S, Scott MS. Identification of discrete classes of small nucleolar RNA featuring different ends and RNA binding protein dependency. Nucleic Acids Res. 2014;42:10073–10085. doi: 10.1093/nar/gku664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taft RJ, Kaplan CD, Simons C, Mattick JS. Evolution, biogenesis and function of promoter-associated RNAs. Cell Cycle. 2009;8:2332–2338. doi: 10.4161/cc.8.15.9154. [DOI] [PubMed] [Google Scholar]
- 14.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morceau F, Chateauvieux S, Gaigneaux A, Dicato M, Diederich M. Long and short non-coding RNAs as regulators of hematopoietic differentiation. Int J Mol Sci. 2013;14:14744–14770. doi: 10.3390/ijms140714744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 17.Scaruffi P, Stigliani S, Moretti S, Coco S, De Vecchi C, Valdora F, Garaventa A, Bonassi S, Tonini GP. Transcribed-Ultra Conserved Region expression is associated with outcome in high-risk neuroblastoma. BMC Cancer. 2009;9:441. doi: 10.1186/1471-2407-9-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Klerk E, ‘t Hoen PA. Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet. 2015;31:128–139. doi: 10.1016/j.tig.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 22.Horvitz HR, Sulston JE. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics. 1980;96:435–454. doi: 10.1093/genetics/96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee R, Feinbaum R, Ambros V. A short history of a short RNA. Cell. 2004;116:S89–S92, 1 p following S96. doi: 10.1016/s0092-8674(04)00035-2. [DOI] [PubMed] [Google Scholar]
- 24.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 25.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 26.Wightman B, Bürglin TR, Gatto J, Arasu P, Ruvkun G. Negative regulatory sequences in the lin-14 3’-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans development. Genes Dev. 1991;5:1813–1824. doi: 10.1101/gad.5.10.1813. [DOI] [PubMed] [Google Scholar]
- 27.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 28.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3’UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 30.Li Z, Rana TM. Molecular mechanisms of RNA-triggered gene silencing machineries. Acc Chem Res. 2012;45:1122–1131. doi: 10.1021/ar200253u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 32.Kim YK, Kim B, Kim VN. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc Natl Acad Sci USA. 2016;113:E1881–E1889. doi: 10.1073/pnas.1602532113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Triboulet R, Chang HM, Lapierre RJ, Gregory RI. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA. 2009;15:1005–1011. doi: 10.1261/rna.1591709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 36.Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 39.Roberts TC. The MicroRNA Biology of the Mammalian Nucleus. Mol Ther Nucleic Acids. 2014;3:e188. doi: 10.1038/mtna.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y, et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das S, Bedja D, Campbell N, Dunkerly B, Chenna V, Maitra A, Steenbergen C. miR-181c regulates the mitochondrial genome, bioenergetics, and propensity for heart failure in vivo. PLoS One. 2014;9:e96820. doi: 10.1371/journal.pone.0096820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X, Liang H, Zhang CY, Zen K. miRNA regulates noncoding RNA: a noncanonical function model. Trends Biochem Sci. 2012;37:457–459. doi: 10.1016/j.tibs.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Iorio MV, Croce CM. Causes and consequences of microRNA dysregulation. Cancer J. 2012;18:215–222. doi: 10.1097/PPO.0b013e318250c001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shu J, Xia Z, Li L, Liang ET, Slipek N, Shen D, Foo J, Subramanian S, Steer CJ. Dose-dependent differential mRNA target selection and regulation by let-7a-7f and miR-17-92 cluster microRNAs. RNA Biol. 2012;9:1275–1287. doi: 10.4161/rna.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lutter D, Marr C, Krumsiek J, Lang EW, Theis FJ. Intronic microRNAs support their host genes by mediating synergistic and antagonistic regulatory effects. BMC Genomics. 2010;11:224. doi: 10.1186/1471-2164-11-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moretti F, Thermann R, Hentze MW. Mechanism of translational regulation by miR-2 from sites in the 5’ untranslated region or the open reading frame. RNA. 2010;16:2493–2502. doi: 10.1261/rna.2384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 49.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 50.Vasudevan S, Tong Y, Steitz JA. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545–1549. doi: 10.4161/cc.7.11.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamanu TK, Radovanovic A, Archer JA, Bajic VB. Exploration of miRNA families for hypotheses generation. Sci Rep. 2013;3:2940. doi: 10.1038/srep02940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mathelier A, Carbone A. Large scale chromosomal mapping of human microRNA structural clusters. Nucleic Acids Res. 2013;41:4392–4408. doi: 10.1093/nar/gkt112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Londin E, Loher P, Telonis AG, Quann K, Clark P, Jing Y, Hatzimichael E, Kirino Y, Honda S, Lally M, et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc Natl Acad Sci USA. 2015;112:E1106–E1115. doi: 10.1073/pnas.1420955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 56.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 57.Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 58.Kinch LN, Grishin NV. The human Ago2 MC region does not contain an eIF4E-like mRNA cap binding motif. Biol Direct. 2009;4:2. doi: 10.1186/1745-6150-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 61.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 63.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuzuoğlu-Öztürk D, Bhandari D, Huntzinger E, Fauser M, Helms S, Izaurralde E. miRISC and the CCR4-NOT complex silence mRNA targets independently of 43S ribosomal scanning. EMBO J. 2016;35:1186–1203. doi: 10.15252/embj.201592901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tat TT, Maroney PA, Chamnongpol S, Coller J, Nilsen TW. Cotranslational microRNA mediated messenger RNA destabilization. Elife. 2016;5:pii: e12880. doi: 10.7554/eLife.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bawankar P, Loh B, Wohlbold L, Schmidt S, Izaurralde E. NOT10 and C2orf29/NOT11 form a conserved module of the CCR4-NOT complex that docks onto the NOT1 N-terminal domain. RNA Biol. 2013;10:228–244. doi: 10.4161/rna.23018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zekri L, Kuzuoğlu-Öztürk D, Izaurralde E. GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. EMBO J. 2013;32:1052–1065. doi: 10.1038/emboj.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fukao A, Mishima Y, Takizawa N, Oka S, Imataka H, Pelletier J, Sonenberg N, Thoma C, Fujiwara T. MicroRNAs trigger dissociation of eIF4AI and eIF4AII from target mRNAs in humans. Mol Cell. 2014;56:79–89. doi: 10.1016/j.molcel.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 70.Ricci EP, Limousin T, Soto-Rifo R, Rubilar PS, Decimo D, Ohlmann T. miRNA repression of translation in vitro takes place during 43S ribosomal scanning. Nucleic Acids Res. 2013;41:586–598. doi: 10.1093/nar/gks1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Béthune J, Artus-Revel CG, Filipowicz W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012;13:716–723. doi: 10.1038/embor.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 73.Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 74.Brina D, Miluzio A, Ricciardi S, Biffo S. eIF6 anti-association activity is required for ribosome biogenesis, translational control and tumor progression. Biochim Biophys Acta. 2015;1849:830–835. doi: 10.1016/j.bbagrm.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 75.Sanvito F, Piatti S, Villa A, Bossi M, Lucchini G, Marchisio PC, Biffo S. The beta4 integrin interactor p27(BBP/eIF6) is an essential nuclear matrix protein involved in 60S ribosomal subunit assembly. J Cell Biol. 1999;144:823–837. doi: 10.1083/jcb.144.5.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gallo S, Manfrini N. Working hard at the nexus between cell signaling and the ribosomal machinery: An insight into the roles of RACK1 in translational regulation. Translation (Austin) 2015;3:e1120382. doi: 10.1080/21690731.2015.1120382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhäuser N, Marchisio PC, Biffo S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- 78.Miluzio A, Beugnet A, Grosso S, Brina D, Mancino M, Campaner S, Amati B, de Marco A, Biffo S. Impairment of cytoplasmic eIF6 activity restricts lymphomagenesis and tumor progression without affecting normal growth. Cancer Cell. 2011;19:765–775. doi: 10.1016/j.ccr.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 79.Miluzio A, Beugnet A, Volta V, Biffo S. Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation. EMBO Rep. 2009;10:459–465. doi: 10.1038/embor.2009.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miluzio A, Oliveto S, Pesce E, Mutti L, Murer B, Grosso S, Ricciardi S, Brina D, Biffo S. Expression and activity of eIF6 trigger malignant pleural mesothelioma growth in vivo. Oncotarget. 2015;6:37471–37485. doi: 10.18632/oncotarget.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 82.Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol. 2006;13:1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- 83.Molotski N, Soen Y. Differential association of microRNAs with polysomes reflects distinct strengths of interactions with their mRNA targets. RNA. 2012;18:1612–1623. doi: 10.1261/rna.033142.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Buchan JR, Parker R. Molecular biology. The two faces of miRNA. Science. 2007;318:1877–1878. doi: 10.1126/science.1152623. [DOI] [PubMed] [Google Scholar]
- 85.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 86.Truesdell SS, Mortensen RD, Seo M, Schroeder JC, Lee JH, LeTonqueze O, Vasudevan S. MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci Rep. 2012;2:842. doi: 10.1038/srep00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 88.Brümmer A, Hausser J. MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. Bioessays. 2014;36:617–626. doi: 10.1002/bies.201300104. [DOI] [PubMed] [Google Scholar]
- 89.Cipolla GA. A non-canonical landscape of the microRNA system. Front Genet. 2014;5:337. doi: 10.3389/fgene.2014.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716–7729. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou H, Rigoutsos I. MiR-103a-3p targets the 5’ UTR of GPRC5A in pancreatic cells. RNA. 2014;20:1431–1439. doi: 10.1261/rna.045757.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Choudhury M, Friedman JE. Epigenetics and microRNAs in preeclampsia. Clin Exp Hypertens. 2012;34:334–341. doi: 10.3109/10641963.2011.649931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stahlhut C, Slack FJ. MicroRNAs and the cancer phenotype: profiling, signatures and clinical implications. Genome Med. 2013;5:111. doi: 10.1186/gm516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gasparini P, Cascione L, Fassan M, Lovat F, Guler G, Balci S, Irkkan C, Morrison C, Croce CM, Shapiro CL, et al. microRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget. 2014;5:1174–1184. doi: 10.18632/oncotarget.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yan W, Li R, Liu Y, Yang P, Wang Z, Zhang C, Bao Z, Zhang W, You Y, Jiang T. MicroRNA expression patterns in the malignant progression of gliomas and a 5-microRNA signature for prognosis. Oncotarget. 2014;5:12908–12915. doi: 10.18632/oncotarget.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, Fernandez AF, Davalos V, Villanueva A, Montoya G, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 98.Ziebarth JD, Bhattacharya A, Cui Y. Integrative analysis of somatic mutations altering microRNA targeting in cancer genomes. PLoS One. 2012;7:e47137. doi: 10.1371/journal.pone.0047137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sun G, Yan J, Noltner K, Feng J, Li H, Sarkis DA, Sommer SS, Rossi JJ. SNPs in human miRNA genes affect biogenesis and function. RNA. 2009;15:1640–1651. doi: 10.1261/rna.1560209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Svoronos AA, Engelman DM, Slack FJ. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016;76:3666–3670. doi: 10.1158/0008-5472.CAN-16-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pekarsky Y, Croce CM. Role of miR-15/16 in CLL. Cell Death Differ. 2015;22:6–11. doi: 10.1038/cdd.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Reid G. MicroRNAs in mesothelioma: from tumour suppressors and biomarkers to therapeutic targets. J Thorac Dis. 2015;7:1031–1040. doi: 10.3978/j.issn.2072-1439.2015.04.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17:215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 105.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 106.Wang X, Cao L, Wang Y, Wang X, Liu N, You Y. Regulation of let-7 and its target oncogenes (Review) Oncol Lett. 2012;3:955–960. doi: 10.3892/ol.2012.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu ZL, Wang H, Liu J, Wang ZX. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol Cell Biochem. 2013;372:35–45. doi: 10.1007/s11010-012-1443-3. [DOI] [PubMed] [Google Scholar]
- 108.Sandhu SK, Fassan M, Volinia S, Lovat F, Balatti V, Pekarsky Y, Croce CM. B-cell malignancies in microRNA Eμ-miR-17~92 transgenic mice. Proc Natl Acad Sci USA. 2013;110:18208–18213. doi: 10.1073/pnas.1315365110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mavrakis KJ, Van Der Meulen J, Wolfe AL, Liu X, Mets E, Taghon T, Khan AA, Setty M, Rondou P, Vandenberghe P, et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL) Nat Genet. 2011;43:673–678. doi: 10.1038/ng.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 111.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tian Y, Luo A, Cai Y, Su Q, Ding F, Chen H, Liu Z. MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. J Biol Chem. 2010;285:7986–7994. doi: 10.1074/jbc.M109.062877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang CZ, Yuan P, Li Y. MiR-126 regulated breast cancer cell invasion by targeting ADAM9. Int J Clin Exp Pathol. 2015;8:6547–6553. [PMC free article] [PubMed] [Google Scholar]
- 115.Gong M, Ma J, Guillemette R, Zhou M, Yang Y, Yang Y, Hock JM, Yu X. miR-335 inhibits small cell lung cancer bone metastases via IGF-IR and RANKL pathways. Mol Cancer Res. 2014;12:101–110. doi: 10.1158/1541-7786.MCR-13-0136. [DOI] [PubMed] [Google Scholar]
- 116.Rokavec M, Li H, Jiang L, Hermeking H. The p53/miR-34 axis in development and disease. J Mol Cell Biol. 2014;6:214–230. doi: 10.1093/jmcb/mju003. [DOI] [PubMed] [Google Scholar]
- 117.Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, Zheng X. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44–53. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 118.Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM, Slack FJ. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci USA. 2012;109:E1695–E1704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, Cheng JQ. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem. 2010;285:17869–17879. doi: 10.1074/jbc.M110.101055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 120.Levati L, Pagani E, Romani S, Castiglia D, Piccinni E, Covaciu C, Caporaso P, Bondanza S, Antonetti FR, Bonmassar E, et al. MicroRNA-155 targets the SKI gene in human melanoma cell lines. Pigment Cell Melanoma Res. 2011;24:538–550. doi: 10.1111/j.1755-148X.2011.00857.x. [DOI] [PubMed] [Google Scholar]
- 121.Li CL, Nie H, Wang M, Su LP, Li JF, Yu YY, Yan M, Qu QL, Zhu ZG, Liu BY. microRNA-155 is downregulated in gastric cancer cells and involved in cell metastasis. Oncol Rep. 2012;27:1960–1966. doi: 10.3892/or.2012.1719. [DOI] [PubMed] [Google Scholar]
- 122.Shaham L, Binder V, Gefen N, Borkhardt A, Izraeli S. MiR-125 in normal and malignant hematopoiesis. Leukemia. 2012;26:2011–2018. doi: 10.1038/leu.2012.90. [DOI] [PubMed] [Google Scholar]
- 123.Sun YM, Lin KY, Chen YQ. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol. 2013;6:6. doi: 10.1186/1756-8722-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 125.Almeida MI, Reis RM, Calin GA. MicroRNA history: discovery, recent applications, and next frontiers. Mutat Res. 2011;717:1–8. doi: 10.1016/j.mrfmmm.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 126.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 127.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 128.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol. 2015;40:41–51. doi: 10.1016/j.semcdb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O’Connor ST, Chin AR, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hou X, Zhang M, Qiao H. Diagnostic significance of miR-106a in gastric cancer. Int J Clin Exp Pathol. 2015;8:13096–13101. [PMC free article] [PubMed] [Google Scholar]
- 134.Nagpal N, Ahmad HM, Chameettachal S, Sundar D, Ghosh S, Kulshreshtha R. HIF-inducible miR-191 promotes migration in breast cancer through complex regulation of TGFβ-signaling in hypoxic microenvironment. Sci Rep. 2015;5:9650. doi: 10.1038/srep09650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang XF, Li KK, Gao L, Li SZ, Chen K, Zhang JB, Wang D, Tu RF, Zhang JX, Tao KX, et al. miR-191 promotes tumorigenesis of human colorectal cancer through targeting C/EBPβ. Oncotarget. 2015;6:4144–4158. doi: 10.18632/oncotarget.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Muniyappa MK, Dowling P, Henry M, Meleady P, Doolan P, Gammell P, Clynes M, Barron N. MiRNA-29a regulates the expression of numerous proteins and reduces the invasiveness and proliferation of human carcinoma cell lines. Eur J Cancer. 2009;45:3104–3118. doi: 10.1016/j.ejca.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 137.Guo Y, Fu W, Chen H, Shang C, Zhong M. miR-24 functions as a tumor suppressor in Hep2 laryngeal carcinoma cells partly through down-regulation of the S100A8 protein. Oncol Rep. 2012;27:1097–1103. doi: 10.3892/or.2011.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhao G, Liu L, Zhao T, Jin S, Jiang S, Cao S, Han J, Xin Y, Dong Q, Liu X, et al. Upregulation of miR-24 promotes cell proliferation by targeting NAIF1 in non-small cell lung cancer. Tumour Biol. 2015;36:3693–3701. doi: 10.1007/s13277-014-3008-4. [DOI] [PubMed] [Google Scholar]
- 139.Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med. 2012;12:27–33. doi: 10.2174/156652412798376170. [DOI] [PMC free article] [PubMed] [Google Scholar]


