Abstract
The association between GSTP1 A>G polymorphism and chemosensitivity of osteosarcoma is controversial according to previously published studies. We conducted this meta-analysis to further investigate the role of GSTP1 A>G genetic variation in response to chemotherapy resistance in patients with osteosarcoma. Using the electronic databases of Pubmed, Wanfang and CNIK were searched to find the studies related to the GSTP1 A>G polymorphism and chemosensitivity of osteosarcoma. The genotype of AA, AG and GG were extracted from the chemotherapy sensitivity and chemotherapy resistance group. The association between GSTP1 A>G polymorphism and chemosensitivity was calculated by STATA11.0 software. The correlation between GSTP1 A>G polymorphism and chemotherapy response was assessed by odds ratio (OR) and its 95% confidence interval (95%CI). Four studies with 681 cases were finally included in this meta-analysis. The pooled data indicated that there was no significant association between GSTP1 A>G polymorphism and chemosensitivity in patients with osteosarcoma [Homozygous genetic model (GG vs AA): OR=0.53, 95%CI: 0.25-1.12, P=0.10; recessive genetic model (GG vs GA+AA): OR=0.61, 95%CI:0.34-1.11,P=0.11; and dominant genetic model (GG+AG vs AA): OR=0.67, 95%CI:0.42-1.07,P=0.10]. No correlation between GSTP1 A>G polymorphism and chemosensitivity was found according to this present meta-analysis. However, the small number of cases in each included study and significant statistical heterogeneity among the trials means the conclusion should be regarded as conservative.
Keywords: GSTP1, Polymorphism, Chemosensitivity, Meta-analysis
1. Introduction
Osteosarcoma is one of the leading causes of malignant tumor related death for children and adolescents [1]. Epidemiology studies show that osteosarcoma is the eighth-most common form of childhood cancer, comprising 2.4% of all malignancies in pediatric patients, and about 20% of all primary bone cancers [2]. The prognosis of osteosarcoma is relative poor due to its preponderance for lung metastasis. The standard treatment for osteosarcoma is complete radical, surgical, en bloc resection of the cancer with adjuvant chemotherapy after operation or neoadjuvant chemotheray before surgical resection of the primary tumor [3]. Despite combined therapy, more than 30% of the patients showed the recurrence or metastatic disease during the first five years after diagnosis [2]. The clinical response to chemotherapeutics is a complex trait that is influenced by genetic and environmental factors [4]. The glutathione S-transferase pi gene (GSTP1) is a polymorphic gene encoding active, functionally different GSTP1 variant proteins that are thought to function in xenobiotic metabolism and play a role in susceptibility to cancer [5,6], and other diseases. Several studies have discussed the association between chemotherapy sensitivity of osteosarcoma and GSTP1 genetic polymorphism [7,8] but without conclusive results. Thus, we performed this meta-analysis in order to further evaluate the association between GSTP1 A>G polymorphism and chemosensitivity in patients with osteosarcoma.
2. Material and methods
2.1. Studies searching and inclusion
The electronic search was performed in the databases of Pubmed, Wanfang and CNKI to find the studies related to the GSTP1 A>G genetic variation in response to chemotherapy resistance in patients with osteosarcoma. The searching words were (GSTP1 OR glutathione S-transferase pi gene) AND osteosarcoma AND (polymorphisms OR variant). The search was last performed in November 11, 2015. The inclusion criteria were as follows: (1) The diagnosis of osteosarcoma should be confirmed by pathology or cytology; (2) The distribution of genotype can be extracted from each included study;(3) The papers were published in English or Chinese; (4) The patients in the included studies have not received neoadjuvant chemoradiation.
2.2. Data extraction
The full text of the included studies were carefully read. The related data and information for each included study were extracted by two reviewers (WU Fengfeng & YE Ruqing) independently. (1) The name of the first authors; (2) The year the research performed; (3) The distribution of genotype AA, AG and GG; (4) The methods of genotyping. (5) The chemotherapy regimen. The data were extracted independently by two reviewers and checked by a third reviewer as described in the Cochrane Handbook for systematic reviews.
2.3. Statistical analysis
All the analyses were performed by the STATA/SE 11.0 (StataCorp LP, http://www.stata.com) software. The odds of GG, GA and AA genotype in chemotherapy sensitivity group versus chemotherapy resistance group was demonstrated by the odds ratio (OR) and its 95% confidence intervals (CI). The statistical heterogeneity among the included articles was investigated by chi-square (χ2) test [9], and its inconsistency was tested by I2 [10]. The OR was pooled by random effect model if the statistical heterogeneity existed otherwise the fixed effect model was performed. The Begger’s funnel plot and Egger’s line regression tests were used to investigate the potential publication bias for the included studies [11].
3. Results
3.1. General information of the included studies
We electronically searched the databases of Pubmed, Wanfang and CNKI to find the suitable studies. Finally 4 studies with 681 cases were included in this meta-analysis according to the inclusion and exclusion criteria. All of the 4 papers were written in English. 3 studies use the PCR as the genotyping assay and 1 paper use the PCR-RFLP assay. The general characteristics for the included 4 papers is demonstrated in Table 1.
Table 1.
General information of the included 4 studies
| First author | Publication (year) | n | Genotyping | Chemotherapy regimen | Sensitivity | Resistance | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | AA | AG | GG | |||||
| Li[12] | 2014 | 162 | PCR | Doxorubicin+ methotrexate+ cisplatin | 44 | 33 | 11 | 24 | 30 | 20 |
| Liu[7] | 2014 | 186 | PCR | Doxorubicin+ methotrexate+ cisplatin | 48 | 37 | 13 | 30 | 36 | 22 |
| Teng[8] | 2013 | 146 | PCR | Adriamycin+ methotrexate+ cisplatin | 35 | 29 | 13 | 22 | 28 | 19 |
| Yang[13] | 2012 | 187 | PCR-RFLP | Doxorubicin+ methotrexate+ cisplatin | 54 | 26 | 17 | 56 | 23 | 11 |
4. Meta-analysis
4.1. Homozygous genetic model (GG vs AA)
In the homozygous genetic model (GG vs AA), significant statistical heterogeneity was found among the studies (I2=67.1%, P=0.03). So, the odds ratio was calculated by random effects model. The pooled results showed that patients with GG homozygous gene type was not associated with chemotherapy sensitivity (OR=0.53, 95%CI: 0.25-1.12, P=0.10), Figure 1.
Figure 1.
Forest plot of GSTP1 A>G polymorphisms and chemosensitivity of osteosarcoma in omozygous genetic model (GG vs AA)
4.2. Recessive genetic model (GG vs GA+AA)
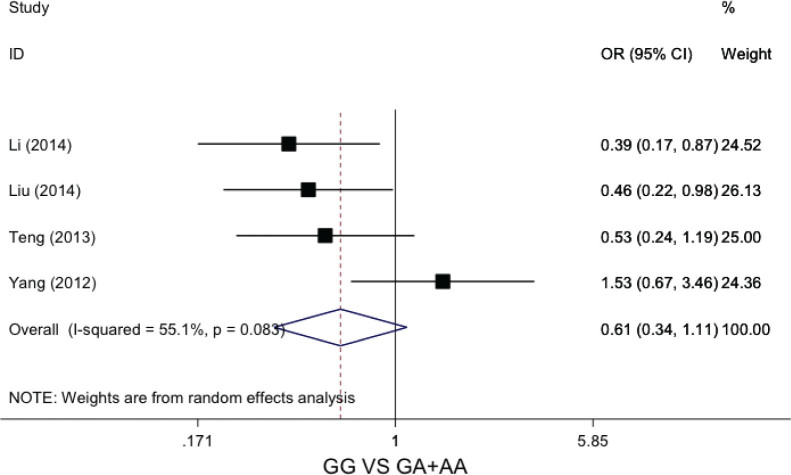
In the recessive genetic model (GG vs GA+AA), the statistical heterogeneity was not significant (I2=55.1%, P=0.08). We pooled the data by fixed effects model. The calculated data indicated that patients with GG genotype did not increase the chemotherapy sensitivity compared to GA or AA genotype (OR=0.61, 95%CI: 0.34-1.11, P=0.11), Figure 2.
Figure 2.
Forest plot of GSTP1 A>G polymorphisms and chemosensitivity of osteosarcoma in omozygous genetic model (GG vs GA+AA)
4.3. Dominant genetic model (GG+AG vs AA)
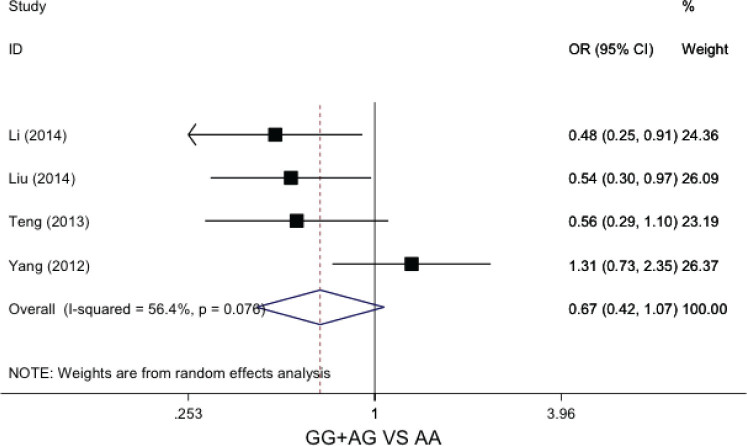
In the dominant genetic model (GG+AG vs AA), no significant statistical heterogeneity was not found across the included studies (I2=56.4%, P=0.07). The data was calculated by fixed effects model. The pooled results showed that patients with GG or AG genotype was not sensitive to chemotherapy (OR=0.67, 95%CI: 0.42-1.07, P=0.10), Figure 3.
Figure 3.
Forest plot of GSTP1 A>G polymorphisms and chemosensitivity of osteosarcoma in dominant genetic model (GG+AG vs AA)
4.4. Publication bias
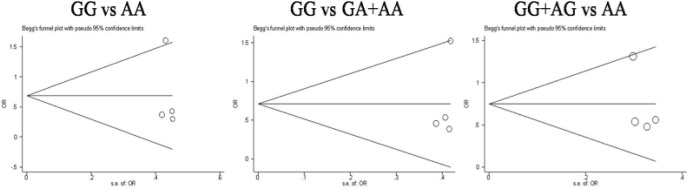
The publication bias for the three genetic models were evaluated by Egger’s line regression test and begger’s funnel plot. No significant publication bias was found in the three genetic models by Egger test (PGG vs AA =0.71; P GG vs GA+AA=0.49; PGG+AG vs AA =0.42) and begger’s funnel plot, Figure 4.
Figure 4.
The funnel plot for publication bias evaluation
5. Discussion
Generally, GSTP1 plays a critical role in the development of carcinogenesis for several malignant tumors [14-17]. It was indicated that the polymorphisms of GSTP1 was associated with endogenous DNA damage. Some epidemiological researches revealed the association between polymorphisms of GSTP1 and cancer risk [17-19].
Recently, some studies showed that the GSTP1A>G polymorphism can affect the chemotherapy sensitivity in patients with osteosarcoma [8,20]. Teng et al [8] have performed a comprehensive study investigating the association between GSTP1 genetic variation and chemotherapy sensitivity or clinical outcome in patients with osteosarcoma. In their study, the author found that subjects with the GSTP 1 GG single nucleotide were more likely to be resistant to chemotherapy(P<0.05). But another study performed by Yang included 187 cases of osteosarcoma and discussed the association between GSTP1 GSTP1A>G polymorphism and chemotherapy resistance [13]. They found that the patients with GSTP1 GG/GA genotypes had significantly higher rates of response to chemotherapy, with adjusted OR (95% CI) of 2.19 (1.15-6.21). The results of Teng and Yang were quite different from each other.
Because of the conflicting results showed by previous study, we performed this meta-analysis in order to further evaluate the role of GSTP1 A>G genetic variation in response to chemotherapy sensitivity in patients with osteosarcoma. In our study we found that there was no association between GSTP1 A>G polymorphism and chemosensitivity in patients with osteosarcoma [Homozygous genetic model (GG vs AA): OR=0.53, 95%CI: 0.25-1.12, P=0.10; recessive genetic model (GG vs GA+AA): OR=0.61, 95%CI:0.34-1.11,P=0.11; and dominant genetic model (GG+AG vs AA): OR=0.67, 95%CI:0.42-1.07,P=0.10]. But there were two major limitations in our study. Firstly, significant heterogeneity was found among the included articles which could decrease the statistical power to find the difference between groups. Secondly, only four studies with small number of cases were included in this meta-analysis. The small number of included papers also reduces the statistical power. Therefore, in order to explicate the association between GSTP1 A>G genetic variation in response to chemotherapy resistance in patients with osteosarcoma, further large sample multicenter studies are greatly needed.
Footnotes
Conflict of interest statement: Authors state no conflict of interest.
References
- Faisham WI, Mat SAZ, Alsaigh LN. et al. Prognostic factors and survival rate of osteosarcoma: A single-institution study. Asia Pac J Clin Oncol. 2015 doi: 10.1111/ajco.12346. [DOI] [PubMed] [Google Scholar]
- Ottaviani G, Jaffe N.. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- Hansen AR, Hughes BG, Paul S. et al. Single institution retrospective review of perioperative chemotherapy in adult and adolescent patients with operable osteosarcoma. Asia Pac J Clin Oncol. 2014 doi: 10.1111/ajco.12167. [DOI] [PubMed] [Google Scholar]
- Nilsonne U.. Epidemiology and some prognostic factors in osteosarcoma in Sweden. La semaine des hopitaux : organe fonde par l’Association d’enseignement medical des hopitaux de Paris. 1982;58(30-31):1727–1728. [PubMed] [Google Scholar]
- Oliveira C, Lourenco GJ, Sagarra RA, Derchain SF, Segalla JG, Lima CS.. Polymorphisms of glutathione S-transferase Mu 1 (GSTM1), Theta 1 (GSTT1), and Pi 1 (GSTP1) genes and epithelial ovarian cancer risk. Dis Markers. 2012;33(3):155–159. doi: 10.3233/DMA-2012-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiers I, Shanks JH, Bostwick DG.. Glutathione S-transferase pi (GSTP1) hypermethylation in prostate cancer: review. Pathology. 2007;39(3):299–304. doi: 10.1080/00313020701329906. [DOI] [PubMed] [Google Scholar]
- Liu S, Yi Z, Ling M, Shi J, Qiu Y, Yang S.. Predictive potential of ABCB1, ABCC3, and GSTP1 gene polymorphisms on osteosarcoma survival after chemotherapy. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(10):9897–9904. doi: 10.1007/s13277-014-1917-x. [DOI] [PubMed] [Google Scholar]
- Teng JW, Yang ZM, Li J, Xu B.. Predictive role of Glutathione S-transferases (GSTs) on the prognosis of osteosarcoma patients treated with chemotherapy. Pakistan journal of medical sciences. 2013;29(5):1182–1186. doi: 10.12669/pjms.295.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerSimonian R, Laird N.. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG, Deeks JJ, Altman DG.. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger M, Davey SG, Schneider M, Minder C.. Bias in meta-analysis detected by a simple graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Tian ZQ, Jiang SN, Feng T.. Effect of variation of ABCB1 and GSTP1 on osteosarcoma survival after chemotherapy. Genet Mol Res. 2014;13(2):3186–3192. doi: 10.4238/2014.April.25.3. [DOI] [PubMed] [Google Scholar]
- Yang LM, Li XH, Bao CF.. Glutathione S-transferase P1 and DNA polymorphisms influence response to chemotherapy and prognosis of bone tumors. Asian Pac J Cancer Prev. 2012;13(11):5883–5886. doi: 10.7314/apjcp.2012.13.11.5883. [DOI] [PubMed] [Google Scholar]
- Sugimoto M.. [Glutathione S-transferases (GSTs)]. Nihon rinsho. Japanese journal of clinical medicine. 1995;53(5):1253–1259. [PubMed] [Google Scholar]
- Belchik SM, Xun L.. S-glutathionyl-(chloro)hydroquinone reductases: a new class of glutathione transferases functioning as oxidoreductases. Drug Metab Rev. 2011;43(2):307–316. doi: 10.3109/03602532.2011.552909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Wang Z, Zhang W. et al. Association between glutathione S-transferases M1 and T1 gene polymorphisms and prostate cancer risk: a systematic review and meta-analysis. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(1):247–256. doi: 10.1007/s13277-013-1030-6. [DOI] [PubMed] [Google Scholar]
- Strange RC, Fryer AA.. The glutathione S-transferases: influence of polymorphism on cancer susceptibility. IARC Sci Publ. 1999;(148):231–249. [PubMed] [Google Scholar]
- Zhou CF, Ma T, Zhou DC, Shen T, Zhu QX.. Association of glutathione S-transferase pi (GSTP1) Ile105Val polymorphism with the risk of skin cancer: a meta-analysis. Arch Dermatol Res. 2015;307(6):505–513. doi: 10.1007/s00403-015-1576-9. [DOI] [PubMed] [Google Scholar]
- Park JY, Schantz SP, Stern JC, Kaur T, Lazarus P.. Association between glutathione S-transferase pi genetic polymorphisms and oral cancer risk. Pharmacogenetics. 1999;9(4):497–504. [PubMed] [Google Scholar]
- Zhang SL, Mao NF, Sun JY, Shi ZC, Wang B, Sun YJ.. Predictive potential of glutathione S-transferase polymorphisms for prognosis of osteosarcoma patients on chemotherapy. Asian Pac J Cancer Prev. 2012;13(6):2705–2709. doi: 10.7314/apjcp.2012.13.6.2705. [DOI] [PubMed] [Google Scholar]