Abstract
In vertebrates, the gonad arises as a bipotential primordium that can differentiate as a testis or ovary. Cells are initially primed to adopt either fate by balanced antagonistic signaling pathways and transcription networks. Sexual fate is determined by activating the testis or ovarian pathway and repressing the alternative pathway. A complex, dynamic transcription network underlies this process, as approximately half the genome is being transcribed during this period, and many genes are expressed in a sexually dimorphic manner. This network is highly plastic, however multiple lines of evidence suggest that many elements of the pathway converge on the stabilization or disruption of Sox9 expression. The single gene mutational approach has led to the identification of ~30 additional genes involved in vertebrate sex determination. However,>50% of human disorders of sexual development (DSDs) are not explained by any of these genes, suggesting many critical elements of the system await discovery. Emerging technologies and genetic resources enable the investigation of the sex determination network on a global scale in the context of a variable genetic background or environmental influences. Using these new tools we can investigate how cells establish a bipotential state that is poised to adopt either sexual fate, and how they integrate multiple signaling and transcriptional inputs to drive a cell fate decision. Elucidating the genetic architecture underlying sex determination in model systems can lead to the identification of conserved modules correlated with phenotypic outcomes, and critical pressure points in the network that predict genes involved in DSDs in humans.
Keywords: Sex determination, Sry, Sox9, Sex reversal, Systems biology, Systems genetics, eQTL
Genetic sex is established at conception when an X- or Y-bearing sperm fertilizes an X- bearing ovum. However, male or female sexual development is not initiated until mid-gestation, as the result of the decision within the bipotential gonad to differentiate as a testis or ovary. Sexually dimorphic differentiation of the gonad is the first step in male and female development, occurring prior to the production of hormones, which control subsequent steps of phenotypic sexual development.
Two decades have passed since the discovery that Sry (sex determining region of Chr Y) acts as a dominant genetic determinant of sex in mammals. We have since learned that this “dominant determinant” acts more like a nudge1. It is expressed at low levels in the testis, and may influence the underlying transcriptional network by directly up-regulating only one gene, Sox9 (SRY-box containing gene 9). The idea that the female is a default pathway has been discarded, and a more complex and nuanced view of sex determination is emerging. Cells of the gonad primordium are initially poised between sexual fates by balanced antagonistic intercellular signaling pathways and intracellular transcription networks. Sexual fate is established during a brief temporal window, and requires both the activation of the testis or ovarian pathway and repression of the alternative pathway. A large, dynamic transcription network underlies this process, as evidenced by microarray profiles showing that fully half of all genes in the genome are expressed in XX and XY gonads during the critical window of sex determination.
Approximately ten percent of expressed genes exhibit a sexually dimorphic pattern within 24 hours following the onset of Sry expression. However, mutational analyses in mice have confirmed roles in early gonadogenesis and/or primary sex determination for only approximately 30 genes. More than 50% of human sex reversal cases remain unexplained by variation in SRY or any of these known genes. Moreover, numerous genomic regions that modify sex reversal phenotypes in inbred mouse strains have been mapped, and many of these do not harbor genes with known roles in sex determination. These deficiencies in our understanding call for a more global approach.
It seems likely that an evolutionarily-conserved framework underlies sex determination, but if so, what is its structure? How is sexual plasticity conferred at the level of the underlying transcription network, maintained for several days, and ultimately disrupted to drive the testis or ovarian pathways? How is a high level of expression variability tolerated in the network? If many genes must act in a combinatorial manner to buffer this variability, what are the limits of the system? What are the critical nodes in the network, and how are they regulated? Is there an underlying modularity in the network associated with various aspects of the sexual fate decision?
Answers to these questions will require approaches that provide a systems-level perspective and integrate multiple sources of global molecular data (including genetic variability within and among species, as well as differences at the level of the transcriptome and proteome) and link them to phenotypic outcomes. In this focus article, we take a systems perspective to review the critical first 36 hours of gonadogenesis in mice, culminating in gonadal sex determination. We place special emphasis on the establishment and maintenance of sexual fate within a single cell, as well as the spatiotemporal establishment of sexual fate across the entire field of the gonad. Throughout, we highlight holes in our current understanding and recent studies that seek to address these gaps. We conclude with a discussion of emerging systems-level approaches and genetic resources and how they will provide a broader perspective of the gonad as a differentiating organ system. Readers that seek a more detailed molecular understanding of testis and ovary development are directed to recent comprehensive reviews by Wainwright et al2 and Liu et al3.
Building a Bipotential Developmental Network
In mice, the gonads arise around embryonic day 10 (E10.0) as paired thickenings of the coelomic epithelium covering the ventral-medial surface of the mesonephros. Both the timing and spatial aspects of the initial cell proliferation that gives rise to the gonad appear quite precise, yet surprisingly little is understood about what underlies the specification and boundaries of this gonad field. Mutational analyses have identified a handful of genes that are required for this first phase of proliferation in both sexes, including SF1/Nr5a1 (nuclear receptor subfamily 5, group A, member 1), Wt1 (Wilms tumor 1 homolog), M33/Cbx2 (chromobox homolog 2), Lhx9 (LIM homeobox protein 9), Emx2 (empty spiracles homolog 2), and Igf1r (insulin-like growth factor I receptor)/Insr (insulin receptor)/Insrr (insulin receptor-related receptor). Disruption of any of these genes results in varying degrees of gonad dysgenesis. Microarray analyses of gonad somatic cells at this early stage revealed a vast, active transcription network that is similar in both sexes before E11.54, 5. Thousands of genes are expressed early in the bipotential gonad, including a large number of homeobox, zinc finger, and SOX (Sry-box) transcription factors. Many of these genes may play a role in specifying the spatial boundaries for the initial waves of cell proliferation, or in regulating the plasticity of this cell population.
For the first 36 hours of their development the gonad primordia remain bipotential, competent to develop as testes or ovaries irrespective of genetic sex. The early gonad primordium consists of 2–3 somatic precursor cell types, including supporting cell precursors (so called because of their role in supporting the development of germ cells throughout reproductive life) and steroidogenic cell precursors. In addition, primordial germ cells, which are specified extra-embryonically at the base of the allantois, finish their migration through the hindgut and begin to colonize the gonad as early as E10.5. Surprisingly, all of these precursor cell types are themselves initially bipotential and proceed to adopt sex-specific fates in the testis and ovary.
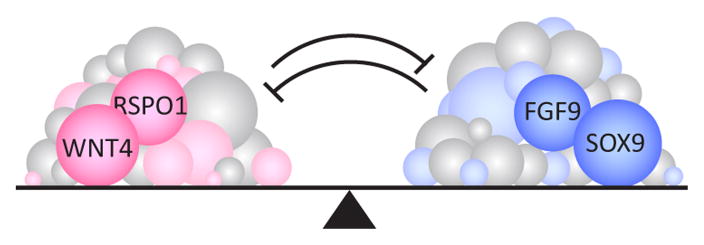
The plasticity of the gonad at the bipotential stage may stem in part from a balanced network state established by the antagonistic intercellular signals WNT4 (wingless-related MMTV integration site 4) and FGF9 (fibroblast growth factor 9)6. Before E11.5, Fgf9 and Wnt4 are expressed in both XY and XX gonads – Fgf9 is expressed along the anteroposterior (AP) axis in the coelomic domain at the surface of the gonad, while Wnt4 is expressed at the gonad-mesonephros border. These opposing signaling pathways may hold gonadal cells in an undifferentiated state. It is not clear how the boundaries of these two expression domains are established and maintained, nor is it known by what mechanism cells process this balanced signaling state to remain undifferentiated. However, based on the large number of genes expressed at this stage, it seems likely that many other factors contribute to this balanced network (Fig. 1).
Figure 1.
Many genes may be involved in establishing a bipotential state in the early gonad. Balanced antagonistic signaling pathways, including FGF9 and WNT4, hold somatic precursor cells in an undifferentiated state. The transcriptome is highly active and complex during this window, which suggests that many more genes and pathways are involved in conferring this plasticity.
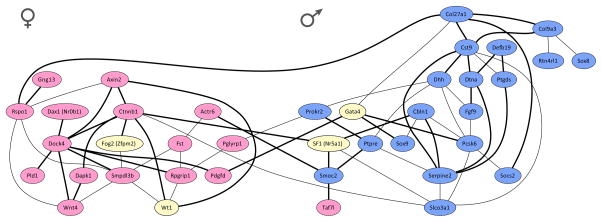
What is the transcription signature of the bipotential network that exists in early XY and XX gonads? One clue from a recent study7 suggests an intracellular battle of the sexes. We quantified gene expression in E11.5 XY gonads from a large genetically heterogeneous population from an intercross of C57BL/6J (B6) and 129S1/SvImJ (129S1) inbred strains, and found an unexpectedly high level of expression variability across individuals, despite the fact that samples were stage-matched and all were at or past the close of the bipotential window. Using this expression variability, which was driven by genetic variation segregating in the population, we identified two distinct clusters of coexpressed genes (Fig. 2). Surprisingly, when these genes were annotated based on their later association with the male or female differentiation pathway (based on known functional data or sexually dimorphic expression in XY/XX microarray experiments), cluster membership was largely restricted to male- or female-associated genes. Thus, at the end of the bipotential window as the male pathway is first being activated, a clear transcriptional signature of the female pathway is still evident in XY gonads.
Figure 2.
Evidence for a transcriptional tug-of-war underlying sex determination. A coexpression network was estimated for a subset of 40 genes quantified in 68 XY gonad samples from a mixed F2 intercross of B6 and 129S1. ‘Male’ genes that are enriched in XY gonads at E11.5 are highlighted in blue, whereas ‘female’-enriched genes are highlighted in pink, and genes with known roles in sex determination that are not expressed in a sexually-dimorphic pattern at E11.5 are highlighted in yellow. Thick black edges represent more robust coexpression relationships (partial correlation coefficient ≥ 0.33), while thinner edges are less robust but still significant (partial correlation coefficient ≥ 0.25).
Permission pending from Cold Spring Harbor Press
At first glance, this suggests that the male pathway is being superimposed on an underlying female pathway. However, there is also evidence for the expression of genes later associated with the male pathway in E11.5 XX gonads, even though Sry is not present8. This finding suggests that the early gonad progenitors in both XY and XX gonads are primed to adopt either fate, and sex determination proceeds as a consequence of a tug-of-war between male and female gene sub-networks. Perhaps this dual lineage priming is a characteristic of “bipotential cells” in general. This idea is consistent with the identification of XX patients that develop a testis in the absence of SRY. The cause of female-to-male sex reversal in some of these cases is disruption of the WNT4/RSPO1 (R-spondin homolog)/β-catenin signaling pathways that oppose male development in XX gonads9, 10, however it is likely that many other genes are involved.
Interestingly, the gonad retains the ability to form a testis or ovary irrespective of genetic sex from its origin at E10.0 until ~E11.5. However, if the male pathway is not engaged by this time, the ovarian pathway is stabilized and resistant to perturbation11. This defines a critical window, and suggests that elucidating the expression dynamics of the network as it evolves will be important.
Disrupting the Bipotential State
Primary sex determination refers to the decision within the bipotential gonad to become a testis or an ovary, a process in mammals that normally hinges on the presence or absence of the Y Chromosome. In a series of experiments analyzing XY-XX chimeric testes12, Palmer and Burgoyne showed that only the somatic supporting cell lineage (i.e. pre-Sertoli cells) exhibited a significant bias for the presence of the Y Chromosome, suggesting that the testis-determining gene on the Y acts autonomously in supporting cell precursors, and sexual differentiation of all other cell types proceeds from intercellular interactions with this population. Shortly thereafter, Sry was identified as the Y-linked genetic switch underlying male sex determination in mammals and shown to be expressed in pre-Sertoli cells as predicted13–15.
Sry encodes a high mobility group (HMG)-box transcription factor that is transiently expressed in pre-Sertoli cells from ~E10.5–12.016. Little is known about the regulatory mechanisms underlying Sry expression in a brief and tightly constrained spatiotemporal pattern. Sry is first expressed in cells at the center of the gonad, and then spreads to the anterior and then posterior poles17, 18. Expression is extinguished following the same pattern, and individual cells are believed to express Sry for <7 hours15. Loss-of-function experiments implicated a few genes as potential regulators of Sry expression, including Wt119, 20, Nr5a1/SF121, Lhx922, Emx223, Cbx2/M3324, Igf1r/Insr/Insrr25, and the combination of Gata4 (GATA binding protein 4)/Fog2/Zfpm2 (zinc finger protein, multitype 2)26. However, as many of these genes are also involved in the formation of the gonad primordium, it is difficult to determine whether they directly regulate Sry or affect the initial proliferation of the pre-Sertoli lineage. In eQTL studies, an unidentified modifier of Sry expression has been mapped on Chromosome 17. ChIP assays will be necessary to identify direct regulators of Sry.
It was initially assumed that Sry would be found to directly regulate numerous genes and pathways, however 20 years later, the consensus view is that the sole function for Sry during sex determination may be to upregulate the expression of one critical downstream target, Sox9 (SRY-box containing gene 9)27. In fact, Sry is not conserved across vertebrates, and appears to act as the male trigger only in eutherian mammals. Other genes can act as the genetic switch, such as DMRT1 (DM-related transcription factor 1) in the chicken28 and its related homolog DMY (DM-related transcription factor on Chr Y) in some species of medaka fish29. In other vertebrate species, environmental mechanisms (such as temperature or even population density) can act as the trigger of male development30. Perhaps most surprisingly, coincident genetic and environmental sex determining mechanisms have been found within a single species, illustrating the flexibility at the top of the male pathway31, 32. This suggests that many genes could potentially assume the role of the genetic sex determination switch. However, Sox9 seems to be a conserved element of the male pathway in all systems that have been investigated. Thus the constraint limiting the evolution of new potential switch genes may be their ability to activate/stabilize Sox9 expression during a critical temporal window of development. Importantly, this emerging model does not preclude the possibility that the genetic or environmental switch in some species activates the female rather than the male pathway, although it does predict that a female switch would directly or indirectly repress Sox9 expression33.
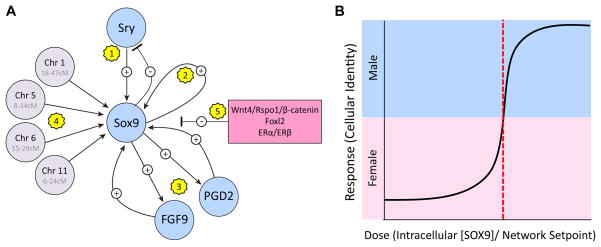
Like Sry, Sox9 encodes a HMG-box transcription factor that is thought to bind and bend DNA. Sox9 is expressed at basal levels in both XY and XX gonads before E11.0, but is thereafter upregulated by SRY in XY gonads and extinguished in XX gonads. Sox9 expression is tightly regulated and buffered against perturbation by a complex network of intra- and intercellular feedforward loops. Indeed, this regulatory complexity provides strong support for the importance of Sox9 to the male differentiation pathway. Following the initial “nudge” by SRY1, Sox9 expression is amplified and then maintained by multiple feed-forward loops (Fig. 3). SOX9 is able to bind its own promoter with higher affinity than SRY27, and consequently displaces SRY and activates its own expression in an autoregulatory loop. In addition, multiple intercellular signaling pathways, including FGF96 and PGD2 (Prostaglandin D2)34, act as independent feedforward loops to amplify Sox9 expression in pre-Sertoli cells. These signaling pathways may act as a failsafe mechanism to recruit cells to the Sertoli fate in extreme cases where too few Sox9-expressing cells are specified at the outset12. We have mapped at least four additional modifiers of Sox9 expression (expression QTLs)7 that do not harbor genes with known functions during sex determination. Two of these regions, on Chromosomes 1 and 11, also mapped as strain-specific modifiers of sex reversal (phenotypic QTLs) in the hemizygous Dax1-/Y (Nr0b1, Nuclear receptor subfamily 0, group b, member 1) and YDOM mouse models, and both QTLs enhanced susceptibility to or protection from sex reversal by affecting Sox9 expression35, 36. Finally, Sox9 expression is regulated directly or indirectly by female pathway genes (see below)6, 37. The complexity in the regulation of SOX9 that has been uncovered by recent experiments reveals many negative and positive feedback loops that may be a more general characteristic of this mutually antagonistic and highly canalized system.
Figure 3.
Sex determination within individual XY supporting cells depends on the level of Sox9 expression.
A. The complexity of Sox9 regulation points to its importance as the conserved regulatory hub of the male differentiation pathway. Sox9 is expressed at basal levels early in both XY and XX gonads. 1) A pulse of Sry expression activates the initial upregulation of Sox9 in XY somatic supporting cell precursors. Sox9 may or may not be involved in extinguishing Sry expression. 2) Following the initial activation by SRY, SOX9 is able to bind its own promoter with higher affinity than SRY and act in an autoregulatory feedforward loop. 3) Intercellular Prostaglandin D2 (PGD2) and FGF9 signaling pathways act as independent feedforward loops to amplify Sox9 expression downstream of Sry-mediated activation. 4) Many other genes are likely to directly or indirectly regulate Sox9 expression or function, including unidentified genes underlying expression QTLs on mouse Chromosomes 1, 5, 6, and 11. 5) Multiple genes associated with the female differentiation pathway, including Wnt4/Rspo1/β-catenin, Foxl2 (forkhead box L2), and the estrogen receptors ERα and ERβ, are likely to repress Sox9 expression to canalize the female pathway. It is important to note that one or more of the genes underlying the Sox9 eQTLs could influence Sox9 expression indirectly by activating or repressing female pathway genes.
B. Within individual gonad supporting cells, sexual fate is likely to be a binary decision. The intracellular concentration of SOX9 most likely must reach a critical threshold (denoted by a red dotted line) relative to the underlying setpoint of the network before E11.5 to impose a male-specific state on the transcription network. Cells that fail to express SOX9 above this threshold adopt the alternative female-specific network state.
Stabilizing the Sex Determination Decision
Within gonad supporting cells, we hypothesize that the sexual fate decision operates as a bistable switch followed by stabilization mechanisms. Despite the high level of expression variability observed during sex determination, individual cells seem to adopt one of only two network states38. Concurrent male- and female-associated network motifs are present in uncommitted supporting cells early during the bipotential period (see Fig. 2). However, as this critical window closes around E11.5, the intracellular transcription network begins to resolve to a male- or female-specific state. These male- and female-specific states appear to be quite stable and robust to a wide range of fluctuations in gene expression levels. State switching (i.e., trans-differentiation from a Sox9-expressing Sertoli cell to a Foxl2-expressing granulosa cell37) is rare. This observation, along with strict temporal constraints39, suggests that the cellular establishment of a sex-specific transcriptional state in response to a lineage-specific differentiation factor is bistable, non-linear, and ultrasensitive.
All current evidence in vertebrates points to SOX9 as the critical differentiation factor in males40 (Fig. 3B). It seems likely that the nuclear concentration of SOX9 must reach a critical threshold (denoted in Figure 3B by a red dotted line) before E11.5 to impose a male-specific state on the underlying transcription network41, 42. Below this threshold, Sox9 expression may be insufficient to drive the male pathway, thus supporting cells adopt a female network state. This is the case in all XX cells where Sox9 is initially expressed at a low level but is never upregulated above the threshold, as well as in XY cells with defects in one or more of the positive feedback pathways outlined in Figure 3A. Indeed, one requirement for bistable switch-like behavior is the presence of a feedforward loop. PGD234 and FGF96 signaling pathways, as well as SOX9 autoregulation of its own enhancer27, fulfill this requirement.
The ability to repress the alternative pathway may be of equal importance for establishing a bistable differentiation switch in gonad supporting cells. In XX or XY gonads, the supporting cell fate is defined as much by the genes that are silenced as by the genes that are expressed8. XX cells silence male pathway genes and XY cells silence female pathway genes. Surprisingly, evidence suggests that the sexual fate decision must be actively maintained and the alternative sex must be repressed well after sex is determined. In a case where repression of Sox9 is not maintained in adult females, XX cells transdifferentiate to the alternative cell fate (i.e. granulosa cells transdifferentiate to Sertoli-like cells)37. Similarly, loss of Dmrt1 leads to loss of Sox9 in the adult testis and reprogramming of XY cells to the female fate43.
In bipotential XY gonads, the levels of female pathway genes likely provide an initial set point for the network and establish a state-switching threshold for SOX9 expression. Evidence from studies of genetic background effects on XY sex reversal suggests that Sox9 must be expressed above some threshold relative to this underlying baseline to adopt and maintain the Sertoli cell fate. The female gene-derived set point may vary among individual supporting cells in the gonad, and consequently each cell may have a unique threshold for establishment and maintenance of the male pathway. New gene expression platforms are sensitive enough to accurately quantify gene expression in single cells44, 45, and used in combination with laser capture microdissection (LCM) to isolate individual fluorescently-labeled early gonad somatic cells (e.g. SF1-eGFP4), now make it possible to map dynamic gene expression changes in individual cells across this gonad field at the earliest stages of differentiation. Importantly, community-effect mechanisms exist that may override the micro-heterogeneity in the population and recruit the entire field to the male or female pathway under normal conditions (see below).
Propagating the Decision Across the Gonad Field
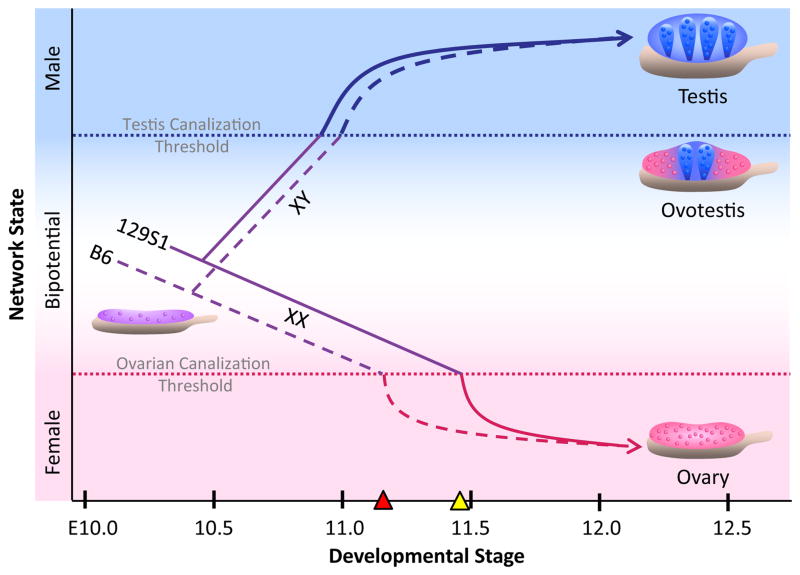
All data to date suggest that sexual fate of the gonad depends on the number of specified pre-Sertoli cells (i.e., cells that are expressing SOX9 above a critical threshold) present in the gonad when the bipotential window closes around E11.5 (Fig. 4). This critical timepoint appears to be set by the expression of female pathway genes in the bipotential XY gonad. Once the female pathway has progressed to a “point of no return” (termed the Ovarian Canalization Threshold, OCT), the number of specified pre-Sertoli cells in the XY gonad must surpass a threshold (termed the Testis Canalization Threshold, TCT) to direct testis development. It is unclear how female pathway genes stably repress the male differentiation pathway after this critical timepoint. Expression of female pathway genes above the OCT may render individual XY cells unable to express SOX9 at high enough levels to activate the male pathway. Alternatively, the female pathway may repress the male pathway by altering chromatin structure, effectively limiting the exposure of downstream male effectors to SOX9, and blocking establishment of the Sertoli fate.
Figure 4.
Sexual fate of the XY gonad depends on the number of specified pre-Sertoli cells (i.e., cells that are expressing SOX9 above a differentiation threshold) present in the gonad when the bipotential window closes. This critical timepoint appears to be set by the expression of female pathway genes in the bipotential XY gonad. Once the female pathway has progressed to a “point of no return” (the Ovarian Canalization Threshold, OCT; denoted as a red dotted line), the number of specified pre-Sertoli cells in the XY gonad must surpass a threshold (the Testis Canalization Threshold; blue dotted line) to direct testis development. If the number of pre-Sertoli cells does not meet this threshold by this critical timepoint, the XY gonad full or partially sex reverses to an ovary or ovotestis. In XX gonads, the female pathway surpasses the OCT and is canalized toward ovarian development. Variation in the expression of female pathway genes in XY gonads among inbred strains may affect the duration of the bipotential window. For example, the higher expression or earlier onset of female pathway genes in B6 XY gonads (dashed purple line) relative to 129S1 (solid purple line) may cause the bipotential window to close earlier in B6 (denoted by red triangle; compare to yellow triangle = critical timepoint in 129S1). In XX gonads, the female pathway proceeds past the OCT and is canalized toward ovarian development.
XY gonads in which the number of pre-Sertoli cells falls below the TCT at the critical timepoint will undergo full or partial sex reversal and develop as ovaries or intersex gonads (i.e. ovotestes), respectively. Ovotestes have a stereotypical arrangement of testicular and ovarian regions, with testicular tissue at the center and ovarian tissue at one or both poles. This order along the AP axis likely reflects the earlier onset of the male pathway in the center relative to the poles17, 18. In many cases (e.g., YDOM-associated sex reversal), an intersex phenotype results from a failure at the top of the genetic cascade, in the level and/or timing of Sry expression39. However, failure to express Sry above a threshold by a specific time cannot fully account for all ovotestis phenotypes, and other cases provide insight into the antagonistic relationship among spatiotemporal constraints, intercellular signaling, and structure-mediated activation of testis development. For example, in Dax1-/Y and Fgf9 mutants on the B6 strain background, supporting cells at the poles express Sry with normal timing and levels, but do not maintain Sox9 expression above the threshold required for testicular development, illustrating the importance of positive feedback downstream of SRY-mediated activation6, 35. Undifferentiated supporting cell precursors are more likely to activate the male pathway if surrounded by differentiated Sertoli cells. Diffusable extracellular ligands (e.g. FGF96, PDGF/platelet derived growth factor46, DHH/desert hedgehog47, PGD234) likely play an important role in conferring this community effect. Surprisingly, an extracellular serine protease, Serpine2 (serine peptidase inhibitor, clade E, member 2), mapped to a central position in the male coexpression network in Figure 27, suggesting that ECM components may also influence the establishment of the testis pathway. Clearly, much work remains to elucidate the mechanisms and constraints controlling the propagation of sexual fate along the AP axis. Ovotestes provide a good model for dissecting these complex interactions38.
Interestingly, strain variation in the expression of female pathway genes in XY gonads may affect the exact timing of this critical “point of no return” (Fig. 4). For example, the higher expression, earlier onset, and/or delayed repression of many female pathway genes in B6 XY gonads relative to 129S1 may cause the bipotential window to close earlier in B6 (denoted by red triangle; compare to yellow triangle = critical timepoint in 129S1). The importance of timing to sex determination is not a new idea. Indeed, 25 years before global gene expression data for the gonad was available, Eva Eicher proposed that strain differences in the timing of the male and female pathways may underlie the sensitivity of B6 to sex reversal48.
Recent experiments defined the boundaries of the bipotential window in the ICR strain background by artificially upregulating SRY in XX gonad explants at different stages11. Extending this experimental approach to other strain backgrounds may expose differences in this temporal window. If this critical timepoint is earlier in B6, this may account for the observed susceptibility of the B6 genetic background to XY sex reversal in response to genetic perturbations that delay the activation of the male pathway (e.g., YDOM). Despite this bias, sex reversal is not observed in wildtype B6 XY mice, and this observation may be accounted for by increased expression of Sox9 in B67. Indeed, there may be strong balancing selection in the gonad for mutations that affect gene expression in compensatory pathways. In fact, in a genetically heterogeneous population of E11.5 XY gonads, expression levels for 50 genes were highly variable across the population (n=82), yet all XY gonads in this cross are predicted to develop as testes. This suggests that the buffering capacity of the network – the ability of the network to compensate for allelic variation – is extensive. Although it is not yet clear what the limits of variability are, this finding predicts that much expression variation can be tolerated. Only certain combinations of discordant alleles will be selected against because they disrupt the ability of the pathway to resolve in both directions.
Modularity in the Transcriptional Network
Testis and ovary structure are well-conserved across vertebrate species, yet the last decade of research has proven that there are many paths to these two destinations (for review, refer to DeFalco and Capel49). Although critical regulatory network hubs (e.g., Sox9) and core cellular processes (e.g., cell proliferation and testis cord formation) are conserved and required for male sex determination across vertebrate species, the relative timing of individual processes varies considerably40. For example, in T.scripta, a temperature-dependent turtle species, SOX9 is expressed at high levels in both male and female gonads at the bipotential stage33. Moreover, cord-like structures that are male specific in mice are initially present in both sexes in turtles. Down-regulation of SOX9 and loss of cord structures occurs at the female producing temperature in turtles, and results in differentiation of an ovary. Thus, although adult testes and ovaries are structurally very similar between turtles and mice, their differentiation occurs differently. The finding that many of the same genes are expressed in the turtle (e.g. Sox9, Amh/anti-Mullerian hormone, SF1/Nr5a1, Dmrt1, Wt1, and Foxl2), but in a different temporal sequence, makes sense in light of the reordering of the steps in differentiation. This interspecific modular heterochrony has been observed for other aspects of gonad development49 and in other developing systems50, and may represent a more general characteristic of developmental networks. From an evolutionary perspective, the gonad is only constrained by its adult function (i.e., reproductive fitness), and therefore much of the variability in gene expression levels and module timing is likely to be neutral.
Variability is also observed among vertebrates in the duration of the bipotential window, and the cause and significance of this variation is not well-understood. Species with genetic sex determination (GSD) mechanisms (e.g. mouse and humans) differ greatly in the length of this period. In the mouse, the undifferentiated stage is short, lasting only ~36 hours from formation of the gonad at E10.0 until ~E11.511, whereas in humans this period likely extends for 4–5 weeks from the initial specification around gestational week 3 to the upregulation of male pathway genes during weeks 7–8. The length of the bipotential window is even more variable among species that utilize environmental mechanisms for sex determination, and can vary significantly between sexes in some species where the growth rate differs. For example, in the temperature-dependent turtle T. scripta, the bipotential (i.e. temperature-sensitive) period spans 10 days for eggs incubated at the female-producing temperature (31°C) but lasts twice as long for eggs incubated at the male-producing temperature (26°C)33. Differences in growth rate, the duration of the bipotential stage, and the sequence in which morphological events occur may account for temporal differences in expression data. In future studies at the global level it may be possible to correlate specific expression modules with morphological differentiation events.
Conclusion
Over the past two decades, the single gene mutational approach (including fortuitous as well as intentional mutations) has been the predominant experimental strategy for identifying and validating sex determination genes, and has successfully elucidated roles for nearly 30 genes, including Sry and Sox9. Loss- and gain-of-function experiments will continue to play an important role in characterizing the function of single genes during development, however a number of studies illustrate the limitations of this approach for sex determination. First, the gonad appears to be highly buffered against most perturbations, and good candidate sex determination genes (identified by sexually dimorphic expression or gene ontology/pathway membership) have been ablated with little or no phenotypic consequence51. In one particularly heroic experiment, all three members of the insulin receptor tyrosine kinase family were knocked out before a sex-reversed phenotype was observed25. This and other studies52, 53 expose the high level of genetic redundancy and transcriptional buffering present in the gonad transcription network at the time of sex determination, and predict that roles for many individual genes during sex determination will be masked. Yet over 50% of human DSDs remain unexplained by variation in known sex determination genes. This suggests that numerous genes with critical roles in the process (i.e. having large effect sizes) await discovery, or that a majority of DSDs result from subtle changes in the dosage or expression of multiple genes. These genes with modest effects would likely escape detection in a single perturbation experiment, however in combination with other variant alleles or environmental insult, may cause a sexual disorder. This is in line with evidence from recent genome-wide association studies (GWAS) showing that tens to hundreds of variants, most of which have small effects, associate with common diseases54.
A wealth of new genetic resources (particularly in the mouse model), genome-wide analysis platforms, and computational methods have appeared on the landscape. These tools will enable the next leap forward in biomedical research by overcoming both the limitations of the single gene approach and the analytical hurdles associated with the identification of numerous small effect variants underlying complex disorders. For example, the emerging Collaborative Cross (CC) recombinant inbred panel and associated Diversity Outbred (DO) heterogeneous stock population model the complexity of the human population (the 8 founder strains capture over 89% of the variation observed in mice55) in a normally distributed56 random manner. The “pre-CC” (ie. nearly inbred) strains and DO stock have already proven to be powerful resources for mapping allelic variants associated with complex phenotypic as well as expression traits56–59. Similarly, mapping studies in other mouse strains are being aided by the recently completed full sequences for 17 common inbred strains60 and accurate imputed full sequences (from high density genotype datasets) for over 100 additional strains61, 62. As genome-wide analysis platforms continue to improve and lower in cost, it is feasible for a single investigator to quantify genetic variation (including alternative splicing and copy number variants63–65) and molecular phenotypes (e.g. mRNA4, 5, 66/protein67 abundance) across the genome at single nucleotide resolution. These systems-level analyses are becoming increasingly common in the field of sex determination. Moreover, by utilizing advanced computational tools56, 68, 69 to integrate multiple types of molecular data, it is now possible to elucidate directed gene interactions at an unprecedented level7. Loss- and gain-of-function experiments will continue to be the gold standard for candidate gene validation - in this regard, the application of gene delivery technologies70–72 to in vitro cell73, 74 and ex vivo gonad explant culture70, 75 models will make it feasible to artificially perturb the expression of many genes at low cost and high-throughput. These assays will facilitate the identification of causative genes underlying QTLs and the elucidation of combinatorial interactions among small effect genes. Finally, much of the transcriptional architecture underlying sex determination is highly conserved between mouse and human. Therefore, a comprehensive understanding of the gonad transcription network in the mouse, coupled to an interspecies-derived template of where variation in the network is tolerated, should be highly informative for predicting causative genes from DNA-sequencing approaches in affected patients and elucidating the etiology of unexplained human disorders of sexual development.
Acknowledgments
Notes
We would like to thank Lindsey Mork for artistic support with figures. We thank Lindsey Mork, Samantha Jameson, and Jonah Cool for their helpful comments on this manuscript.
Contributor Information
Steven C Munger, Department of Cell Biology, Duke University Medical Center.
Blanche Capel, Department of Cell Biology, Duke University Medical Center.
References
- 1.Sekido R, Lovell-Badge R. Sex determination and SRY: down to a wink and a nudge? Trends Genet. 2009;25(1):19–29. doi: 10.1016/j.tig.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Wainwright EN, Wilhelm D. The game plan: cellular and molecular mechanisms of mammalian testis development. Curr Top Dev Biol. 2010;90:231–262. doi: 10.1016/S0070-2153(10)90006-9. [DOI] [PubMed] [Google Scholar]
- 3.Liu CF, Liu C, Yao HH. Building pathways for ovary organogenesis in the mouse embryo. Curr Top Dev Biol. 2010;90:263–290. doi: 10.1016/S0070-2153(10)90007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nef S, Schaad O, Stallings NR, Cederroth CR, Pitetti JL, Schaer G, Malki S, Dubois-Dauphin M, Boizet-Bonhoure B, Descombes P, et al. Gene expression during sex determination reveals a robust female genetic program at the onset of ovarian development. Dev Biol. 2005;287:361–377. doi: 10.1016/j.ydbio.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Beverdam A, Koopman P. Expression profiling of purified mouse gonadal somatic cells during the critical time window of sex determination reveals novel candidate genes for human sexual dysgenesis syndromes. Hum Mol Genet. 2006;15:417–431. doi: 10.1093/hmg/ddi463. [DOI] [PubMed] [Google Scholar]
- 6.Kim Y, Kobayashi A, Sekido R, DiNapoli L, Brennan J, Chaboissier MC, Poulat F, Behringer RR, Lovell-Badge R, Capel B. Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol. 2006;4:e187. doi: 10.1371/journal.pbio.0040187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munger SC, Aylor DL, Syed HA, Magwene PM, Threadgill DW, Capel B. Elucidation of the transcription network governing mammalian sex determination by exploiting strain-specific susceptibility to sex reversal. Genes Dev. 2009;23:2521–2536. doi: 10.1101/gad.1835809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jameson SA, Natarajan A, Cool J, Defalco T, Maatouk DM, Mork L, Munger SC, Capel B. Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet. 2012;8:e1002575. doi: 10.1371/journal.pgen.1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vainio S, Heikkila M, Kispert A, Chin N, McMahon AP. Female development in mammals is regulated by Wnt-4 signalling. Nature. 1999;397:405–409. doi: 10.1038/17068. [DOI] [PubMed] [Google Scholar]
- 10.Parma P, Radi O, Vidal V, Chaboissier MC, Dellambra E, Valentini S, Guerra L, Schedl A, Camerino G. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat Genet. 2006;38:1304–1309. doi: 10.1038/ng1907. [DOI] [PubMed] [Google Scholar]
- 11.Hiramatsu R, Matoba S, Kanai-Azuma M, Tsunekawa N, Katoh-Fukui Y, Kurohmaru M, Morohashi K, Wilhelm D, Koopman P, Kanai Y. A critical time window of Sry action in gonadal sex determination in mice. Development. 2009;136:129–138. doi: 10.1242/dev.029587. [DOI] [PubMed] [Google Scholar]
- 12.Palmer SJ, Burgoyne PS. In situ analysis of fetal, prepuberal and adult XX----XY chimaeric mouse testes: Sertoli cells are predominantly, but not exclusively, XY. Development. 1991;112:265–268. doi: 10.1242/dev.112.1.265. [DOI] [PubMed] [Google Scholar]
- 13.Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- 14.Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 15.Sekido R, Bar I, Narvaez V, Penny G, Lovell-Badge R. SOX9 is up-regulated by the transient expression of SRY specifically in Sertoli cell precursors. Dev Biol. 2004;274:271–279. doi: 10.1016/j.ydbio.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 16.Koopman P, Munsterberg A, Capel B, Vivian N, Lovell-Badge R. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature. 1990;348:450–452. doi: 10.1038/348450a0. [DOI] [PubMed] [Google Scholar]
- 17.Albrecht KH, Eicher EM. Evidence that Sry is expressed in pre-Sertoli cells and Sertoli and granulosa cells have a common precursor. Dev Biol. 2001;240:92–107. doi: 10.1006/dbio.2001.0438. [DOI] [PubMed] [Google Scholar]
- 18.Bullejos M, Koopman P. Spatially dynamic expression of Sry in mouse genital ridges. Dev Dyn. 2001;221:201–205. doi: 10.1002/dvdy.1134. [DOI] [PubMed] [Google Scholar]
- 19.Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- 20.Hammes A, Guo JK, Lutsch G, Leheste JR, Landrock D, Ziegler U, Gubler MC, Schedl A. Two splice variants of the Wilms’ tumor 1 gene have distinct functions during sex determination and nephron formation. Cell. 2001;106:319–329. doi: 10.1016/s0092-8674(01)00453-6. [DOI] [PubMed] [Google Scholar]
- 21.Luo X, Ikeda Y, Parker KL. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 22.Birk OS, Casiano DE, Wassif CA, Cogliati T, Zhao L, Zhao Y, Grinberg A, Huang S, Kreidberg JA, Parker KL, et al. The LIM homeobox gene Lhx9 is essential for mouse gonad formation. Nature. 2000;403:909–913. doi: 10.1038/35002622. [DOI] [PubMed] [Google Scholar]
- 23.Miyamoto N, Yoshida M, Kuratani S, Matsuo I, Aizawa S. Defects of urogenital development in mice lacking Emx2. Development. 1997;124:1653–1664. doi: 10.1242/dev.124.9.1653. [DOI] [PubMed] [Google Scholar]
- 24.Katoh-Fukui Y, Tsuchiya R, Shiroishi T, Nakahara Y, Hashimoto N, Noguchi K, Higashinakagawa T. Male-to-female sex reversal in M33 mutant mice. Nature. 1998;393:688–692. doi: 10.1038/31482. [DOI] [PubMed] [Google Scholar]
- 25.Nef S, Verma-Kurvari S, Merenmies J, Vassalli JD, Efstratiadis A, Accili D, Parada LF. Testis determination requires insulin receptor family function in mice. Nature. 2003;426:291–295. doi: 10.1038/nature02059. [DOI] [PubMed] [Google Scholar]
- 26.Tevosian SG, Albrecht KH, Crispino JD, Fujiwara Y, Eicher EM, Orkin SH. Gonadal differentiation, sex determination and normal Sry expression in mice require direct interaction between transcription partners GATA4 and FOG2. Development. 2002;129:4627–4634. doi: 10.1242/dev.129.19.4627. [DOI] [PubMed] [Google Scholar]
- 27.Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–934. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- 28.Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, Sinclair AH. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- 29.Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, Kobayashi T, Morrey CE, Shibata N, Asakawa S, Shimizu N, et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–563. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- 30.Janzen FJ, Phillips PC. Exploring the evolution of environmental sex determination, especially in reptiles. J Evol Biol. 2006;19:1775–1784. doi: 10.1111/j.1420-9101.2006.01138.x. [DOI] [PubMed] [Google Scholar]
- 31.Quinn AE, Georges A, Sarre SD, Guarino F, Ezaz T, Graves JA. Temperature sex reversal implies sex gene dosage in a reptile. Science. 2007;316:411. doi: 10.1126/science.1135925. [DOI] [PubMed] [Google Scholar]
- 32.Radder RS, Quinn AE, Georges A, Sarre SD, Shine R. Genetic evidence for co-occurrence of chromosomal and thermal sex-determining systems in a lizard. Biol Lett. 2008;4:176–178. doi: 10.1098/rsbl.2007.0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barske LA, Capel B. Estrogen represses SOX9 during sex determination in the red-eared slider turtle Trachemys scripta. Dev Biol. 2010;341:305–314. doi: 10.1016/j.ydbio.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 34.Wilhelm D, Martinson F, Bradford S, Wilson MJ, Combes AN, Beverdam A, Bowles J, Mizusaki H, Koopman P. Sertoli cell differentiation is induced both cell-autonomously and through prostaglandin signaling during mammalian sex determination. Dev Biol. 2005;287:111–124. doi: 10.1016/j.ydbio.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 35.Bouma GJ, Albrecht KH, Washburn LL, Recknagel AK, Churchill GA, Eicher EM. Gonadal sex reversal in mutant Dax1 XY mice: a failure to upregulate Sox9 in pre-Sertoli cells. Development. 2005;132:3045–3054. doi: 10.1242/dev.01890. [DOI] [PubMed] [Google Scholar]
- 36.Nikolova G, Sinsheimer JS, Eicher EM, Vilain E. The Chromosome 11 Region From Strain 129 Provides Protection From Sex Reversal in XYPOS Mice. Genetics. 2008;179:419–427. doi: 10.1534/genetics.108.088088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uhlenhaut NH, Jakob S, Anlag K, Eisenberger T, Sekido R, Kress J, Treier AC, Klugmann C, Klasen C, Holter NI, et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell. 2009;139:1130–1142. doi: 10.1016/j.cell.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 38.Wilhelm D, Washburn LL, Truong V, Fellous M, Eicher EM, Koopman P. Antagonism of the testis- and ovary-determining pathways during ovotestis development in mice. Mech Dev. 2009;126(5–6):324–336. doi: 10.1016/j.mod.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bullejos M, Koopman P. Delayed Sry and Sox9 expression in developing mouse gonads underlies B6-Y(DOM) sex reversal. Dev Biol. 2005;278:473–481. doi: 10.1016/j.ydbio.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 40.Morrish BC, Sinclair AH. Vertebrate sex determination: many means to an end. Reproduction. 2002;124:447–457. doi: 10.1530/rep.0.1240447. [DOI] [PubMed] [Google Scholar]
- 41.Pask AJ, Calatayud NE, Shaw G, Wood WM, Renfree MB. Oestrogen blocks the nuclear entry of SOX9 in the developing gonad of a marsupial mammal. BMC Biol. 2010;8:113. doi: 10.1186/1741-7007-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Santa Barbara P, Moniot B, Poulat F, Berta P. Expression and subcellular localization of SF-1, SOX9, WT1, and AMH proteins during early human testicular development. Dev Dyn. 2000;217:293–298. doi: 10.1002/(SICI)1097-0177(200003)217:3<293::AID-DVDY7>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 43.Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, Zarkower D. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature. 2011;476(7358):101–104. doi: 10.1038/nature10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 45.Tang F, Barbacioru C, Bao S, Lee C, Nordman E, Wang X, Lao K, Surani MA. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brennan J, Tilmann C, Capel B. Pdgfr-alpha mediates testis cord organization and fetal Leydig cell development in the XY gonad. Genes Dev. 2003;17:800–810. doi: 10.1101/gad.1052503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bitgood MJ, Shen L, McMahon AP. Sertoli cell signaling by Desert hedgehog regulates the male germline. Curr Biol. 1996;6:298–304. doi: 10.1016/s0960-9822(02)00480-3. [DOI] [PubMed] [Google Scholar]
- 48.Eicher EM, Washburn LL. Inherited sex reversal in mice: identification of a new primary sex-determining gene. J Exp Zool. 1983;228:297–304. doi: 10.1002/jez.1402280213. [DOI] [PubMed] [Google Scholar]
- 49.DeFalco T, Capel B. Gonad morphogenesis in vertebrates: divergent means to a convergent end. Annu Rev Cell Dev Biol. 2009;25:457–482. doi: 10.1146/annurev.cellbio.042308.13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev Cell. 2011;20:483–496. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tohonen V, Frygelius J, Mohammadieh M, Kvist U, Pelliniemi LJ, O’Brien K, Nordqvist K, Wedell A. Normal sexual development and fertility in testatin knockout mice. Mol Cell Biol. 2005;25:4892–4902. doi: 10.1128/MCB.25.12.4892-4902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ross A, Munger S, Capel B. Bmp7 regulates germ cell proliferation in mouse fetal gonads. Sex Dev. 2007;1:127–137. doi: 10.1159/000100034. [DOI] [PubMed] [Google Scholar]
- 53.Frygelius J, Oscarson M, Nordqvist K, Wedell A, Tohonen V. The reproductive tissue specific cystatin subgroup of genes: expression during gonadal development in wildtype and testatin knockout animals. Sex Dev. 2007;1:363–372. doi: 10.1159/000111768. [DOI] [PubMed] [Google Scholar]
- 54.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roberts A, Pardo-Manuel de Villena F, Wang W, McMillan L, Threadgill DW. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm Genome. 2007;18:473–481. doi: 10.1007/s00335-007-9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS, Ferris MT, Frelinger JA, Heise M, Frieman MB, et al. Genetic analysis of complex traits in the emerging collaborative cross. Genome Res. 2011;21(8):1213–1222. doi: 10.1101/gr.111310.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Durrant C, Tayem H, Yalcin B, Cleak J, Goodstadt L, Pardo-Manuel de Villena F, Mott R, Iraqi F. Collaborative Cross mice and their power to map host susceptibility to Aspergillus fumigatus infection. Genome Res. 2011;21(8):1239–1248. doi: 10.1101/gr.118786.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mathes WF, Aylor DL, Miller DR, Churchill GA, Chesler EJ, Pardo Manuel de Villena F, Threadgill DW, Pomp D. Architecture of energy balance traits in emerging lines of the Collaborative Cross. Am J Physiol Endocrinol Metab. 2011;300(6):E1124–1134. doi: 10.1152/ajpendo.00707.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Svenson K, Gatti D, Valdar W, Welsh CE, Cheng R, Chesler E, Palmer AA, McMillan L, Churchill GA. The Mouse Diversity Outbred Population. Genetics. 2012;190(2):437–447. doi: 10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hutchins LN, Ding Y, Szatkiewicz JP, Von Smith R, Yang H, de Villena FP, Churchill GA, Graber JH. CGDSNPdb: a database resource for error-checked and imputed mouse SNPs. Database (Oxford) 2010:baq008. doi: 10.1093/database/baq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Szatkiewicz JP, Beane GL, Ding Y, Hutchins L, Pardo-Manuel de Villena F, Churchill GA. An imputed genotype resource for the laboratory mouse. Mamm Genome. 2008;19:199–208. doi: 10.1007/s00335-008-9098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ledig S, Hiort O, Scherer G, Hoffmann M, Wolff G, Morlot S, Kuechler A, Wieacker P. Array-CGH analysis in patients with syndromic and non-syndromic XY gonadal dysgenesis: evaluation of array CGH as diagnostic tool and search for new candidate loci. Hum Reprod. 2010;25:2637–2646. doi: 10.1093/humrep/deq167. [DOI] [PubMed] [Google Scholar]
- 64.Tannour-Louet M, Han S, Corbett ST, Louet JF, Yatsenko S, Meyers L, Shaw CA, Kang SH, Cheung SW, Lamb DJ. Identification of de novo copy number variants associated with human disorders of sexual development. PLoS One. 2010;5:e15392. doi: 10.1371/journal.pone.0015392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.White S, Ohnesorg T, Notini A, Roeszler K, Hewitt J, Daggag H, Smith C, Turbitt E, Gustin S, van den Bergen J, et al. Copy Number Variation in Patients with Disorders of Sex Development Due to 46,XY Gonadal Dysgenesis. PLoS One. 2011;6:e17793. doi: 10.1371/journal.pone.0017793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bouma GJ, Hudson QJ, Washburn LL, Eicher EM. New candidate genes identified for controlling mouse gonadal sex determination and the early stages of granulosa and Sertoli cell differentiation. Biol Reprod. 2010;82:380–389. doi: 10.1095/biolreprod.109.079822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ewen K, Baker M, Wilhelm D, Aitken RJ, Koopman P. Global survey of protein expression during gonadal sex determination in mice. Mol Cell Proteomics. 2009;8:2624–2641. doi: 10.1074/mcp.M900108-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim YA, Wuchty S, Przytycka TM. Identifying causal genes and dysregulated pathways in complex diseases. PLoS Comput Biol. 2011;7:e1001095. doi: 10.1371/journal.pcbi.1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hageman RS, Leduc MS, Korstanje R, Paigen B, Churchill GA. A bayesian framework for inference of the genotype-phenotype map for segregating populations. Genetics. 2011;187:1163–1170. doi: 10.1534/genetics.110.123273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ryan J, Ludbrook L, Wilhelm D, Sinclair A, Koopman P, Bernard P, Harley VR. Analysis of gene function in cultured embryonic mouse gonads using nucleofection. Sex Dev. 2011;5:7–15. doi: 10.1159/000322162. [DOI] [PubMed] [Google Scholar]
- 71.Svingen T, Wilhelm D, Combes AN, Hosking B, Harley VR, Sinclair AH, Koopman P. Ex vivo magnetofection: a novel strategy for the study of gene function in mouse organogenesis. Dev Dyn. 2009;238:956–964. doi: 10.1002/dvdy.21919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Parrington J, Coward K, Hibbitt O, Kubota H, Young C, McIlhinney J, Jones O. In vivo gene transfer into the testis by electroporation and viral infection--a novel way to study testis and sperm function. Soc Reprod Fertil Suppl. 2007;65:469–474. [PubMed] [Google Scholar]
- 73.Polanco JC, Wilhelm D, Mizusaki H, Jackson A, Browne C, Davidson T, Harley V, Sinclair A, Koopman P. Functional analysis of the SRY-KRAB interaction in mouse sex determination. Biol Cell. 2009;101:55–67. doi: 10.1042/BC20080061. [DOI] [PubMed] [Google Scholar]
- 74.Knower KC, Sim H, McClive PJ, Bowles J, Koopman P, Sinclair AH, Harley VR. Characterisation of urogenital ridge gene expression in the human embryonal carcinoma cell line NT2/D1. Sex Dev. 2007;1:114–126. doi: 10.1159/000100033. [DOI] [PubMed] [Google Scholar]
- 75.Hiramatsu R, Harikae K, Tsunekawa N, Kurohmaru M, Matsuo I, Kanai Y. FGF signaling directs a center-to-pole expansion of tubulogenesis in mouse testis differentiation. Development. 2010;137:303–312. doi: 10.1242/dev.040519. [DOI] [PubMed] [Google Scholar]