Abstract
Transformed cells adapt metabolism to support tumor initiation and progression. Specific metabolic activities can participate directly in the process of transformation or support the biological processes that enable tumor growth. Exploiting cancer metabolism for clinical benefit requires defining the pathways that are limiting for cancer progression and understanding the context specificity of metabolic preferences and liabilities in malignant cells. Progress towards answering these questions is providing new insight into cancer biology and can guide the more effective targeting of metabolism to help patients.
Changes in cell metabolism can contribute to transformation and tumor progression. Metabolic phenotypes can also be exploited to image tumors, provide prognostic information, and treat cancer. Thus, understanding cancer metabolism has implications for understanding basic cancer pathophysiology and for clinical oncology.
CELL-AUTONOMOUS REPROGRAMMING OF CANCER METABOLISM
Many recent cancer metabolism studies have focused on the cell-autonomous effects of cancer-promoting mutations. This has uncovered new principles in metabolic regulation and in crosstalk between signaling and metabolic networks. No pathway has received more attention than aerobic glycolysis (the Warburg effect) (Vander Heiden et al., 2009). This phenomenon involves the propensity for proliferating cells, including cancer cells, to take up glucose and secrete the carbon as lactate even when oxygen is present. Principles governing glycolytic regulation in cancer cells have been extensively reviewed (Cairns and Mak, 2016).
Warburg interpreted tumor lactate secretion as an indication that oxidative metabolism (i.e. respiration) was damaged. However, numerous studies, including Warburg’s original work, fail to demonstrate defective respiration as a general feature of malignant cells (Koppenol et al., 2011). Instead, respiration and other mitochondrial activities are required for tumor growth (DeBerardinis and Chandel, 2016). Furthermore, in non-transformed cells the Warburg effect is a reversible phenomenon tethered to proliferation, indicating that it reflects proliferation-associated changes in metabolism rather than a unique feature of malignancy.
Proliferating cells tend to express glucose transporters and glycolytic enzymes out of proportion to the machinery required to oxidize pyruvate (Curi et al., 1988), consistent with preferential conversion of glucose to lactate without loss of respiration. This distinction is important because intermediates generated by the tricarboxylic acid (TCA) cycle are precursors for lipids, amino acids, and nucleotides. These precursors complement precursor metabolites from glycolysis and other pathways and are necessary to support proliferation (DeBerardinis et al., 2007).
Fuels besides glucose also contribute to core metabolic functions of cancer cells: energy formation, biomass assimilation and redox control. Glutamine is a prominent example (Altman et al., 2016); however, recent work has revealed that a diversity of nutrients and pathways support these functions. The expanding metabolic repertoire of cancer cells has been reviewed extensively, with acetate and other fatty acids, lactate, branched chain amino acids, serine, and glycine representing some of the nutrients that are needed to fuel different cancers (DeBerardinis and Chandel, 2016; White, 2013).
The growing list of cancer fuels contrasts with prevailing views from just a few years ago, which suggested oncogenic signaling imposed a rigid dependence on specific nutrients (Vander Heiden et al., 2009). The current picture is more logical when one considers that cancer cells must compete for fuels in a crowded, nutrient-limited tissue environment. The ability to use a panoply of fuels is advantageous, with some cancer cells even relying on autophagic degradation or scavenging of macromolecules (Commisso et al., 2013; Guo et al., 2016; Palm et al., 2015).
This review builds on the increasingly sophisticated understanding of cancer metabolism derived from work over the last decade. Our goal is to convey emerging paradigms and questions in order to guide future research so altered metabolism can be exploited to improve patient care.
FUNCTIONAL CLASSIFICATION OF REPROGRAMMED METABOLIC ACTIVITIES IN CANCER CELLS
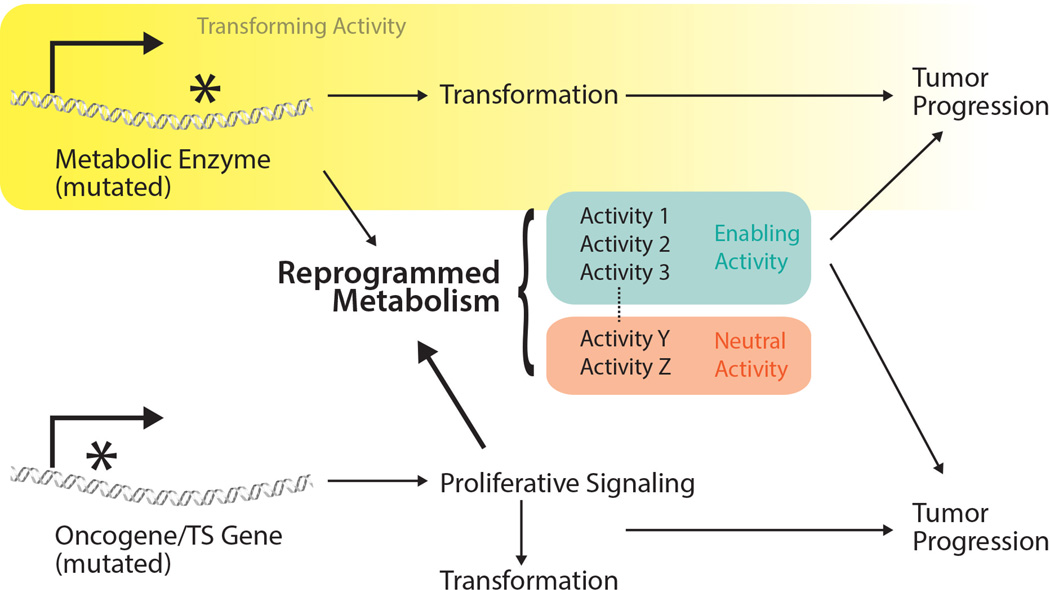
Not all reprogrammed metabolic activities contribute equally to cancer. With many metabolic activities under oncogenic control, categorizing them based on whether they are transforming, enabling, or neutral can clarify the role of each activity in cancer biology and predict how it might be exploited in basic research and clinical oncology (Figure 1).
Figure 1. Classification of reprogrammed metabolic activities.
Some enzyme mutations result in perturbed metabolic activities or metabolite levels that contribute to cancer initiation (transforming activities). Mutations in oncogenes and tumor suppressor genes transform cells, in part, by activating proliferative signaling and/or by inducing broad changes in gene expression that favor cell proliferation, both of which induce metabolic reprogramming. Some metabolic alterations enable cancer progression while others are neutral and not required for cancer cell proliferation or survival. Enzyme mutations resulting in transforming activities may also induce metabolic network changes that are enabling or neutral for tumor progression.
Transforming activities
These activities directly contribute to cell transformation, and blocking them might prevent tumorigenesis in susceptible patients or antagonize disease progression. At present, only a handful of metabolic activities can be considered as transforming based on genetic evidence. These include metabolic perturbations caused by enzyme mutations that either arise somatically in a recurrent fashion; and/or are inherited in the germline of patients with heritable cancer predisposition syndromes. Three examples of these types of mutations have been intensely studied: mutations in the genes encoding isocitrate dehydrogenases-1 and −2 (IDH1, IDH2); mutations in components of the succinate dehydrogenase (SDH) complex; and mutations in fumarate hydratase (FH) (Losman and Kaelin, 2013).
Somatic mutations in IDH1 and IDH2 occur in several tumor types (Losman and Kaelin, 2013). These monoallelic mutations generate an enzyme with neomorphic ability to convert α-ketoglutarate (αKG) to (D)-2-hydroxyglutarate (D-2HG) (Dang et al., 2009). D-2HG accumulates to high levels in IDH-mutant tumors and interferes with the function of several αKG-dependent dioxygenases, including the prolyl hydroxylases that target HIF-a subunits for degradation and epigenetic enzymes that regulate the methylation status of histones and DNA (Losman and Kaelin, 2013). This interferes with expression of genes required for differentiation.
SDH and FH catalyze sequential reactions in the TCA cycle (Figure 2). Both enzymes act as tumor suppressors in familial cancer syndromes where patients inherit one loss-of-function allele and lose expression of the other allele in tumors (Baysal et al., 2000; Tomlinson et al., 2002). Consequently, tumors accumulate high levels of succinate and/or fumarate. Like D-2HG, these dicarboxylic acids interfere with dioxygenase function (Sciacovelli et al., 2016; Selak et al., 2005; Xiao et al., 2012).
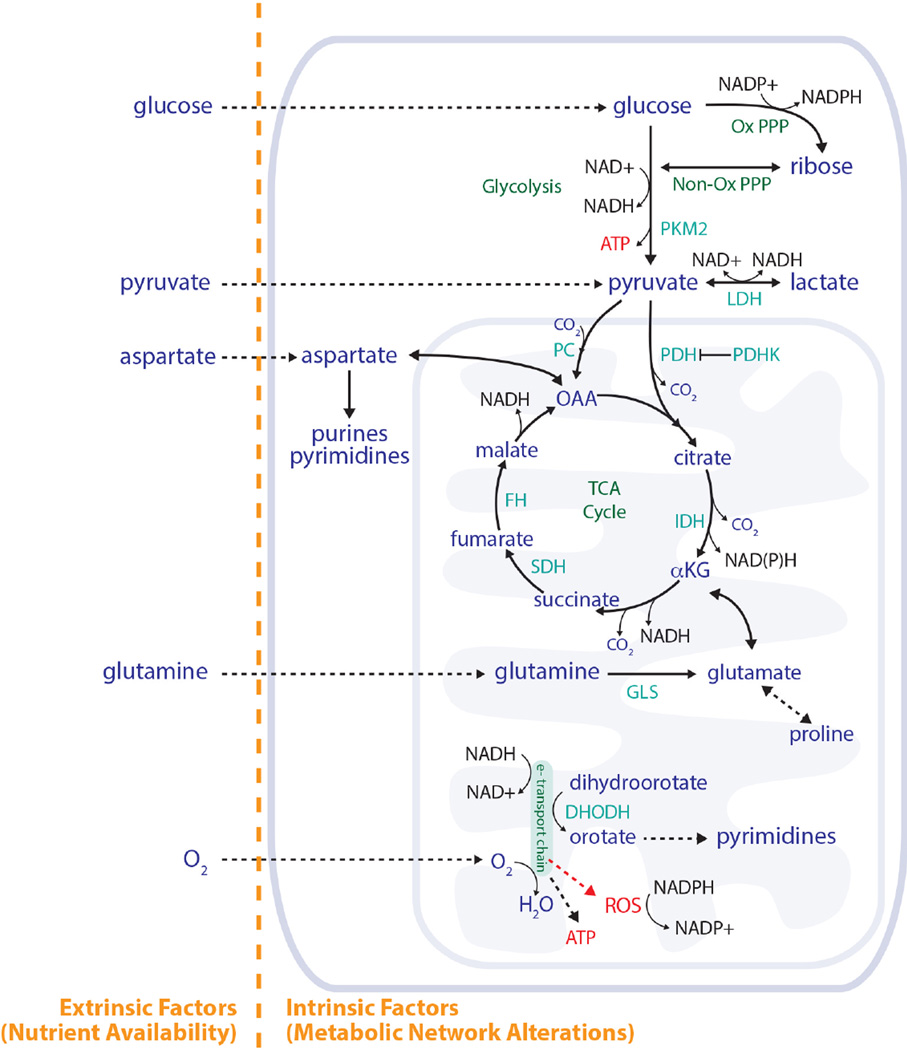
Figure 2. Nutrient availability and the metabolic network both influence metabolic phenotypes.
Both nutrient availability and metabolic network configuration affect how cells use metabolism to produce ATP, generate macromolecules, and regulate redox state. Key reactions in central carbon metabolism are shown, including how the TCA cycle and the electron transport chain are involved in purine and pyrimidine synthesis. Some of the reactions catalyzed by enzymes and metabolites discussed in this review are also shown for reference. PKM2, pyruvate kinase M2; LDH, lactate dehydrogenase; PDH, pyruvate dehydrogenase; PDHK, pyruvate dehydrogenase kinase; PC, pyruvate carboxylase; IDH, isocitrate dehydrogenase; SDH, succinate dehydrogenase; FH, fumarate hydratase; GLS, glutaminase; DHODH, dihydroorotate dehydrogenase; Ox PPP, oxidative pentose phosphate pathway; Non-Ox PPP, non-oxidative pentose phosphate pathway; αKG, α-ketoglutarate; OAA, oxaloacetate; ROS, reactive oxygen species.
In mouse models, mutations in these metabolic enzymes do not transform cells on their own, but cooperate with other mutations to promote neoplasia. Loss of FH in the kidney leads to cyst formation, and hyperproliferation results when p21 is also deleted (Zheng et al., 2015). Idh2 mutations synergize with oncogenic Kras to produce intrahepatic cholangiocarcinoma (Saha et al., 2014). IDH mutations increase the number of hematopoietic progenitor cells (Sasaki et al., 2012), and cooperate with increased HoxA9 and Meis1a expression or with mutant Flt3 or Nras to cause AML (Chen et al., 2013; Kats et al., 2014).
An important concept emerged from these studies. In all cases, an accumulated metabolite inhibits enzymes using αKG as a co-substrate to promote tumorigenesis (Kaelin and McKnight, 2013). Thus, a unifying feature of D-2HG, succinate and fumarate as “oncometabolites” is causing non-metabolic effects that promote transformation in receptive contexts. Interestingly, modestly elevated levels of the L-enantiomer of 2HG might also contribute to malignancy (Shim et al., 2014). Given the importance of dioxygenases in regulating epigenetics, fluctuations in metabolite levels may also play functional roles in normal development. Consistent with this idea, extreme abnormalities in metabolite levels observed in monogenic metabolic diseases, such as deficiency in the dehydrogenases converting L- and D-2HG to αKG, are associated with developmental abnormalities (Erez and DeBerardinis, 2015).
Enabling activities
These activities are altered in cancer cells but are not involved in transformation. They carry out conventional metabolic tasks like supporting energetics, generating macromolecules and maintaining redox state and are required for tumor progression (Figure 1). In many cases they are effectors of oncogenes and tumor suppressor genes. For example, oncogene expression is sufficient to enhance glucose uptake in fibroblasts (Flier et al., 1987). Transcriptional targets of c-MYC include metabolite transporters and enzymes required for glucose metabolism, glutaminolysis and biosynthesis, and inhibiting these activities suppresses growth of c-MYC-driven tumors (Altman et al., 2016). Oncogenic KRAS also regulates nutrient acquisition, macromolecular synthesis and redox homeostasis, and inhibiting these pathways suppresses oncogenic KRAS-driven tumor growth (White, 2013). Activation of mTORC1, an essential component of growth factor signaling networks, enables protein, lipid and nucleic acid synthesis through a variety of mechanisms (Howell et al., 2013). Genetic suppression of metabolic activities may also enable tumor growth. For example, somatic deletion of genes involved in arginine synthesis allows aspartate to be diverted towards nucleotide synthesis, thereby supporting cell proliferation (Rabinovich et al., 2015). Most existing and investigational metabolic therapies are directed against enabling activities (Table 1).
Table 1.
Drugs targeting metabolic enzymes to treat malignancies or suppress immune cell proliferation.
| DRUG | TARGET(S) | EXAMPLE INDICATIONS |
|---|---|---|
| Nucleotide Metabolism | ||
| Methotrexate | Dihydrofolate Reductase (DHFR) | Lymphoma Breast Cancer Choriocarcinoma Osteosarcoma |
| Pemetrexed | DHFR Glycinamide ribonucleotide formyl- transferase (GARFT) Thymidylate synthase |
Lung cancer Mesothelioma |
| 5-Fluorouracil | Thymidylate synthase | Colon cancer Pancreatic Cancer Breast Cancer |
| Gemcitabine Cytarabine Fludarabine |
Ribonucleotide reductase DNA synthesis |
Pancreatic, lung cancer Leukemia Lymphoma |
| Hydroxyurea | Ribonucleotide reductase | AML |
| Leflunomide | DHODH | Rheumatoid arthritis psoriatic arthritis |
| Azathioprine | DNA synthesis | Immunosuppression for transplant |
| Amino Acid Metabolism | ||
| L-asparginase | Asparagine depletion | ALL |
| Select Drugs in Trials | ||
| AG120, AG221, AG881, IDH305, BAY1436032, FT-2102 |
IDH1 and/or IDH2 | Clinical trials for AML, MDS, and solid tumors with mutant IDH1 or IDH2 |
| Epacadostat | Indoleamine 2,3-dioxygenase-1 (IDO1) | Clinical trials for CLL, ovarian cancer, melanoma, other cancers |
| CB-839 | Glutaminase | Clinical trials for AML, ALL, solid tumors |
| TVB-2640 | Fatty acid synthase | Clinical trials for solid tumors |
| PEG-BCT-100 (ADI-PEG20) AEB-1102 |
Arginine depletion | Clinical trials for hepatocellular carcinoma and other hematological and solid tumors including those with ASS1 deficiency |
The cell target(s) for several drugs as well as some approved indications are shown. Some drugs targeting metabolism in clinical trials are also shown.
Neutral activities
Many metabolic features of cancer cells are dispensable for tumor growth (Figure 1). In a given context, these activities are predicted to be poor therapeutic targets. Fluctuating nutrient access may cause activities to be required in some contexts and dispensable in others. Thus, confidently classifying an activity as neutral is challenging and requires definitive proof that loss of the activity does not impair tumor progression.
Pyruvate kinase M2 (PKM2) provides an example of an enzyme dispensable for tumor growth. PKM2 is an isoform of the glycolytic enzyme pyruvate kinase that is expressed in most cancers (Figure 2) and is regulated by oncogenic signaling (Israelsen and Vander Heiden, 2015). PKM2 expression is not required for growth of breast tumors, liver tumors, leukemia, or xenograft tumors from various human cancer cell lines in mice (Dayton et al., 2016; Israelsen and Vander Heiden, 2015). However, studies of PKM2 still provide insight into cancer metabolic regulation. PKM2 loss promotes cancer in some mouse models, and characterizing these models has argued that how glucose is metabolized can influence tumor progression (Dayton et al., 2016; Israelsen et al., 2013). In fact, activating PKM2 can slow growth of some tumors, but the ability of tumors to lose pyruvate kinase expression argues against PKM2 being a good therapeutic target for most cancers (Israelsen and Vander Heiden, 2015).
WHAT PRODUCTS OF METABOLISM ARE LIMITING FOR PROLIFERATION?
One approach to identify enabling activities is to determine which aspects of metabolism are limiting for cancer cell proliferation. Targeting activities that supply limiting materials for proliferation is therapeutically attractive, especially if the pathways used are less important in normal proliferative tissues. Although several metabolic products have been proposed as critical outputs of cancer metabolism, which are rate limiting for proliferation remains controversial.
Is ATP limiting for proliferating cells?
ATP plays a critical role in all cells to enable otherwise unfavorable processes. Aerobic glycolysis has been historically interpreted as a shift from oxidative phosphorylation toward fermentation to generate ATP (Gatenby and Gillies, 2004; Vander Heiden et al., 2009). This route of ATP production yields less ATP per mole of glucose, but allows fast ATP generation even in low oxygen (Koppenol et al., 2011). Energy is derived from ATP hydrolysis to ADP (or AMP), and thus depends on the ATP/ADP (or ATP/AMP) ratio. Therefore, cells must continuously oxidize nutrients to regenerate ATP to maintain homeostasis. Although many proliferative processes also require ATP hydrolysis, the additional ATP needed for proliferation is small relative to the requirements for homeostasis (Vander Heiden et al., 2009). Thus, while producing ATP is critical for survival, it may not always be limiting for proliferation.
Little evidence supports ATP limitation as a reason cells use aerobic glycolysis. First, many proliferating cells use aerobic glycolysis regardless of nutrient and oxygen availability (Vander Heiden et al., 2009). Second, cancer cells retain the capacity to increase fermentation when respiration is inhibited, and also can increase respiration when mitochondria are uncoupled (Andrzejewski et al., 2014; Birsoy et al., 2014). The existence of “spare respiratory capacity” argues that the ATP/ADP ratio is sufficiently high to limit electron transport, and substrates are available in excess of the demand for ATP synthesis. Finally, increasing ATP consumption can promote proliferation (Fang et al., 2010), providing further evidence that the ability to generate ATP is greater than what is needed by proliferating cells.
Is NADPH limiting for proliferating cells?
Cells store energy as reduced carbon in carbohydrates and lipids, and the anabolic reactions used to synthesize these materials requires electrons from NADPH (Figure 3), leading to the hypothesis that NADPH may be limiting for proliferation (Vander Heiden et al., 2009). However, the fact that some cancers like clear cell renal cell carcinoma accumulate lipids (Qiu et al., 2015) argues that these cancers have sufficient NADPH to produce lipids in excess of those needed for cell membranes. Most macromolecular biosynthesis occurs in the cytosol where several enzymes can produce NADPH including malic enzyme 1, IDH1, and folate metabolism; however, the oxidative pentose phosphate pathway (oxPPP) is a major source of cytosolic NAPDH (Fan et al., 2014; Lewis et al., 2014) (Figure 2). Despite this, most ribose is produced via the non-oxidative pentose phosphate pathway, which does not produce NAPDH (Boros et al., 2000; Ying et al., 2012). A major regulator of oxPPP flux is the NADP+/NADPH ratio, and exposing cells to oxidative stress increases oxPPP flux (Kuehne et al., 2015) arguing cells have excess capacity to produce NADPH even when ROS levels increase. Thus, while NADPH is important to help cancer cells cope with ROS, generating enough cytosolic NADPH for competing metabolic pathways may not be limiting for proliferation. This assertion is supported by both classic literature (Reitzer et al., 1980) and the fact that reduced oxPPP flux resulting from G6PD deficiency has no impact on human cancer risk (Cocco, 1987).
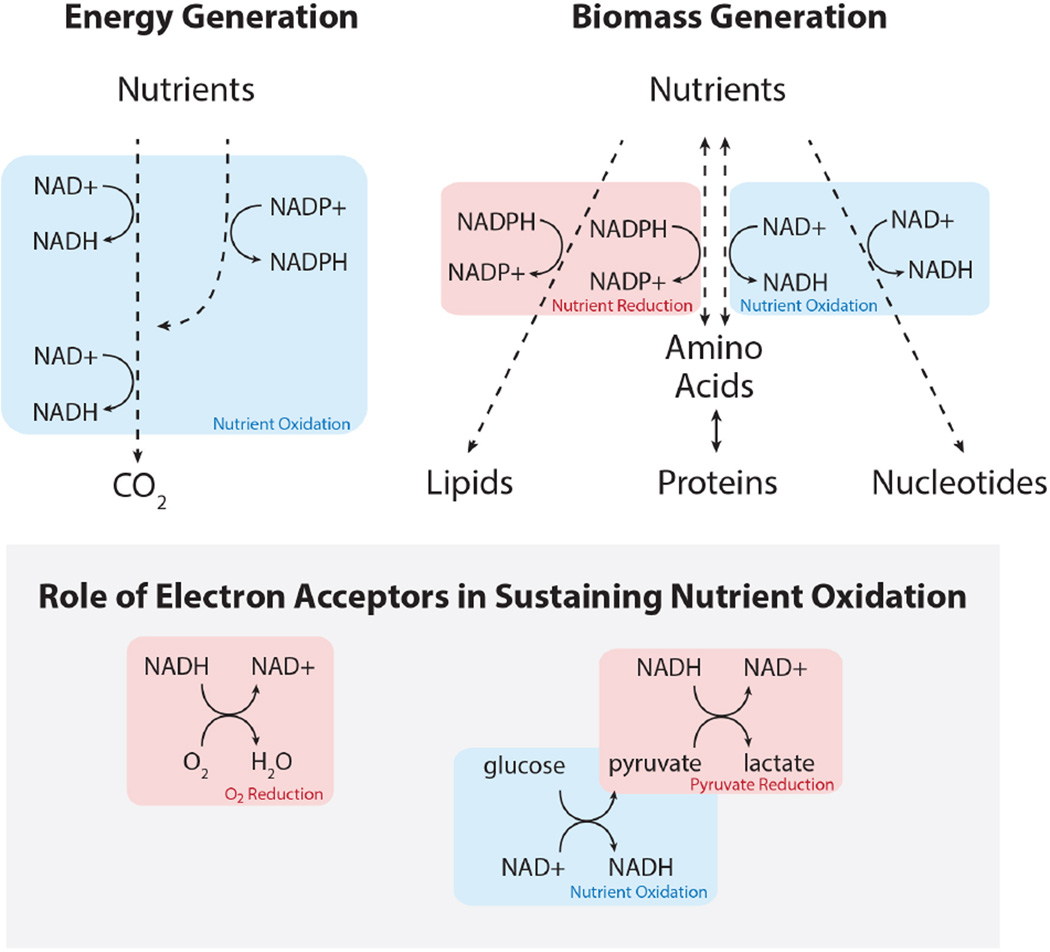
Figure 3. Role of redox cofactors in energy generation and biosynthesis.
Cells rely on nutrient oxidation to generate ATP (energy), either through glycolysis or via NADH generation to fuel oxidative phosphorylation. Generating biomass can involve either nutrient reduction or nutrient oxidation. Continued nutrient oxidation requires cycling of NADH back to NAD+, which necessitates transfer of electrons to an electron acceptor such as oxygen.
Products of the TCA cycle can be limiting for proliferation
Clues to which aspects of metabolism are most limiting for proliferation come from studies where proliferation arrest from metabolic disruption is rescued by re-supplying metabolites. A classic example is glutamine metabolism for TCA cycle anaplerosis (DeBerardinis et al., 2007; Yuneva et al., 2007). Most cultured cells require glutamine, and providing cells with αKG or other TCA cycle intermediates can rescue proliferation in low glutamine conditions (Yuneva et al., 2007). Activating pyruvate carboxylase to allow anaplerosis from glucose can also enable proliferation in low glutamine and contexts where glutamine is not used (Figure 2) (Cheng et al., 2011; Davidson et al., 2016b; Sellers et al., 2015).
The TCA cycle carbon requirement suggests a product of this cycle can be limiting for proliferation. TCA cycle intermediates are precursors for several biosynthetic intermediates, but production of amino acids accounts for the dominant disposition of TCA cycle carbon in biomass (Hosios et al., 2016). Aspartate, asparagine, proline and glutamate can all be produced from TCA cycle intermediates, and cells synthesize these amino acids from glutamine even when they are present in the environment (Hosios et al., 2016). Indeed, synthesis of both aspartate (Birsoy et al., 2015; Sullivan et al., 2015) and proline (Loayza-Puch et al., 2016) can be limiting under some conditions.
Nucleotide synthesis can be limiting for proliferation
Supplying nucleotide bases can rescue proliferation of glutamine-deprived cells and cells with high pyruvate kinase activity suggesting nucleotide base production is important downstream of major nutrient pathways (Gaglio et al., 2009; Lunt et al., 2015). Acquiring deoxyribonucleotide bases is limiting in other cancer contexts (Aird et al., 2013), and nucleotide synthesis is the target of several chemotherapeutics (Table 1), arguing that acquiring nucleotides might be a metabolic bottleneck for diverse cancers. Why might nucleotide base synthesis be limiting on a biochemical level? One important factor is that unlike lipids and proteins, nucleotides may not be provided in sufficient quantities or proportions by the tumor microenvironment and synthesis of these complex molecules requires the integrated metabolism of non-essential amino acids, ribose and one-carbon donors.
The DHODH step of pyrimidine biosynthesis is directly coupled to the mitochondrial electron transport chain and oxygen consumption (Figure 2) and is important for both AML and melanoma progression (Sykes et al., 2016; White et al., 2011). While oxygen is abundant in standard tissue culture, oxygen levels in even the best perfused tissues are 9% or less (Bertout et al., 2008), a value closer to the oxygen levels used to study hypoxia responses in culture. Oxygen delivery can be more limiting than glucose for normal tissue physiology, as evidenced by lactic acid build-up in muscle during exercise, by increased hematocrit enhancing the performance of elite athletes, and by the fact that angiogenesis is regulated by oxygen levels rather than by nutrients even though tissue delivery of both relies on the vasculature. Oxygen can also be limiting for tumor growth. Individuals living at high altitude have lower cancer incidence (Burtscher, 2014), and increasing oxygen delivery to tissues with erythropoietin promotes cancer progression (Szenajch et al., 2010).
Cancer cells require oxidative metabolism to form tumors (Davidson et al., 2016b; Weinberg et al., 2010). Oxygen consumption is coupled to ATP synthesis, but maintaining an adequate ATP/ADP ratio may not be why respiration is limiting for proliferation. Respiration also regenerates NAD+ for oxidation reactions (Figure 3), which are required for aspartate synthesis (Birsoy et al., 2015; Sullivan et al., 2015), providing another link between oxygen consumption and nucleotide synthesis. Nucleotide base carbon is more oxidized than the carbon in most nutrients. Aspartate is a key oxidized precursor for both purines and pyrimidines (Figure 2), blood aspartate levels are low, and most cells are unable to take up aspartate from the environment (Birsoy et al., 2015). Thus, stoichiometric reduction of oxygen or another molecule is required for aspartate and nucleotide base synthesis (Sullivan et al., 2015). The ability to produce aspartate can be limiting for some tumors (Gui et al., 2016), and respiration also supports production of folate species for purine synthesis (Bao et al., 2016; Meiser et al., 2016). Taken together, these studies argue access to oxygen or alternative electron acceptors limits nucleotide synthesis for some cancer cells.
Consequences of electron acceptor limitation in cancer
The notion that chemical disposal of excess electrons to synthesize nucleotides is limiting for proliferation is attractive because it explains several cancer metabolism phenotypes. First, it provides an explanation for the propensity of cancer cells to produce lactate. NAD+ regeneration from NADH requires electron disposal (Figure 3). Conversion of pyruvate to lactate is driven by the NAD+/NADH ratio, and decreasing this ratio in proliferating cells because of increased nucleotide production may be sufficient to drive increased lactate production. All proliferating cells must replicate DNA, so the accompanying reduction in NAD+/NADH ratio could also explain the use of fermentation by many proliferating cells across organisms and conditions. Cancer cells have a very low NAD+/NADH ratio (Hung et al., 2011), and changes in NAD+/NADH ratio correlate better with tumor growth rate than changes in ATP/ADP ratio (Gui et al., 2016). Regenerating NAD+ via orthogonal pathways can also increase cell proliferation when respiration is impaired (Birsoy et al., 2015; Sullivan et al., 2015; Titov et al., 2016), and disposal of excess electrons can limit proliferation in prokaryotic systems (Dietrich et al., 2013).
A low NAD+/NADH ratio also could explain increased ROS in cancer. ROS levels might increase because cells are deficient in electron donors to detoxify ROS. However, electron acceptor deficiency leading to inefficient mitochondrial electron transport chain function will also lead to increased ROS (Figure 2). The latter explanation is consistent with angiogenesis and oxygen delivery being a limitation for tumor growth (Gatenby and Gillies, 2004).
WHAT DETERMINES HOW DIFFERENT TUMORS USE METABOLISM?
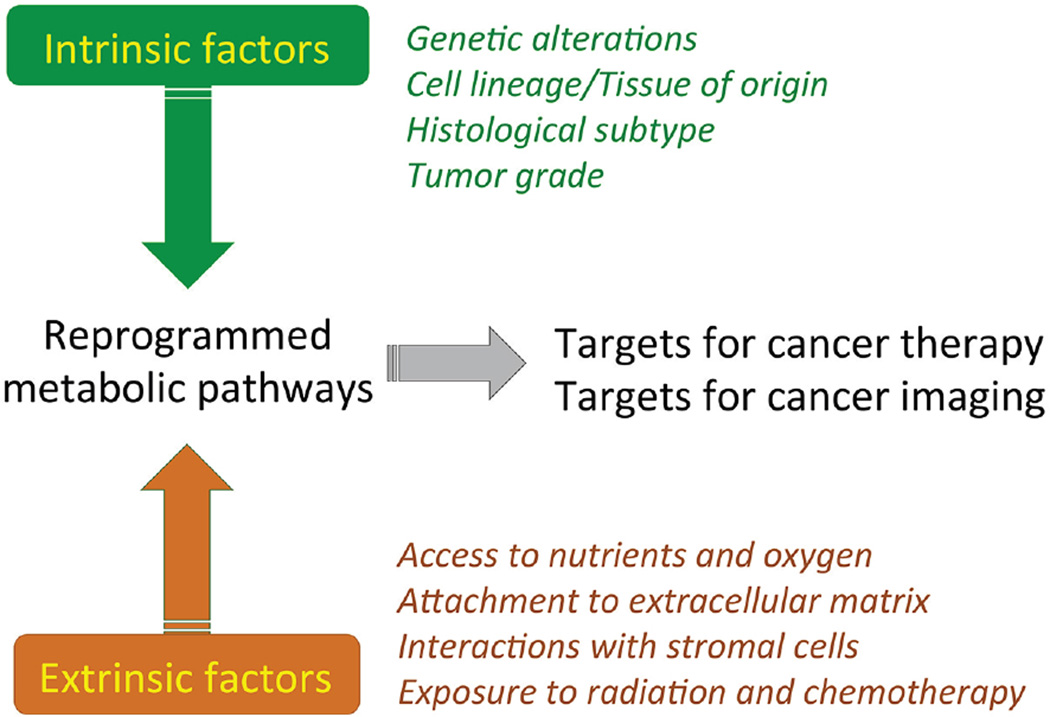
Pathways downstream of oncogenes and tumor suppressors regulate cancer cell metabolism. Genomic alterations can also result in copy number gains and losses of genes encoding metabolic enzymes, and this may induce vulnerabilities (Li et al., 2014; Locasale et al., 2011; Possemato et al., 2011). However, the extent to which metabolic preferences are hard-wired by the tumor genotype is less clear, because many non-genetic factors also influence tumor metabolism (Figure 2). As in all tissues, tumor metabolism is dictated by a variety of intrinsic and extrinsic factors (Figure 4). We need to understand how these factors are integrated to create metabolic dependencies.
Figure 4. Intrinsic and extrinsic influences on cancer cell metabolic reprogramming.
Reprogrammed metabolic pathways, including pathways involved in bioenergetics, anabolism, and redox homeostasis are common features of tumor tissue. The metabolic phenotype of cancer cells is the cumulative result of a variety of processes both intrinsic and extrinsic to the malignant cell. An integrated understanding of the relative contributions of all processes, in the context of intact tumors, will be necessary to exploit metabolic reprogramming in the clinic. Both intrinsically and extrinsically regulated pathways provide opportunities for clinical translation, including new targets for therapy and for non-invasive imaging to detect and monitor cancer.
The environment can affect cancer cell metabolism
Cancer cells in culture have a different metabolic phenotype from tumors. While many cancer cell lines quantitatively convert glucose to lactate, glucose oxidation is prevalent in tumors (Davidson et al., 2016b; Hensley et al., 2016; Maher et al., 2012; Marin-Valencia et al., 2012). Cultured lung cancer cells use glutamine to supply TCA cycle carbon, while lung tumors in mice prefer glucose as a TCA cycle fuel (Davidson et al., 2016b; Hensley et al., 2016; Sellers et al., 2015). These differences translate into altered vulnerabilities, as lung cancer cell lines require glutaminase for proliferation while tumors derived from these same cells do not; the converse is true for enzymes involved in glucose oxidation (Davidson et al., 2016b).
The environment can also affect the efficacy of drugs targeting metabolism. Metformin and other biguanides are mitochondrial complex I inhibitors that slow tumor growth by preventing complex I-mediated NAD+ regeneration (Wheaton et al., 2014). Thus, alternative NAD+ regeneration pathways decrease complex I dependence and promote metformin resistance (Gui et al., 2016). Lipid depletion potentiates the effect of acetyl-coA carboxylase inhibitors in culture, but the same drugs affect lung tumor growth in vivo despite the presumed availability of fatty acids (Svensson et al., 2016). Tumors vary in the fraction of actively proliferating cells, which influences metabolic dependency (Coloff et al., 2016). Both genetic and drug screens have identified context-specific metabolic liabilities, implying that the tumor environment can influence sensitivity to metabolic inhibitors (Possemato et al., 2011; Wenzel et al., 2014).
Cell lineage can also affect cancer metabolism
Most cytotoxic chemotherapies, including those that target nucleotide metabolism, are only effective against select cancers. A genetic predictor of response is lacking for most of these drugs, with sensitive cancer types defined instead by the cancer tissue of origin. This suggests some metabolic dependencies might be defined by tumor lineage. Indeed, acute lymphoblastic leukemia (ALL) cells are auxotrophic for asparagine and are sensitive to asparagine depletion by L-asparaginase. DHODH activity is a lineage-specific requirement of myeloid cells that impacts differentiation (Sykes et al., 2016). MYC induces distinct metabolic phenotypes in mouse liver and lung tumors, with liver tumors displaying enhanced glutamine catabolism and lung tumors retaining the ability to synthesize glutamine from glucose (Yuneva et al., 2012). Mouse lung and pancreatic tumors initiated by the same driver mutations also exhibit differences in amino acid metabolism with lung tumors relying on uptake of free amino acids from the blood (Mayers et al., 2016), while pancreatic tumors obtain amino acids from extracellular protein (Davidson et al., 2016a). These differences in amino acid metabolism translate into a differential dependency of lung and pancreatic tumors on branched chain amino acid nitrogen for nucleotide synthesis (Mayers et al., 2016).
An explanation for how lineage influences tumor metabolism is suggested by metabolic gene expression studies. The metabolic network of tumors is more similar to the normal tissue from where the cancer arose than it is to the metabolic network of tumors arising from another tissue (Hu et al., 2013), and normal tissues wire metabolism differently to support their unique functions. Mechanistically, tissue identity is established through epigenetic regulation of gene expression, and despite altered gene expression in cancer, many aspects of tissue identity are maintained (Gaude and Frezza, 2016). Metabolism can also influence epigenetic state, but the fact that tumors retain metabolic features from their parental normal tissue argues cancers adapt existing tissue metabolism to support abnormal proliferation rather than converging on a universal proliferation program. This distinction is critical, because it suggests that drugs targeting proliferative metabolism may not be effective against all tumors.
Tumor lineage also modifies the penetrance of transforming mutations. The restricted tumor spectrum in patients with familial cancer syndromes caused by SDH and FH mutations indicates an exquisite context-dependence for these events to cause malignancy. Why specific mutations cause lineage-restricted cancers is not understood, but one might speculate that some metabolic alterations are only tolerated in the context of a specific metabolic network.
Tissue-specific differences in nutrient availability impose further constraints on metabolism. How cancer cells generate aspartate in different contexts illustrates how such constraints can affect production of a key metabolite for nucleotide synthesis. Cells in culture where oxygen is in excess produce aspartate from glutamine via the TCA cycle, a pathway that involves multiple oxidation steps (Birsoy et al., 2015; DeBerardinis et al., 2007; Sullivan et al., 2015)(Figure 2). However, in lung tumors where oxygen is less abundant, aspartate is produced from glucose via a series of reactions with fewer oxidation steps (Davidson et al., 2016b; Hensley et al., 2016; Sellers et al., 2015). Pancreatic tumors are particularly oxygen-limited and use yet another strategy, scavenging amino acids from extracellular protein through KRAS-dependent macropinocytosis and circumventing any need to synthesize aspartate from other nutrients (Commisso et al., 2013; Davidson et al., 2016a; Kamphorst et al., 2015).
Interactions with benign cells can affect cancer cell metabolism
Tumors consist of a complex milieu of malignant and non-malignant cell types with distinct metabolic preferences. Specific nutrients might be available to cancer cells because they are produced in a given tissue, and differential consumption of nutrients by cancer and non-cancer cells might further affect metabolite levels. In the brain, astrocytes metabolize glucose and secrete lactate to be used by neurons as a source of energy (Bittar et al., 1996). A similar symbiotic relationship may exist within tumors, where some cells ferment glucose to lactate, which is then used as a respiratory substrate for other cells (Sonveaux et al., 2008). Alanine produced by pancreatic stellate cells can promote pancreatic cancer cell proliferation (Sousa et al., 2016) and bone marrow stromal cells provide cysteine to promote survival of chronic lymphocytic leukemia cells (Zhang et al., 2012). Similar relationships likely exist in all cancers, but dissecting how cell populations share nutrients within tumors remains a challenge.
ROLES FOR METABOLIC REPROGRAMMING DURING CANCER PROGRESSION
Most cancer-related deaths occur after therapy resistance has emerged and/or after tumor cells have spread from the primary site. Whether specific metabolic activities acquired during tumor progression promote therapy resistance or metastasis is being investigated.
Role of metabolism in metastasis
Epithelial-mesenchymal transition (EMT) is a transdifferentiation program associated with metastasis and chemotherapeutic resistance (Ye and Weinberg, 2015). Enforcing an EMT-like phenotype in lung cancer cells reduces lipogenesis and increases respiration (Jiang et al., 2015). Silencing fatty acid synthase in these cells induces EMT and enhances metastatic seeding in mice. Similarly, in a breast cancer model cellular invasion, migration and metastasis correlate with enhanced respiration and mitochondrial biogenesis driven by the transcriptional co-activator PGC1α (LeBleu et al., 2014). Silencing PGC1α reduced metastasis without altering tumor growth at the primary site. In this model, PGC1α was required for cancer cell invasion but not proliferation, perhaps explaining its role in metastasis and dispensability for primary tumor growth.
Several studies have examined metabolic transitions accompanying detachment from extracellular matrix, an early step in metastasis. In breast epithelial cells, loss of matrix attachment results in oxidative stress and cell death (Schafer et al., 2009). Treating detached cells with antioxidants or enhancing NAPDH production by the oxPPP promotes survival. In lung cancer cells, detachment suppresses pathways required to maximize cell growth and induced shuttling of cytosolic reducing equivalents into the mitochondria to combat ROS (Jiang et al., 2016). Specifically, reductive carboxylation of cytosolic αKG is induced to produce citrate, which then enters the mitochondria to produce NADPH. This pathway is dispensable in monolayer culture but required in the anchorage-independent state.
Additional bottlenecks can prevent metastasis after cells enter the circulation. Circulating melanoma cells and metastases have elevated ROS compared to primary tumors (Piskounova et al., 2015). Highly metastatic tumors undergo reversible metabolic changes allowing them to increase mitochondrial NADPH and combat ROS stress. Inhibiting this response reduces metastasis, whereas systemic treatment with antioxidants enhances metastasis. Exacerbating oxidative stress potentiates some chemotherapeutic responses (Harris et al., 2015; Raj et al., 2011), and enhances response to radiation therapy (Robbins and Zhao, 2004). Altogether, these studies underscore the importance of managing ROS during tumor progression, and suggest that this might be exploited therapeutically.
Metabolism and therapy resistance
Therapy-resistant tumors have altered metabolic phenotypes relative to treatment-naïve tumors, with enhanced reliance on mitochondrial metabolism in the resistant cancers. Genetic inactivation of oncogenic KRASG12D in pancreatic cancer leads to tumor regression; however, a small number of cancer cells survive in a dormant state and regenerate tumors if KRASG12D is reactivated (Viale et al., 2014). Compared to cells with constitutive KRASG12D expression, the surviving cells have enhanced respiration, reduced glycolysis, and an impaired ability to increase glycolysis when respiration is inhibited. This renders surviving cells susceptible to respiration inhibitors, and treating mice with an ATP synthase inhibitor during tumor regression delayed relapse after KRASG12D reactivation.
Mitochondria also contribute to melanoma therapy resistance. Higher PGC1α expression portends poor outcome in human melanoma (Vazquez et al., 2013). In melanoma cells, PGC1α expression is regulated by the melanocyte-lineage transcription factor MITF and is required to maintain high levels of respiration, low ROS, and resistance to the oxidizing agent piperlongumine. Silencing PGC1α synergizes with piperlongumine to reduce melanoma xenograft growth. In human melanoma with mutant BRAF, gene expression patterns associated with mitochondrial biogenesis predict reduced survival and tumors that relapse after MAPK inhibitor treatment have increased mitochondrial biogenesis gene expression (Zhang et al., 2016). Inhibiting mitochondrial biogenesis using a mitochondrial-targeted Hsp90 inhibitor decreases MAPK inhibitor resistance in xenografts.
In autochthonous models of breast and lung cancer, anti-angiogenic kinase inhibitors enhance AMPK signaling and shift metabolism toward a more oxidative phenotype. Treated cells are more sensitive to inhibitors of oxidative metabolism, and the same drugs synergize with anti-angiogenics to suppress tumor growth (Navarro et al., 2016). Mitochondrial inhibition with metformin can kill chemotherapy-resistant breast tumor stem cells (Janzer et al., 2014). Taken together, these studies demonstrate the importance of mitochondrial function in enabling therapeutic resistance and suggest mitochondrial inhibitors may help curtail cancer progression.
CAN CANCER METABOLISM BE EXPLOITED TO IMPROVE THERAPY?
To target metabolism for therapy, limiting metabolic processes must be identified and understood sufficiently to target the process safely and select responsive patients. Using the classifications described above, transforming and/or enabling activities must be identified with an adequate therapeutic index. Clinical experience with cytotoxic chemotherapy highlights the challenges that will likely confront new metabolic therapies. Many chemotherapies inhibit nucleotide metabolism (Table 1). These drugs are highly effective in some tumors, but efficacy is not solely determined by proliferative index. Tumor cells acquire mutations in cell cycle checkpoint genes that contribute to therapeutic window, but the differential sensitivity of different cancers to these drugs is incompletely understood. Evidence suggests that tumors use diverse mechanisms to maintain nucleotide pools, and this may contribute to efficacy. However, a better understanding of differential sensitivity to metabolism-targeted chemotherapy may help use these drugs more effectively and suggest new targets.
Recent challenges associated with targeting cancer metabolism
Whether studies of cell-autonomous metabolic reprogramming will identify the best therapeutic targets is unproven. Success targeting glycolysis has been limited. Exposing patients to high levels of 2-deoxyglucose generated tumor responses but caused hypoglycemia symptoms, and more tolerable doses in modern trials have been disappointing (Vander Heiden, 2011). Attempts to target the Warburg effect directly have also been met with limited success. Dependence on the enzyme lactate dehydrogenase (LDH) was demonstrated both genetically and pharmacologically (Fantin et al., 2006; Le et al., 2010; Shim et al., 1997), leading to development of LDHA inhibitors (Boudreau et al., 2016), but none progressed to clinical trials, suggesting either unacceptable toxicity, insufficient drug exposure or a lack of LDHA dependence in human tumors. Lactate transport inhibition is currently being tested as an alternative approach. Activating pyruvate dehydrogenase (PDH) by targeting its inhibitory kinase (PDHK) has been proposed as a therapeutic strategy. The PDHK inhibitor dichloroacetate (DCA) elicits metabolic effects in human tumors (Michelakis et al., 2010), but minimal efficacy data has been reported. Recent data indicate that PDH is active and required by some tumors (Davidson et al., 2016b; Hensley et al., 2016), tempering hope that further activating the enzyme will be therapeutically useful.
One success in targeting cancer metabolism is the reversal of 2HG-mediated reprogramming in IDH mutant tumors (Losman and Kaelin, 2013). Drugs targeting mutant IDH can elicit responses in patients with hematological malignancies, but have been less effective in glioma models (Tateishi et al., 2015). The mechanism of 2HG-mediated transformation may differ in different cancers; however, liabilities caused by high 2HG might be exploited as an alternative approach to treat these cancers.
Identifying tumors susceptible to metabolic therapy
Transforming mutations in metabolic enzymes present clinical opportunities where patients can be selected based on tumor genetics. However, most metabolic changes are not driven by metabolic enzyme mutations, and even in the case of IDH mutations, a durable state of mutant enzyme dependence may not always be present (Tateishi et al., 2015). Thus, how to select patients for drugs targeting metabolic enzymes remains a critical therapeutic question.
Based on successes developing protein kinase inhibitors, one approach has been to screen cell line panels to identify genetically susceptible cancers, and then to validate these hypotheses in animal models. However, differential nutrient use in culture and in tumors suggests that the use of cell lines to identify sensitive cancers could identify activities that are neutral in vivo. Conversely, enabling pathways in vivo may be less important in culture. A requirement for glutamine metabolism has been observed in many cancer types (Altman et al., 2016), but both tissue of origin and genetics contribute to this phenotype (Yuneva et al., 2012). The auxotrophy of some cancers for individual amino acids is also influenced by a combination of genetic and non-genetic factors. Auxotrophy for arginine or asparagine can be explained in some cases by decreased expression of synthesis enzymes; however, treatment of ALL with asparaginase is effective even when asparagine synthase is expressed, suggesting that additional factors underlie the need for exogenous asparagine (Krall et al., 2016; Stams et al., 2003). Increased biguanide sensitivity has been attributed to various mutations (Buzzai et al., 2007; Shackelford et al., 2013), but tissue environment also affects response to these drugs (Gui et al., 2016). Selecting patients for chemotherapy based on tumor type argues cell lineage can be an effective tool to stratify patients and argues that in addition to genetics, tumor type and location should be considered when developing metabolism-targeted drugs.
Genetically defined metabolic targets
Sensitivity to some enabling metabolic targets is determined by genetics, including passenger deletion of enzymes that prevent metabolic compensation in tumors. For example, most glioblastoma cells express two isoforms of the glycolytic enzyme enolase encoded by the genes ENO1 and ENO2. ENO1 resides on chromosome 1p36 at a tumor suppressor locus that is homozygously deleted in 1–5% of glioblastomas (Muller et al., 2012). This reduces metabolic redundancy and creates a therapeutic opportunity to target ENO2. The gene encoding the enzyme methylthioadenosine phosphorylase (MTAP) can be deleted with the p16/CDKN2A tumor suppressor gene. The resulting accumulation of MTA reduces PRMT5 methyltransferase activity, creating another therapeutic opportunity (Kryukov et al., 2016; Marjon et al., 2016; Mavrakis et al., 2016). Gene expression changes that create auxotrophies for specific amino acids can also be targeted, with L-asparaginase providing an example of success. Drugs that deplete arginine are being tested in patients with ASS1-deficient tumors (Phillips et al., 2013). Limiting serine in the diet can be efficacious in some p53 mutant cancers (Maddocks et al., 2013).
Non-cell autonomous targeting of tumor metabolism
Understanding the interactions between cancer cell and immune cell metabolism will be critical for combining metabolism targeted therapies with immunotherapies. Nutrient availability and metabolism affect T cell and macrophage differentiation, raising the possibility that targeting tumor cell metabolism may either promote or prevent anti-cancer immune responses.
Cancer cells compete with T cells for glucose in tumors, and restricting T cell glucose metabolism causes lymphocyte exhaustion (Chang et al., 2015; Ho et al., 2015). Thus, therapies that decrease glucose use by cancer cells could make glucose available for T cells and enhance immune effector functions to further limit tumor growth. However, if the same therapies inhibit T cell glucose metabolism, they may limit anti-tumor immunity. Other nutrient levels can affect both cancer and immune cells. High arginine levels promote enhanced T cell survival and anti-tumor activity (Geiger et al., 2016), and therapies that deplete tumor arginine promote immune suppressor cell accumulation (Fletcher et al., 2015). Lactate can blunt T and NK cell function (Brand et al., 2016). Kynurenine, a breakdown product of tryptophan, can modulate both the innate and adaptive immune system and has been implicated in cancer-associated immunosuppression (Platten et al., 2014) and indoleamine-2,3-dioxygenase (IDO) inhibitors that decrease tryptophan metabolism can help break immune tolerance and potentiate chemotherapy (Muller et al., 2005). Increased MTA levels in MTAP-null tumors can also suppress T cell activation (Henrich et al., 2016) and suggests some metabolic interventions might enhance anti-cancer immune responses.
The relationship between tumors and other organ systems is not limited to the immune system. Endothelial cells also undergo factor-induced metabolic reprogramming, and metabolic therapies can limit endothelial cell proliferation and angiogenesis (De Bock et al., 2013). Cancer also causes alterations in whole body metabolism that may influence tumor nutrient availability. Modulating the amino acid composition of the diet can slow cancer growth (Maddocks et al., 2013) and investigations into how diet affects tumor growth remains an underexplored area.
Clinical utility of neutral metabolic activities
By definition, neutral metabolic activities are not therapeutic targets, but some may still present useful opportunities in clinical oncology, particularly as predictive biomarkers or for tumor imaging. PKM2 expression in tumors is the basis for a positron emission tomography (PET) imaging agent. N,N-diarylsulfonamide (DASA) compounds bind to PKM2, and an 11C-labeled analog of DASA-23 is taken up by PKM2-expressing orthotopic gliomas in mice, providing imaging contrast with the normal brain (Witney et al., 2015). Other investigational PET agents exploit the ability of some tumors to take up and retain glutamine (Venneti et al., 2015). Conversion of 13C-labeled pyruvate to lactate can be imaged in patients by hyperpolarized MRI (Nelson et al., 2013). Some targetable activities are also the basis of new imaging techniques, with the detection of 2HG by magnetic resonance spectroscopy a prominent recent example (Andronesi et al., 2012; Choi et al., 2012). Finally, the heterogeneity of metabolic phenotypes among human cancer patients may provide biomarkers that correlate with disease progression (Hakimi et al., 2016).
FUTURE PERSPECTIVES
Analysis of cell-intrinsic metabolic preferences imposed by the oncogenotype has been informative to uncover new paradigms in proliferating cell metabolic regulation. However, the next phase of cancer metabolism research will need to address increasingly complex questions about how intrinsic and extrinsic influences integrate to create exploitable metabolic phenotypes in cancer. This will require consideration of the metabolic preferences hard-wired into cancer cells by tissue-of-origin, interactions between benign and malignant cells within the microenvironment, and influences of the diet and microbiome on the host. Cell culture systems must be improved to better reflect the metabolic limitations in tumors, and these studies will be propelled by improvements in quantitative assessment of metabolic fluxes in different contexts. Ultimately this will enable matching of the right therapies with the right patients to exploit altered metabolism and improve cancer outcomes.
Acknowledgments
We regret that due to space limitations we were unable to cite many excellent studies that shaped our modern understanding of cancer metabolism. We thank members of the DeBerardinis and Vander Heiden laboratories for insight and extensive critique of the manuscript, and Brooke Bevis for help with figures. M.G.V.H. is supported by the NCI (CA168653, CA201276), the Ludwig Center at MIT, SU2C, the Lustgarten Foundation and the Howard Hughes Medical Institute (Faculty Scholars Award). R.J.D. is supported by grants from the NCI (CA157996), Cancer Prevention and Research Institute of Texas (RP160089), V Foundation (Translational Research Award), Robert A. Welch Foundation (I-1733) and the Howard Hughes Medical Institute (Faculty Scholars Award).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aird KM, Zhang G, Li H, Tu Z, Bitler BG, Garipov A, Wu H, Wei Z, Wagner SN, Herlyn M, et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Rep. 2013;3:1252–1265. doi: 10.1016/j.celrep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andronesi OC, Kim GS, Gerstner E, Batchelor T, Tzika AA, Fantin VR, Vander Heiden MG, Sorensen AG. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Science translational medicine. 2012;4:116ra114. doi: 10.1126/scitranslmed.3002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrzejewski S, Gravel SP, Pollak M, St-Pierre J. Metformin directly acts on mitochondria to alter cellular bioenergetics. Cancer Metab. 2014;2:12. doi: 10.1186/2049-3002-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao XR, Ong SE, Goldberger O, Peng J, Sharma R, Thompson DA, Vafai SB, Cox AG, Marutani E, Ichinose F, et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. Elife. 2016:5. doi: 10.7554/eLife.10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PE, Rubinstein WS, Myers EN, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8:967–975. doi: 10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Possemato R, Lorbeer FK, Bayraktar EC, Thiru P, Yucel B, Wang T, Chen WW, Clish CB, Sabatini DM. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature. 2014;508:108–112. doi: 10.1038/nature13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittar PG, Charnay Y, Pellerin L, Bouras C, Magistretti PJ. Selective distribution of lactate dehydrogenase isoenzymes in neurons and astrocytes of human brain. J Cereb Blood Flow Metab. 1996;16:1079–1089. doi: 10.1097/00004647-199611000-00001. [DOI] [PubMed] [Google Scholar]
- Boros LG, Torday JS, Lim S, Bassilian S, Cascante M, Lee WN. Transforming growth factor beta2 promotes glucose carbon incorporation into nucleic acid ribose through the nonoxidative pentose cycle in lung epithelial carcinoma cells. Cancer Res. 2000;60:1183–1185. [PubMed] [Google Scholar]
- Boudreau A, Purkey HE, Hitz A, Robarge K, Peterson D, Labadie S, Kwong M, Hong R, Gao M, Del Nagro C, et al. Metabolic plasticity underpins innate and acquired resistance to LDHA inhibition. Nat Chem Biol. 2016;12:779–786. doi: 10.1038/nchembio.2143. [DOI] [PubMed] [Google Scholar]
- Brand A, Singer K, Koehl GE, Kolitzus M, Schoenhammer G, Thiel A, Matos C, Bruss C, Klobuch S, Peter K, et al. LDHA-Associated Lactic Acid Production Blunts Tumor Immunosurveillance by T and NK Cells. Cell Metab. 2016;24:657–671. doi: 10.1016/j.cmet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- Burtscher M. Effects of living at higher altitudes on mortality: a narrative review. Aging Dis. 2014;5:274–280. doi: 10.14336/AD.2014.0500274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzai M, Jones RG, Amaravadi RK, Lum JJ, DeBerardinis RJ, Zhao F, Viollet B, Thompson CB. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 2007;67:6745–6752. doi: 10.1158/0008-5472.CAN-06-4447. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Mak TW. The current state of cancer metabolism. Nat Rev Cancer. 2016;16:613–614. [Google Scholar]
- Chang CH, Qiu J, O’Sullivan D, Buck MD, Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ, et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Liu Y, Lu C, Cross JR, Morris JPt, Shroff AS, Ward PS, Bradner JE, Thompson C, Lowe SW. Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes Dev. 2013;27:1974–1985. doi: 10.1101/gad.226613.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T, Sudderth J, Yang C, Mullen AR, Jin ES, Mates JM, DeBerardinis RJ. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc Natl Acad Sci U S A. 2011;108:8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, Yang XL, Mashimo T, Raisanen JM, Marin-Valencia I, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18:624–629. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco P. Does G6PD deficiency protect against cancer? A critical review. J Epidemiol Community Health. 1987;41:89–93. doi: 10.1136/jech.41.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coloff JL, Murphy JP, Braun CR, Harris IS, Shelton LM, Kami K, Gygi SP, Selfors LM, Brugge JS. Differential Glutamate Metabolism in Proliferating and Quiescent Mammary Epithelial Cells. Cell Metab. 2016;23:867–880. doi: 10.1016/j.cmet.2016.03.016. [DOI] [PubMed] [Google Scholar]
- Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curi R, Newsholme P, Newsholme EA. Metabolism of pyruvate by isolated rat mesenteric lymphocytes, lymphocyte mitochondria and isolated mouse macrophages. Biochem J. 1988;250:383–388. doi: 10.1042/bj2500383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SM, Jonas O, Keibler MA, Hou H, Luengo A, Mayers JM, Wyckoff J, Del Rosario A, Whitman M, Chin CR, et al. Direct evidence for cancer cell-autonomous extracellular protein catabolism in pancreatic tumors. Nat Med. 2016a doi: 10.1038/nm.4256. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, Bauer MR, Jha AK, O’Brien JP, Pierce KA, et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab. 2016b;23:517–528. doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayton TL, Gocheva V, Miller KM, Israelsen WJ, Bhutkar A, Clish CB, Davidson SM, Luengo A, Bronson RT, Jacks T, et al. Germline loss of PKM2 promotes metabolic distress and hepatocellular carcinoma. Genes Dev. 2016;30:1020–1033. doi: 10.1101/gad.278549.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bock K, Georgiadou M, Schoors S, Kuchnio A, Wong BW, Cantelmo AR, Quaegebeur A, Ghesquiere B, Cauwenberghs S, Eelen G, et al. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell. 2013;154:651–663. doi: 10.1016/j.cell.2013.06.037. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich LE, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC, Newman DK. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol. 2013;195:1371–1380. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erez A, DeBerardinis RJ. Metabolic dysregulation in monogenic disorders and cancer - finding method in madness. Nat Rev Cancer. 2015;15:440–448. doi: 10.1038/nrc3949. [DOI] [PubMed] [Google Scholar]
- Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Shen Z, Huang S, Zhao L, Chen S, Mak TW, Wang X. The ER UDPase ENTPD5 promotes protein N-glycosylation, the Warburg effect, and proliferation in the PTEN pathway. Cell. 2010;143:711–724. doi: 10.1016/j.cell.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- Fletcher M, Ramirez ME, Sierra RA, Raber P, Thevenot P, Al-Khami AA, Sanchez-Pino D, Hernandez C, Wyczechowska DD, Ochoa AC, et al. l-Arginine depletion blunts antitumor T-cell responses by inducing myeloid-derived suppressor cells. Cancer Res. 2015;75:275–283. doi: 10.1158/0008-5472.CAN-14-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flier JS, Mueckler MM, Usher P, Lodish HF. Elevated levels of glucose transport and transporter messenger RNA are induced by ras or src oncogenes. Science. 1987;235:1492–1495. doi: 10.1126/science.3103217. [DOI] [PubMed] [Google Scholar]
- Gaglio D, Soldati C, Vanoni M, Alberghina L, Chiaradonna F. Glutamine deprivation induces abortive s-phase rescued by deoxyribonucleotides in k-ras transformed fibroblasts. PLoS One. 2009;4:e4715. doi: 10.1371/journal.pone.0004715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- Gaude E, Frezza C. Tissue-specific and convergent metabolic transformation of cancer correlates with metastatic potential and patient survival. Nat Commun. 2016;7:13041. doi: 10.1038/ncomms13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger R, Rieckmann JC, Wolf T, Basso C, Feng Y, Fuhrer T, Kogadeeva M, Picotti P, Meissner F, Mann M, et al. L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell. 2016;167:829–842. doi: 10.1016/j.cell.2016.09.031. e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui DY, Sullivan LB, Luengo A, Hosios AM, Bush LN, Gitego N, Davidson SM, Freinkman E, Thomas CJ, Vander Heiden MG. Environment Dictates Dependence on Mitochondrial Complex I for NAD+ and Aspartate Production and Determines Cancer Cell Sensitivity to Metformin. Cell Metab. 2016;24:716–727. doi: 10.1016/j.cmet.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Teng X, Laddha SV, Ma S, Van Nostrand SC, Yang Y, Khor S, Chan CS, Rabinowitz JD, White E. Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev. 2016;30:1704–1717. doi: 10.1101/gad.283416.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi AA, Reznik E, Lee CH, Creighton CJ, Brannon AR, Luna A, Aksoy BA, Liu EM, Shen R, Lee W, et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer Cell. 2016;29:104–116. doi: 10.1016/j.ccell.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris IS, Treloar AE, Inoue S, Sasaki M, Gorrini C, Lee KC, Yung KY, Brenner D, Knobbe-Thomsen CB, Cox MA, et al. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell. 2015;27:211–222. doi: 10.1016/j.ccell.2014.11.019. [DOI] [PubMed] [Google Scholar]
- Henrich FC, Singer K, Poller K, Bernhardt L, Strobl CD, Limm K, Ritter AP, Gottfried E, Volkl S, Jacobs B, et al. Suppressive effects of tumor cell-derived 5’-deoxy-5’-methylthioadenosine on human T cells. Oncoimmunology. 2016;5:e1184802. doi: 10.1080/2162402X.2016.1184802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, Jiang L, Ko B, Skelton R, Loudat L, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC, et al. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosios AM, Hecht VC, Danai LV, Johnson MO, Rathmell JC, Steinhauser ML, Manalis SR, Vander Heiden MG. Amino Acids Rather than Glucose Account for the Majority of Cell Mass in Proliferating Mammalian Cells. Developmental cell. 2016;36:540–549. doi: 10.1016/j.devcel.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell JJ, Ricoult SJ, Ben-Sahra I, Manning BD. A growing role for mTOR in promoting anabolic metabolism. Biochem Soc Trans. 2013;41:906–912. doi: 10.1042/BST20130041. [DOI] [PubMed] [Google Scholar]
- Hu J, Locasale JW, Bielas JH, O’Sullivan J, Sheahan K, Cantley LC, Vander Heiden MG, Vitkup D. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat Biotechnol. 2013;31:522–529. doi: 10.1038/nbt.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung YP, Albeck JG, Tantama M, Yellen G. Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab. 2011;14:545–554. doi: 10.1016/j.cmet.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, Li J, Yu Y, Sasaki M, Horner JW, et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155:397–409. doi: 10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsen WJ, Vander Heiden MG. Pyruvate kinase: Function, regulation and role in cancer. Semin Cell Dev Biol. 2015;43:43–51. doi: 10.1016/j.semcdb.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzer A, German NJ, Gonzalez-Herrera KN, Asara JM, Haigis MC, Struhl K. Metformin and phenformin deplete tricarboxylic acid cycle and glycolytic intermediates during cell transformation and NTPs in cancer stem cells. Proc Natl Acad Sci U S A. 2014;111:10574–10579. doi: 10.1073/pnas.1409844111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA, Terada LS, Adams ND, McCabe MT, Pietrak B, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532:255–258. doi: 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Xiao L, Sugiura H, Huang X, Ali A, Kuro-o M, Deberardinis RJ, Boothman DA. Metabolic reprogramming during TGFbeta1-induced epithelial-to-mesenchymal transition. Oncogene. 2015;34:3908–3916. doi: 10.1038/onc.2014.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, Vander Heiden MG, Miller G, Drebin JA, Bar-Sagi D, et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015;75:544–553. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kats LM, Reschke M, Taulli R, Pozdnyakova O, Burgess K, Bhargava P, Straley K, Karnik R, Meissner A, Small D, et al. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell. 2014;14:329–341. doi: 10.1016/j.stem.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- Krall AS, Xu S, Graeber TG, Braas D, Christofk HR. Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nat Commun. 2016;7:11457. doi: 10.1038/ncomms11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Wilson FH, Ruth JR, Paulk J, Tsherniak A, Marlow SE, Vazquez F, Weir BA, Fitzgerald ME, Tanaka M, et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science. 2016;351:1214–1218. doi: 10.1126/science.aad5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehne A, Emmert H, Soehle J, Winnefeld M, Fischer F, Wenck H, Gallinat S, Terstegen L, Lucius R, Hildebrand J, et al. Acute Activation of Oxidative Pentose Phosphate Pathway as First-Line Response to Oxidative Stress in Human Skin Cells. Mol Cell. 2015;59:359–371. doi: 10.1016/j.molcel.2015.06.017. [DOI] [PubMed] [Google Scholar]
- Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM, et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16:992–1003. doi: 10.1038/ncb3039. 1001–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Qiu B, Lee DS, Walton ZE, Ochocki JD, Mathew LK, Mancuso A, Gade TP, Keith B, Nissim I, et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513:251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza-Puch F, Rooijers K, Buil LC, Zijlstra J, Oude Vrielink JF, Lopes R, Ugalde AP, van Breugel P, Hofland I, Wesseling J, et al. Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature. 2016;530:490–494. doi: 10.1038/nature16982. [DOI] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losman JA, Kaelin WG., Jr What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27:836–852. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt SY, Muralidhar V, Hosios AM, Israelsen WJ, Gui DY, Newhouse L, Ogrodzinski M, Hecht V, Xu K, Acevedo PN, et al. Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol Cell. 2015;57:95–107. doi: 10.1016/j.molcel.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher EA, Marin-Valencia I, Bachoo RM, Mashimo T, Raisanen J, Hatanpaa KJ, Jindal A, Jeffrey FM, Choi C, Madden C, et al. Metabolism of [U-13 C]glucose in human brain tumors in vivo. NMR in biomedicine. 2012;25:1234–1244. doi: 10.1002/nbm.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XL, Rajagopalan KN, Maddie M, Vemireddy V, Zhao Z, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15:827–837. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjon K, Cameron MJ, Quang P, Clasquin MF, Mandley E, Kunii K, McVay M, Choe S, Kernytsky A, Gross S, et al. MTAP Deletions in Cancer Create Vulnerability to Targeting of the MAT2A/PRMT5/RIOK1 Axis. Cell Rep. 2016;15:574–587. doi: 10.1016/j.celrep.2016.03.043. [DOI] [PubMed] [Google Scholar]
- Mavrakis KJ, McDonald ER, 3rd, Schlabach MR, Billy E, Hoffman GR, deWeck A, Ruddy DA, Venkatesan K, Yu J, McAllister G, et al. Disordered methionine metabolism in MTAP/CDKN2A–deleted cancers leads to dependence on PRMT5. Science. 2016;351:1208–1213. doi: 10.1126/science.aad5944. [DOI] [PubMed] [Google Scholar]
- Mayers JR, Torrence ME, Danai LV, Papagiannakopoulos T, Davidson SM, Bauer MR, Lau AN, Ji BW, Dixit PD, Hosios AM, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science. 2016;353:1161–1165. doi: 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiser J, Tumanov S, Maddocks O, Labuschagne CF, Athineos D, Van Den Broek N, Mackay GM, Gottlieb E, Blyth K, Vousden K, et al. Serine one-carbon catabolism with formate overflow. Sci Adv. 2016;2:e1601273. doi: 10.1126/sciadv.1601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelakis ED, Sutendra G, Dromparis P, Webster L, Haromy A, Niven E, Maguire C, Gammer TL, Mackey JR, Fulton D, et al. Metabolic modulation of glioblastoma with dichloroacetate. Sci Transl Med. 2010;2:31ra34. doi: 10.1126/scitranslmed.3000677. [DOI] [PubMed] [Google Scholar]
- Muller AJ, DuHadaway JB, Donover PS, Sutanto-Ward E, Prendergast GC. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nat Med. 2005;11:312–319. doi: 10.1038/nm1196. [DOI] [PubMed] [Google Scholar]
- Muller FL, Colla S, Aquilanti E, Manzo VE, Genovese G, Lee J, Eisenson D, Narurkar R, Deng P, Nezi L, et al. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature. 2012;488:337–342. doi: 10.1038/nature11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P, Bueno MJ, Zagorac I, Mondejar T, Sanchez J, Mouron S, Munoz J, Gomez-Lopez G, Jimenez-Renard V, Mulero F, et al. Targeting Tumor Mitochondrial Metabolism Overcomes Resistance to Antiangiogenics. Cell Rep. 2016;15:2705–2718. doi: 10.1016/j.celrep.2016.05.052. [DOI] [PubMed] [Google Scholar]
- Nelson SJ, Kurhanewicz J, Vigneron DB, Larson PE, Harzstark AL, Ferrone M, van Criekinge M, Chang JW, Bok R, Park I, et al. Metabolic imaging of patients with prostate cancer using hyperpolarized [1-(1)(3)C]pyruvate. Sci Transl Med. 2013;5:198ra108. doi: 10.1126/scitranslmed.3006070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, Park Y, Wright K, Pavlova NN, Tuveson DA, Thompson CB. The Utilization of Extracellular Proteins as Nutrients Is Suppressed by mTORC1. Cell. 2015;162:259–270. doi: 10.1016/j.cell.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MM, Sheaff MT, Szlosarek PW. Targeting arginine-dependent cancers with arginine-degrading enzymes: opportunities and challenges. Cancer Res Treat. 2013;45:251–262. doi: 10.4143/crt.2013.45.4.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, Leitch AM, Johnson TM, DeBerardinis RJ, Morrison SJ. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527:186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten M, von Knebel Doeberitz N, Oezen I, Wick W, Ochs K. Cancer Immunotherapy by Targeting IDO1/TDO and Their Downstream Effectors. Front Immunol. 2014;5:673. doi: 10.3389/fimmu.2014.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu B, Ackerman D, Sanchez DJ, Li B, Ochocki JD, Grazioli A, Bobrovnikova-Marjon E, Diehl JA, Keith B, Simon MC. HIF2alpha-Dependent Lipid Storage Promotes Endoplasmic Reticulum Homeostasis in Clear-Cell Renal Cell Carcinoma. Cancer Discov. 2015;5:652–667. doi: 10.1158/2159-8290.CD-14-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich S, Adler L, Yizhak K, Sarver A, Silberman A, Agron S, Stettner N, Sun Q, Brandis A, Helbling D, et al. Diversion of aspartate in ASS1-deficient tumours fosters de novo pyrimidine synthesis. Nature. 2015;527:379–383. doi: 10.1038/nature15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj L, Ide T, Gurkar AU, Foley M, Schenone M, Li X, Tolliday NJ, Golub TR, Carr SA, Shamji AF, et al. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature. 2011;475:231–234. doi: 10.1038/nature10167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Reitzer LJ, Wice BM, Kennell D. The pentose cycle. Control and essential function in HeLa cell nucleic acid synthesis. J Biol Chem. 1980;255:5616–5626. [PubMed] [Google Scholar]
- Robbins ME, Zhao W. Chronic oxidative stress and radiation-induced late normal tissue injury: a review. Int J Radiat Biol. 2004;80:251–259. doi: 10.1080/09553000410001692726. [DOI] [PubMed] [Google Scholar]
- Saha SK, Parachoniak CA, Ghanta KS, Fitamant J, Ross KN, Najem MS, Gurumurthy S, Akbay EA, Sia D, Cornella H, et al. Mutant IDH inhibits HNF-4alpha to block hepatocyte differentiation and promote biliary cancer. Nature. 2014;513:110–114. doi: 10.1038/nature13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, Harris IS, Holmes R, Wakeham A, Haight J, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, Brugge JS. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciacovelli M, Goncalves E, Johnson TI, Zecchini VR, da Costa AS, Gaude E, Drubbel AV, Theobald SJ, Abbo SR, Tran MG, et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature. 2016;537:544–547. doi: 10.1038/nature19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Sellers K, Fox MP, Bousamra M, 2nd, Slone SP, Higashi RM, Miller DM, Wang Y, Yan J, Yuneva MO, Deshpande R, et al. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J Clin Invest. 2015;125:687–698. doi: 10.1172/JCI72873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, Wei L, Fishbein MC, Czernin J, Mischel PS, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer cell. 2013;23:143–158. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EH, Livi CB, Rakheja D, Tan J, Benson D, Parekh V, Kho EY, Ghosh AP, Kirkman R, Velu S, et al. L-2-Hydroxyglutarate: an epigenetic modifier and putative oncometabolite in renal cancer. Cancer Discov. 2014;4:1290–1298. doi: 10.1158/2159-8290.CD-13-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, De Saedeleer CJ, Kennedy KM, Diepart C, Jordan BF, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118:3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, Kremer D, Hwang RF, Witkiewicz AK, Ying H, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536:479–483. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stams WA, den Boer ML, Beverloo HB, Meijerink JP, Stigter RL, van Wering ER, Janka-Schaub GE, Slater R, Pieters R. Sensitivity to L-asparaginase is not associated with expression levels of asparagine synthetase in t(12;21)+ pediatric ALL. Blood. 2003;101:2743–2747. doi: 10.1182/blood-2002-08-2446. [DOI] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson RU, Parker SJ, Eichner LJ, Kolar MJ, Wallace M, Brun SN, Lombardo PS, Van Nostrand JL, Hutchins A, Vera L, et al. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat Med. 2016;22:1108–1119. doi: 10.1038/nm.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes DB, Kfoury YS, Mercier FE, Wawer MJ, Law JM, Haynes MK, Lewis TA, Schajnovitz A, Jain E, Lee D, et al. Inhibition of Dihydroorotate Dehydrogenase Overcomes Differentiation Blockade in Acute Myeloid Leukemia. Cell. 2016;167:171–186. doi: 10.1016/j.cell.2016.08.057. e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szenajch J, Wcislo G, Jeong JY, Szczylik C, Feldman L. The role of erythropoietin and its receptor in growth, survival and therapeutic response of human tumor cells From clinic to bench - a critical review. Biochim Biophys Acta. 2010;1806:82–95. doi: 10.1016/j.bbcan.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Tateishi K, Wakimoto H, Iafrate AJ, Tanaka S, Loebel F, Lelic N, Wiederschain D, Bedel O, Deng G, Zhang B, et al. Extreme Vulnerability of IDH1 Mutant Cancers to NAD+ Depletion. Cancer Cell. 2015;28:773–784. doi: 10.1016/j.ccell.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titov DV, Cracan V, Goodman RP, Peng J, Grabarek Z, Mootha VK. Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science. 2016;352:231–235. doi: 10.1126/science.aad4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Lim JH, Chim H, Bhalla K, Girnun G, Pierce K, Clish CB, Granter SR, Widlund HR, Spiegelman BM, et al. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venneti S, Dunphy MP, Zhang H, Pitter KL, Zanzonico P, Campos C, Carlin SD, La Rocca G, Lyashchenko S, Ploessl K, et al. Glutamine-based PET imaging facilitates enhanced metabolic evaluation of gliomas in vivo. Sci Transl Med. 2015;7:274ra217. doi: 10.1126/scitranslmed.aaa1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel C, Riefke B, Grundemann S, Krebs A, Christian S, Prinz F, Osterland M, Golfier S, Rase S, Ansari N, et al. 3D high-content screening for the identification of compounds that target cells in dormant tumor spheroid regions. Exp Cell Res. 2014;323:131–143. doi: 10.1016/j.yexcr.2014.01.017. [DOI] [PubMed] [Google Scholar]
- Wheaton WW, Weinberg SE, Hamanaka RB, Soberanes S, Sullivan LB, Anso E, Glasauer A, Dufour E, Mutlu GM, Budigner GS, et al. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. Elife. 2014;3:e02242. doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. Exploiting the bad eating habits of Ras-driven cancers. Genes Dev. 2013;27:2065–2071. doi: 10.1101/gad.228122.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471:518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witney TH, James ML, Shen B, Chang E, Pohling C, Arksey N, Hoehne A, Shuhendler A, Park JH, Bodapati D, et al. PET imaging of tumor glycolysis downstream of hexokinase through noninvasive measurement of pyruvate kinase M2. Sci Transl Med. 2015;7:310ra169. doi: 10.1126/scitranslmed.aac6117. [DOI] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Weinberg RA. Epithelial-Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Mates JM, Alonso FJ, Wang C, Seo Y, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell metabolism. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Frederick DT, Wu L, Wei Z, Krepler C, Srinivasan S, Chae YC, Xu X, Choi H, Dimwamwa E, et al. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J Clin Invest. 2016;126:1834–1856. doi: 10.1172/JCI82661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Trachootham D, Liu J, Chen G, Pelicano H, Garcia-Prieto C, Lu W, Burger JA, Croce CM, Plunkett W, et al. Stromal control of cystine metabolism promotes cancer cell survival in chronic lymphocytic leukaemia. Nat Cell Biol. 2012;14:276–286. doi: 10.1038/ncb2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Cardaci S, Jerby L, MacKenzie ED, Sciacovelli M, Johnson TI, Gaude E, King A, Leach JD, Edrada-Ebel R, et al. Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nat Commun. 2015;6:6001. doi: 10.1038/ncomms7001. [DOI] [PMC free article] [PubMed] [Google Scholar]