Abstract
The low number of envelope (Env) spikes presented on native HIV-1 particles is a major impediment for HIV-1 prophylactic vaccine development. We designed virus-like particle encoding adenoviral vectors utilizing SIVmac239 Gag as an anchor for full length and truncated HIV-1 M consensus Env. Truncated Env overexpressed VRC01 and 17b binding antigen on the surface of transduced cells while the full length Env vaccine presented more and similar amounts of antigen binding to the trimer conformation sensitive antibodies PGT151 and PGT145, respectively. The adenoviral vectors were used to prime Balb/c mice followed by sequential boosting with chimpanzee type 63, and chimpanzee type 3 adenoviral vectors encoding SIVmac239 Gag and full length consensus Env. Both vaccine regimens induced increasing titers of binding antibody responses after each immunization, and significant differences in immune responses between the two groups were observed after the final immunization. Full length Env priming skewed antibody responses towards gp41, while truncated Env priming induced responses primarily targeting gp120 containing and derived antigens. Importantly, no differences in neutralizing antibody responses were found between the different priming regimens as both induced high titered tier 1 neutralizing antibodies, but no tier 2 antibodies, possibly reflecting the similar presentation of trimer specific antibody epitopes. The described vaccine regimens provide insight into the effects of the HIV-1 Env cytoplasmic tail on epitope presentation and subsequent immune responses, which is relevant for the interpretation of current clinical trials that are using truncated Env as an immunogen. The regimens described here provide similar neutralization titers, and thus are useful for investigating the importance of specificity in non-neutralizing antibody mediated protection against viral challenge.
1. Introduction
The envelope glycoprotein (Env) is the major focus of antibody-inducing prophylactic vaccine development against HIV, and has proven to be an exceptionally tough target [1]. Functional virion associated Env is folded as a compact trimer and contains heavily glycosylated surfaces that partially obstruct access to the trimer [2, 3]. Epitopes that are exposed frequently display significant sequence variation and are termed variable loops (V1–V5). However, Env contains a number of structurally conserved and exposed regions that are vulnerable to broadly neutralizing antibodies [4, 5], antibody dependent cellular cytotoxicity (ADCC) mediating antibodies, and have been described as correlates of risk in the RV144 vaccine trial [6].
We focused on virus-like particles (VLPs) encoded within viral vectors based on the antibody response elicited in in the RV144 trial (canarypox based vaccine vector prime [7]), and the clinical development of VLP based vaccines delivered by modified vaccinia Ankara vectors (MVA) [8]. Viral vector encoded VLPs simultaneously provide the ability to present an antigen oligomerized in a VLP context in addition to the display of Env on the cell surface of vaccine transduced cells and in this manner stimulate antibody responses that could exhibit ADCC activity. Early studies using encoded Env faced problems with genetic instability of the full length cytoplasmic tail in MVA vectors, but it was observed that truncation of the cytoplasmic tail improved poxviral replication and transgene expression [9]. Consequently, clinical development has progressed using an Env sequence with a truncated tail in the MVA vectors and a full length tail in an optional DNA prime [8], but testing the importance of the cytoplasmic tail in a side-by-side experiment has not been possible with MVA vectors. Adenoviral vectors with expression cassettes including a tetracycline operator in proximity to the antigen promoter have recently been developed [10], and now allow us to explore the unique features of the HIV-1 Env cytoplasmic tail. The HIV-1 Env cytoplasmic tail is not believed to be targeted by protective antibodies, but has been recognized as a determinant of the quantity of Env incorporated into HIV-1 VLPs [11]. Further interest in the cytoplasmic tail has been instigated by the recent publication by Chen et al. [12], indicated the importance of the full length cytoplasmic tail for occluding induction of non-neutralizing antibody responses. These findings suggest a major role of the length of the tail in maintenance of a specific Env structure which could be important for virus-encoded Env priming of vaccine responses. Here we show that a simple expression cassette encoding Gag and Env separated by a self-cleavable peptide efficiently directs the secretion of VLPs incorporating Env, with more total Env protein incorporated using the truncated tail. Surface staining of vaccine transduced cells with the gp120 monomer binding VRC01 antibody demonstrated that the tail truncation led to a major increased presentation of the CD4 binding site (bs) epitope, and an increase in the presentation of CD4 inducible epitopes (17b), whereas the gp120/gp41 interface sensitive and trimer specific antibodies showed less (PGT151) or similar (PGT145) cell surface expression using the truncated tail. Sequential administration of up to three heterologous vectors primed with either antigen gave rise to increasing antibody responses after each immunization. Whereas higher incorporation of Env in a virus-encoded VLP vaccine did not have any direct effect on the initial humoral response, there was a significant increase in the total Env trimer-specific humoral response following boosting with vectors encoding full length Env. These responses were also paralleled with significantly higher binding to CON-S gp140 in a binding antibody multiplex assay, and a tendency for higher V3 loop specific responses. Conversely, the full length Env primed group showed a tendency to have higher gp41 responses and a significant bias towards gp41 binding antibodies relatively to gp120 binding antibodies. Notably, the increase in CON-S binding antibodies obtained with the truncated Env priming did not translate into differences in neutralizing antibody responses.
2. Materials and Methods
2.1 Mice
Female Balb/c mice aged 6–8 weeks were obtained from Taconic M&B (Ry, Denmark). The mice were allowed to acclimatize for one week prior to the initiation of an experiment. All experiments were approved by the national animal experiments inspectorate (Dyreforsøgstilsynet) and performed according to national guidelines.
2.2 Adenoviral vaccine production
Replication-deficient E1 deleted human adenovirus type 5 (huAd5) [13] and chimpanzee adenovirus type 3 (chAd3) and 63 (chAd63) [14] vectors with deleted E1 and E3 genes were used in this study. The adenoviral vectors expressed SIVmac239 Gag and HIV-1 M CON-S gp160 (CON-S gp160) [15], linked to Gag via a Glycine/Serine/Glycine (GSG) linker followed by a self-cleaving porcine teschovirus-1 2A peptide (P2A) [16], and were produced in Procell 92 cells [17]. A truncated CON-S gp160 version (CON-S gp160Δ139) in a huAd5 vector was produced expressing SIVmac239 Gag linked via P2A to CON-S gp160 truncated after amino acid (aa) 721 (HXB2 numbering, GenBank accession number K03455). The huAd5 vectors and chAd vectors were produced by homologous recombination in Procell 92 cells [18] and BJ5183 [19], respectively. The antigens were inserted in the E1 region for all constructs as described for huAd5 [18] and the chAd vectors [19] and all vectors encoded tetracycline operator (TetO) sequences [10] downstream of the huCMV promoter. After amplification of clonal adenoviral genomes, the particles were purified using a caesium chloride gradient as described [18].
2.3 Immunizations
Mice were injected intramuscularly in the quadriceps muscle (i.m.) (10 mice per group) with 3×109 adenoviral particles in a total volume of 50 µl PBS changing leg at each vaccination time-point.
2.4 Humoral responses
CON-S gp160 responses were measured by enzyme linked immunosorbent assay (ELISA) as adapted from [20]. CON-S gp160 was produced in 293FT cells using a plasmid encoding CON-S gp160 and the pSG3Δenv plasmid [21] (NARRP) by transient transfection. ELISA was performed coating MaxiSorp plates (NUNC) with concanavalin A (ConA) (Sigma-Aldrich) captured CON-S gp160 cell supernatant. Serum samples were added in a series of 2-fold dilutions and a standard anti-Env serum sample was included on all plates in all runs. Binding was detected with HRP labelled polyclonal rabbit anti-mouse immunoglobulins (Dako) used for detection of bound antibody. The anti-Env responses at the final time-point were also measured against CON-S gp140 CFI avi protein (2 µg/ml in carbonate buffer). Gag specific responses were determined using p27 SIVmac239 Gag (Immune Technology) as a coating antigen (2 µg/ml in PBS). Samples were added in 3-fold dilutions, and the mAb 55-2F12 [22] (NARRP) served as a positive control. Binding was detected as above. Area under the curve (AUC) and end-point dilution titers were used to assess the humoral responses. AUC and titers were calculated from either 3-fold (Gag responses) or 2-fold (Env responses) titration series of the samples.
2.5 Binding antibody multiplex assay (BAMA) for determination of the specificity of antibody responses
The HIV-1 binding antibody multiplex assay was performed as previously described [23–26]. Briefly, the HIV-1 antigens, gp41 (clade B HIV-1MN recombinant gp41 protein, ImmunoDiagnostics), Con6 gp120 (Consensus gp120 Env), CON-S gp140 CFI (Group M consensus gp140 CFI Env), gp41 ID epitope (Biotin - CRVLAVERYLRDQQLLGIWGCSGKLICTTAV) [23], murine leukemia virus (MuLV) gp70 B.CaseA2 V1–V2, MuLV gp70, and clade B V3 (Biotin – KKKNNTRKSIHIGPGRAFYATGDIIGDIRQAHC, JPT Peptide Technologies) were conjugated to polystyrene beads (Bio-Rad) and binding was detected using goat anti-mouse IgG-PE or mouse anti-human IgG-PE (Southern Biotech). MuLV gp70 coupled beads were used to account for MuLV gp70 background binding in the V1–V2 antigen coupled to gp70 (gp70 B.CaseA2 V1–V2). Antigen specific binding was measured as mean fluorescent intensity (MFI). Positive controls included HIV-1 immunoglobulin (HIVIG) (NARRP), and the 7B2 and 3B3 IgG monoclonal antibodies (mAbs). Negative controls were blank beads, normal human serum (Sigma), and pre-immune pooled mouse serum. Responses were considered positive if they met antigen-specific positivity criteria of the mean +3 standard deviations of pooled seronegative mice (10) or a minimum of 100 MFI.
2.6 Cell surface expression analysis
Env expression was analysed on the surface of vero cells two days after infection with 50 PFU/cell of either huAd5 vaccine. Cells were stained with the VRC01 [27] (NARRP), PGT145 (IAVI) (specific for quaternary epitopes in V2) [28], PGT151 (IAVI) (specific for epitopes at the interface between gp120/gp41) [29], and 17b (specific for CD4 inducible epitopes overlapping the co-receptor binding site) [30] mAbs. Binding of the mAbs was detected using anti-human IgG Fc-APC antibody (BioLegend), and the cells were acquired using an LSRII or Fortessa instrument (BD Biosciences) and analysed with FlowJo software (Tree Star, Ashland, OR).
2.7 Measurement of neutralizing responses
Pseudovirus production and titration in TZM-bl cells has been described elsewhere [31]. For these analysis serum samples were grouped in two and analysed for their neutralization capability against MN.3 (clade B tier 1), MW965.26 (clade C tier 1) and CON-S (tier 2) in TZM-bl cells. MuLV pseudotyped virus was used as a negative control for each pooled sample together with a pool of naïve serum samples as a negative control for each virus.
2.8 Methods to characterize VLPs
2.8.1 VLP production and purification
Vero cells were infected with 50 PFU/cell of either huAd5 vaccine. 48 hours post infection, supernatants were harvested, clarified and filtered. The VLPs were pelleted at 82.700 g in a Beckman Coulter Ti 70.1 rotor for 2h at 4°C. The pellets were resuspended in 1 ml PBS and left over night at 4°C. The following day, residual medium was removed and pellets were resuspended in PBS at 1:140 of the original volume.
2.8.2 SDS-PAGE
For Western blot analysis and quantification, VLP samples were prepared as previously described [32]. SIVmac239 Gag was detected using either anti-SIV mac251 p27 Gag antibody [33] (MyBioSource) followed by HRP coupled polyclonal rabbit anti-mouse immunoglobulins (Dako) or SIV mac251 antiserum [34] (NARRP) followed by HRP coupled polyclonal rabbit anti-human immunoglobulins (Dako). HIV-1 M CON-S Env variants were probed with an antibody cocktail consisting of VRC01 (1 µg/ml) [27] and b12 (1 µg/ml) [35], and 2G12 [36, 37] (1 µg/ml) (NARRP), and then detected with HRP coupled polyclonal rabbit anti-human immunoglobulins (Dako). The density of Gag bands were quantified using Image Studio Lite software (LI-COR Biosciences).
2.8.3 Env capture
For Env quantification, the HIV-1 M CON-S gp160 ELISA was adapted. Purified VLPs were prepared in lysing buffer (Zeptometrix) and added in two-fold dilutions to a ConA coated MaxiSorp plate. HIV-1 M CON-S gp140 CFI avi was used as a standard. After blocking, Env was detected with an anti-gp120 cocktail at 1 µg/ml consisting of VRC01, b12 and 2G12 (NARRP). HRP coupled polyclonal rabbit anti-human immunoglobulins (Dako) were used to detect the mAbs.
2.8.4 Electron Microscopy
Infected vero cells were fixed with 2% v/v glutaraldehyde in 0.05 M sodium phosphate buffer (pH 7.2). Samples were prepared as described previously using 80 nm thick sections [38].
2.9 Statistical analysis
Exact Wilcoxon Two-Sample tests were performed for comparison between the immunization regimens. Signed rank test was performed for comparison of end point titers within the same immunization group. Control for False Discovery Rate (FDR) was performed using the Benjamini and Hochberg method [39]. The statistical analysis was performed using SAS v9.4 (SAS Institute). p-values < 0.05 were considered significant.
3. Results
3.1. Production of adenoviral vectors expressing chimeric VLPs
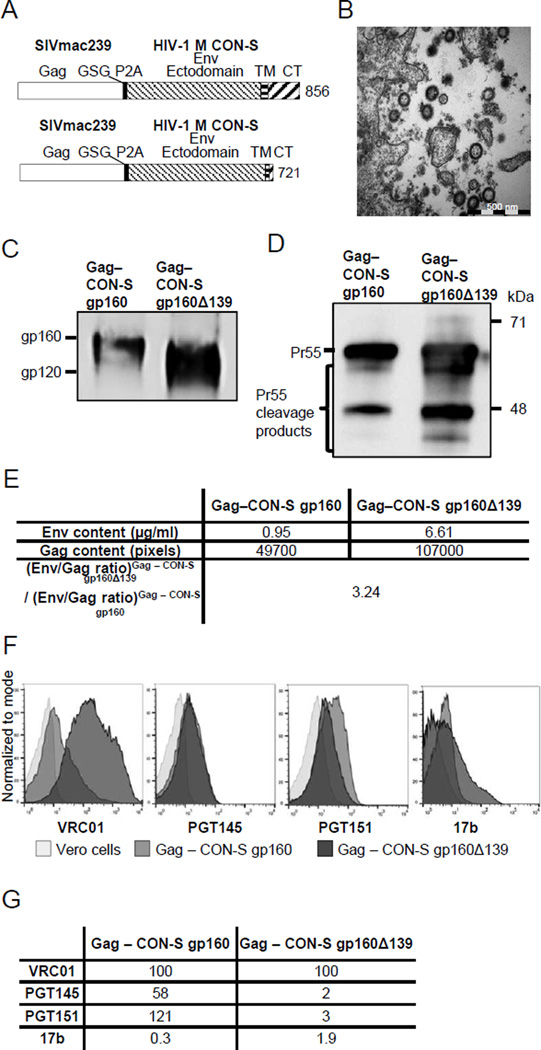
Gag from SIVmac239 and Env from HIV-1 M CON-S (CON-S) linked via a self-cleaving peptide (P2A), were encoded in three adenoviral vectors huAd5, chAd3, and chAd63 to enable heterologous prime-boost immunizations (Fig. 1A, upper schematic). The selection of the CON-S Env sequence followed its established use in macaque and rodent studies [15, 40] along with its thoroughly defined antibody binding sites for neutralizing and non-neutralizing epitopes [41]. With the intention to investigate the effects of potentially increased Env incorporation, we further designed a huAd5 construct encoding a truncated CON-S sequence (Fig. 1A, lower schematic).
Figure 1.
Characteristics of adenovirus encoded VLPs. (A) Vaccine products as encoded in huAd5, chAd63 and chAd3 (top schematic). Lower schematic depicts vaccine products encoded in a second huAd5 vector expressing a truncated form of the HIV-1 Env. TM is transmembrane; CT is cytoplasmic tail. Numbering according to HXB2, GenBank accession number K03455. (B) Vero cells were infected with huAd5 encoding SIVmac239 Gag and HIV-1 CON-S gp160, followed by fixation, embedding, sectioning and staining for transmission electron microscopy. (C) Purified VLPs from huAd5 infected vero cells were analysed by SDS-PAGE and immunoblotting using an anti-gp120 mAb cocktail for detection of Env expression and anti-SIV mac251 Gag p27 antibody for Gag expression (D). (E) Quantifications of Env (by capture ELISA) and Gag (by Western Blot) in purified VLPs were used to calculate the difference in Env to Gag ratio between the constructs. (F) Surface staining of infected vero cells with either huAd5 vaccine (50 PFU/cell) and uninfected cells with the mAbs VRC01, PGT145, PGT151, and 17b as analysed by flow cytometry. (G) Analysis of antigenic properties by comparison of binding index normalized to VRC01 binding between the two huAd5 vaccines after infection of vero cells. Values are ratio of maximum MFI by indicated mAb and the maximum MFI of VRC01 binding for the same vaccine.
VLP expression from the huAd5 expression cassette was analysed by infecting vero cells with the purified vector. To establish if the adenovirus transduction led to formation and secretion of VLPs, we analysed fixed cells by transmission electron microscopy (TEM). Transduced cells secreted large numbers of spherical particles at a size consistent with HIV-1 Gag based VLPs [42, 43] (Fig. 1B). The expression of Gag and Env was confirmed with Western blot using a gp120 mAb cocktail (Fig. 1C) and rhesus macaque SIVmac251 antiserum (not shown). Two bands corresponding to gp160/truncated gp160 and gp120 were visible for Env, while Gag could be identified with bands for pr55 and pr55 cleavage products. Env content quantified by ELISA was normalized to the relative quantity of Gag as determined by Western blot (Fig. 1D). Truncation of the Env cytoplasmic tail increased the ratio of Env to Gag expression by approximately 3-fold as compared to the full length Env (Fig. 1E).
Surface staining of vero cells confirmed the expression of Env from both huAd5 vaccines. The four antibodies, VRC01, 17b, PGT145, and PGT151, were all positive for binding to Env expressed on cells infected with either huAd5 vaccine. VRC01 binding reached a much higher MFI with the CON-S gp160Δ139 vaccine, and an increase in MFI was also observed for 17b with the CON-S gp160Δ139 vaccine. The MFI for PGT145 binding was found to be similar, whereas the PGT151 mAb had an increased MFI for cells infected with CON-S gp160 (Fig. 1F and 1G). Collectively these stainings demonstrate that the CON-S gp160Δ139 vaccine presents more epitope (VRC01), and that the conformation is more open (17b), while there is similar or increased amounts of epitopes displayed for more conformation specific antibodies (PGT145 and PGT151).
3.2. Immunogenicity of adenovirus encoded VLP vaccines
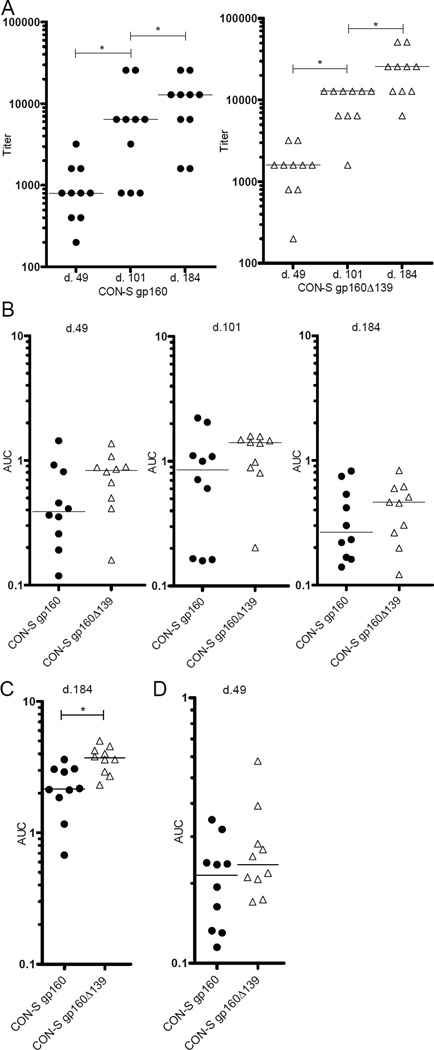
Balb/c mice (10/group) were immunized with huAd5 vectors encoding SIVmac239 Gag and either CON-S gp160 or CON-S gp160Δ139. At week 8, mice were boosted with chAd63 followed by chAd3 at week 16; both vaccines encoding SIVmac239 Gag and CON-S gp160. Serum was harvested 7 weeks post the first and second vaccination, and 10 weeks post the third and final immunization. Total Env specific immunoglobulin responses were analysed by ELISA using ConA captured VLP incorporated Env (Fig. 2A and 2B) (d.49, d.101, d.184) and CON-S gp140 CFI protein (Fig. 2C) (d.184). From analysis of responses with the ConA captured VLP ELISA, we found that the mice in both groups developed antibody responses after the first immunization (d.49). Endpoint titer results were compared by assigning each endpoint titer a score, calculating the difference score between the respective time-points and then performing a Signed Rank test by group. FDR correction was performed using the Benjamini and Hochberg method [39]. Significance is indicated by p < 0.05 after FDR correction (Fig. 2A). The CON-S gp160Δ139 primed group had continuously higher responses than the CON-S gp160 primed group although these differences did not reach statistical significance when calculated based on AUC values (Fig. 2B). To explore this trend further, we also analysed the responses against the CON-S gp140 CFI protein on samples taken 10 weeks after the final immunization (d. 184). Here we found a significant difference between the groups based on AUC values (p-value=0.014, Wilcoxon Exact Test, FDR corrected p-value [39]).
Figure 2.
Serum ELISA Env and Gag responses. Groups of 10 Balb/c mice were primed with huAd5 vectors encoding SIVmac239 Gag and either CON-S gp160 or CON-S gp160Δ139, followed with boosting at week 8 and at week 16 with chAd63 and chAd3 respectively, both booster vaccines encoding SIVmac239 Gag and CON-S gp160. Serum was harvested 7 weeks after the 1st and 2nd immunization (d.49 and d.101 respectively), and 10 weeks after the final immunization (d.184), and total Env immunoglobulin responses were determined either by ConA captured VLP ELISA (A) and (B), or using CON-S CFI gp140 avi protein coating (d. 184) (C), while Gag responses were measured against the p27 protein at d.49 (D). AUC values were determined from either 2-fold (Env responses) or 3-fold (Gag responses) series of endpoint titrations of individual mice. Statistical significance is indicated with (*) (p-value < 0.05). Horizontal lines indicate median of values.
As Env conformation clearly varied between the CON-S gp160Δ139 and the CON-S gp160 transduced cells, we also wanted to measure responses towards a similar epitope from the same expression cassette, Thus, p27 Gag specific responses were analysed in samples harvested 7 weeks after the first immunization. We did not observe any differences in the p27 specific responses (Fig. 2D).
3.3. HIV-1 Specific Binding Antibody Responses
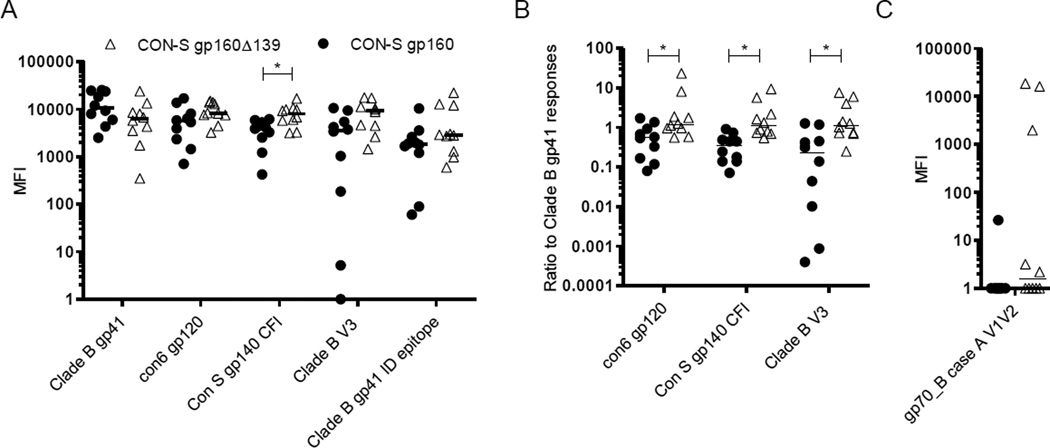
The Env specificity of the antibody responses after final immunization was determined by the BAMA assay. As predicted from the ELISA data using pseudovirus-derived Env and the CON-S CFI gp140 avi protein, the CON-S gp160Δ139 primed group had a significantly higher response against CON-S gp140 CFI than the CON-S gp160 primed group (p-value=0.029, Wilcoxon Exact Test, FDR corrected p-value [39]). In addition, the CON-S gp160Δ139 priming vaccine regimen elicited stronger responses against the V3 loop from clade B, and against con6 gp120; although these differences did not reach significance levels (Fig. 3A). Notably, the CON-S gp160 primed grouped exhibited higher responses towards gp41 which were not significant, but nevertheless quite prominent given the stronger responses towards the gp120, V3 loop, and gp140 antigens in the CON-S gp160Δ139 priming vaccine regimen. Thus, we examined the ratio of the gp41 responses versus the CON-S gp140 CFI, con6 gp120 and V3 loop specific responses. These analyses clearly separated the two groups and demonstrated that relative to each other, the CON-S gp160 priming vaccine is gp41 weighted whereas the CON-S gp160Δ139 priming vaccine regimen induces stronger responses towards antigens containing gp120 or fragments thereof (Fig. 3B). Reactivity towards V1V2 was also assessed, but the responses were modest and only found in the vaccination group primed with CON-S gp160Δ139 (Fig. 3C).
Figure 3.
Specificities of immune responses after final immunizations. (A) Serum samples harvested after the final immunization (d.184) were analysed for binding to the indicated HIV-1 Env antigens. Binding is shown as MFI. (B) Ratio of indicated HIV-1 Env antigens to Clade B gp41 antigen. (C) Serum samples from (A) were analysed for binding to the V1V2 loop antigen fused to the MuLV gp70 leader sequence and plotted with the MuLV gp70 background binding value subtracted. Statistical significance is indicated with (*) (p-value < 0.05). Horizontal lines indicate median of values.
3.4. Neutralization
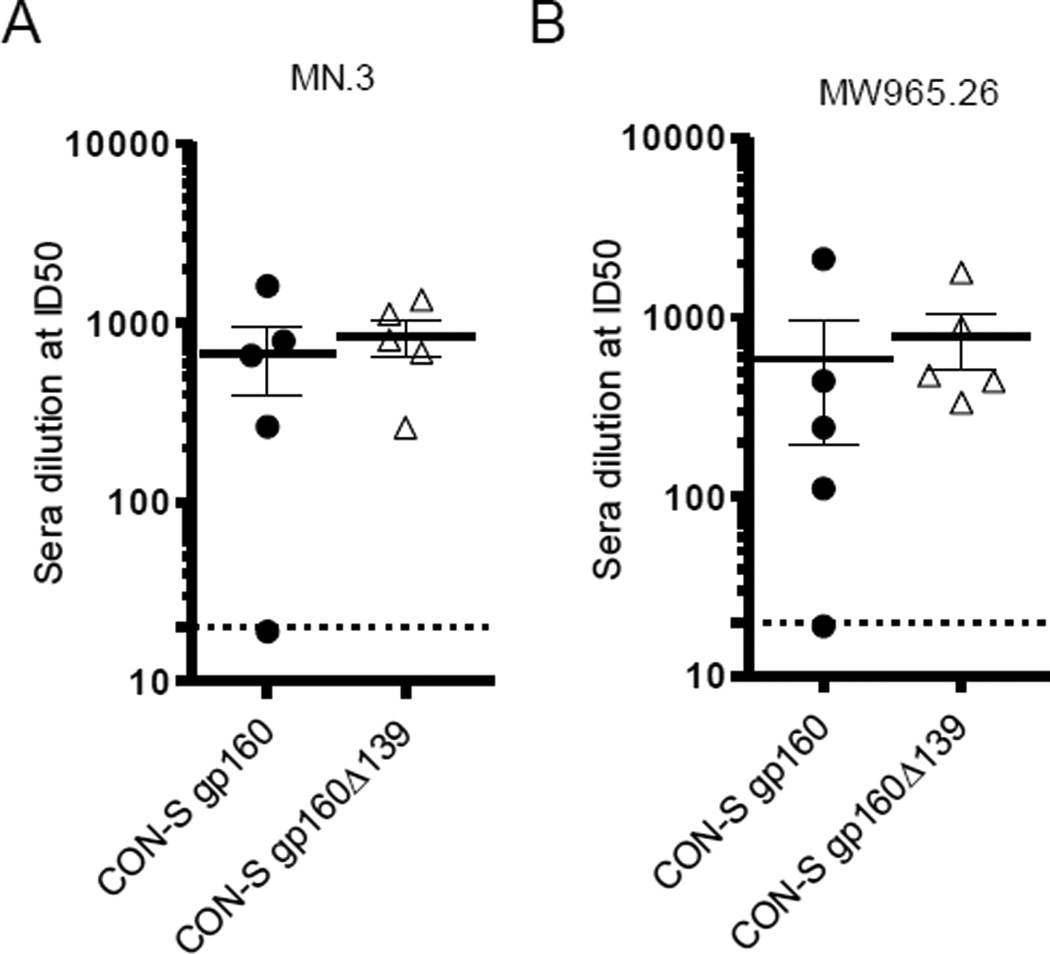
Neutralizing responses were determined against the tier 1 strains MN.3 and MW965.26 and the homologous CON-S strain using the luciferase assay with TZM-bl cells [31]. Serum samples from 10 weeks after the final immunization were pooled in groups of two. All pooled samples had neutralizing antibodies except for one from the CON-S gp160 group (Fig. 4A and B). Similar neutralization capabilities were observed for both vaccination groups against both of the tier 1 strains. Notably, neutralizing responses were not detected against the autologous tier 2 CON-S pseudovirus.
Figure 4.
Neutralizing antibody responses after final immunizations. Neutralization titers were measured in serum samples from d.184 against MN.3 (clade B tier 1) (A) and MW965.26 (clade C tier 1) (B) in the TZM-bl cell based assay. Error bars represent standard error of mean. Dotted lines indicate detection limit.
4. Discussion
We designed two cassettes encoding SIVmac239 Gag and either CON-S gp160 or a truncated CON-S gp160 leaving a cytoplasmic tail of 16 amino acids, and inserted these cassettes into huAd5 vectors. The cassette encoding SIVmac239 Gag and CON-S gp160 was also encoded in two heterologous adenoviral vectors: chAd3 and chAd63. The four vectors were used successfully in mouse immunization regimens where the priming vaccine differed by the use of either the CON-S gp160 or the truncated CON-S gp160Δ139 in huAd5 vectors. These results describe a heterologous vectored adenovirus prime-boost-boost regimen and an investigation into the importance of the Env cytoplasmic tail in the priming of such an immunization regimen.
Following immunization, the antibody titers increased with each immunization irrespectively of the prime. Comparing the humoral responses in the two immunization regimens, the group primed with truncated Env demonstrated a preference for responses against gp120, gp140, and the V3 loop. In contrast, priming with full length Env elicited a relatively higher gp41 response. In a human immunogenicity trial Goepfert et al. found a skewing towards gp41 responses independently if a person was primed with either DNA encoding VLPs with gp160 or MVA encoding gp150 (truncated Env) [8]. However, they also observed that the group primed with MVA encoding a truncated Env had higher titer gp120 responses versus the DNA primed group. In accordance with these and our results, studies have shown that various Env truncations lead to modified antibody binding to the ectodomains of gp120 and gp41 [12, 44]. Thus, it is possible that the skewing towards gp120 responses in the CON-S gp160Δ139 regimen is due to structural effects of the cytoplasmic tail truncation, leading to exposure of epitopes normally hidden in the gp120 of the full length Env and thus more immunodominance of the gp120 protein relative to the gp41 that is more similar between the constructs. Another factor contributing to the specificity of vaccine induced antibodies is the number of Env trimers incorporated in each VLP. Virus particles displaying full length Env and only incorporating few Env trimers would have the Env trimers situated sparsely on the particle, providing broad access to the ectodomain of gp41 and to the region of gp120 in closest proximity to gp41. In contrast, cytoplasmic tail truncations leading to an increase in particle Env incorporation would lead to tighter packaging of the incorporated Env and hypothetically limit the access to the gp41 Env domains as recently suggested [45]. In support of this, we observed stronger PGT151 binding in full length Env and similar PGT145 binding using a full length Env construct. Increased access to gp41 could potentially explain preferential elicitation of gp41 antibodies in the full length Env primed regimen. The higher variation in the elicited responses observed within the CON-S gp160 primed group compared to the CON-S gp160Δ139 group could possibly also be due to the decreased diversity of epitopes presented on the full length Env (cf. Fig. 1F).
The complex nature of Env has made it a difficult target for induction of neutralizing antibodies. Both immunization regimens elicited tier 1a neutralizing antibodies, but failed to elicit neutralizing responses against the autologous tier 2 virus. The observed neutralization titers were similar despite the truncated Env displaying an increased VRC01 binding, and eliciting stronger binding antibody responses. Thus, it can be suggested that induction of neutralizing responses primarily benefit from the presentation of trimer specific epitopes. Such epitopes were presented at roughly similar levels on the surface of vector transduced cells. However, the observed increase in 17b binding to truncated Env suggests that the truncated Env has a more open quaternary conformation. While we were unable to induce tier 2 neutralizing antibodies, the recent demonstration of tier 2 neutralizing antibody induction using well organized SOSIP trimers has intrigued the field [46]. Whereas the SOSIP trimers demonstrated that a correctly folded Env can select for neutralization, such antigens cannot as yet select for breadth [46]. With this in mind it is unclear if attempts at inducing tier 2 neutralization offer a more feasible path towards a clinically useful vaccine, as compared to attempts of inducing protective and binding non-neutralizing antibodies that have already been partially effective in the RV144 trial [6].
Here we have shown that Env truncation can select for stronger gp120 binding antibody responses, which may be a useful component in an HIV-1 vaccine, but it comes at the expense of displaying disproportionally high numbers of gp120 monomeric epitopes in comparison to trimer specific and conformation specific epitopes. In contrast, the full length Env avoided the excess in gp120 monomer epitope display. While our regimen was not independently capable of inducing tier 2 neutralization, this information suggest that the full length Env structure may be preferable in other prime-boost regimens involving VLPs when attempting to induce neutralizing responses.
In summary, this study shows the potential of using heterologous adenoviral vectors encoding VLPs as HIV-1 immunogens. The adenoviral vectors could be designed to modify epitope display and induced continuously increasing humoral responses and cross-clade neutralization of tier 1 viruses.
Highlights.
Heterologous adenoviral vectors encoding HIV-1/SIV VLPs can boost responses in prime-double boost immunization regimens
Tail truncation of HIV CON-S increases antigen presentation with a bias towards gp120 epitopes and CD4 induced epitopes relatively to full length Env
Priming with an Env CT truncated VLP vaccine skews antibody responses towards gp120 specificities, compared to full-length Env priming responses being relatively skewed towards gp41 specificities
Acknowledgments
We thank Birita Fritleifsdóttir Kjærbæk and Bang Nguyen for their technical assistance. We thank Dr. Barton Haynes and Dr. Larry Liao (Duke Human Vaccine Institute, Durham, NC) for contributing essential reagents. We thank Drs. John C. Kappes and Xiaoyun Wu (pSG3Δenv plasmid), Dr. John Mascola (VRC01 monoclonal antibody), Dr. Dennis Burton and Carlos Barbas (IgG1 b12 monoclonal antibody), Dr. Hermann Katinger (2G12 monoclonal antibody), Dr. James E. Robinson (17b monoclonal antibody), Dr. Niels Pedersen (55-2F12 monoclonal antibody), for providing reagents through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH. SIVmac251 antiserum was obtained through the NIH AIDS Reagent Program.
Funding
This work was supported by the Lundbeck foundation, the AIDS foundation, the Arvid Nilssons foundation, the foundation for the Advancement of Medical Sciences, the Axel Muusfeldt’s foundation and the Duke Center for AIDS Research Immunology Core (NIH/DAIDS AI064518).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Boutwell CL, Rolland MM, Herbeck JT, Mullins JI, Allen TM. Viral evolution and escape during acute HIV-1 infection. J Infect Dis. 2010;202(Suppl 2):S309–S314. doi: 10.1086/655653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma BJ, Alam SM, Go EP, Lu X, Desaire H, Tomaras GD, et al. Envelope deglycosylation enhances antigenicity of HIV-1 gp41 epitopes for both broad neutralizing antibodies and their unmutated ancestor antibodies. PLoS Pathog. 2011;7:e1002200. doi: 10.1371/journal.ppat.1002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med. 2013;210:655–663. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwong PD, Mascola JR, Nabel GJ. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat Rev Immunol. 2013;13:693–701. doi: 10.1038/nri3516. [DOI] [PubMed] [Google Scholar]
- 5.Wibmer CK, Moore PL, Morris L. HIV broadly neutralizing antibody targets. Curr Opin HIV AIDS. 2015;10:135–143. doi: 10.1097/COH.0000000000000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corey L, Gilbert PB, Tomaras GD, Haynes BF, Pantaleo G, Fauci AS. Immune correlates of vaccine protection against HIV-1 acquisition. Sci Transl Med. 2015;7:310rv7. doi: 10.1126/scitranslmed.aac7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 8.Goepfert PA, Elizaga ML, Seaton K, Tomaras GD, Montefiori DC, Sato A, et al. Specificity and 6-month durability of immune responses induced by DNA and recombinant modified vaccinia Ankara vaccines expressing HIV-1 virus-like particles. J Infect Dis. 2014;210:99–110. doi: 10.1093/infdis/jiu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyatt LS, Belyakov IM, Earl PL, Berzofsky JA, Moss B. Enhanced cell surface expression, immunogenicity and genetic stability resulting from a spontaneous truncation of HIV Env expressed by a recombinant MVA. Virology. 2008;372:260–272. doi: 10.1016/j.virol.2007.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cottingham MG, Carroll F, Morris SJ, Turner AV, Vaughan AM, Kapulu MC, et al. Preventing spontaneous genetic rearrangements in the transgene cassettes of adenovirus vectors. Biotechnol Bioeng. 2012;109:719–728. doi: 10.1002/bit.24342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang BZ, Liu W, Kang SM, Alam M, Huang C, Ye L, et al. Incorporation of high levels of chimeric human immunodeficiency virus envelope glycoproteins into virus-like particles. J Virol. 2007;81:10869–10878. doi: 10.1128/JVI.00542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Kovacs JM, Peng H, Rits-Volloch S, Lu J, Park D, et al. HIV-1 ENVELOPE. Effect of the cytoplasmic domain on antigenic characteristics of HIV-1 envelope glycoprotein. Science. 2015;349:191–195. doi: 10.1126/science.aaa9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holst PJ, Bartholdy C, Stryhn A, Thomsen AR, Christensen JP. Rapid and sustained CD4(+) T-cell-independent immunity from adenovirus-encoded vaccine antigens. J Gen Virol. 2007;88:1708–1716. doi: 10.1099/vir.0.82727-0. [DOI] [PubMed] [Google Scholar]
- 14.Colloca S, Barnes E, Folgori A, Ammendola V, Capone S, Cirillo A, et al. Vaccine vectors derived from a large collection of simian adenoviruses induce potent cellular immunity across multiple species. Sci Transl Med. 2012;4:115ra2. doi: 10.1126/scitranslmed.3002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao HX, Sutherland LL, Xia SM, Brock ME, Scearce RM, Vanleeuwen S, et al. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology. 2006;353:268–282. doi: 10.1016/j.virol.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vitelli A, Quirion MR, Lo CY, Misplon JA, Grabowska AK, Pierantoni A, et al. Vaccination to conserved influenza antigens in mice using a novel Simian adenovirus vector, PanAd3, derived from the bonobo Pan paniscus. PLoS One. 2013;8:e55435. doi: 10.1371/journal.pone.0055435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker TC, Noel RJ, Coats WS, Gomez-Foix AM, Alam T, Gerard RD, et al. Use of recombinant adenovirus for metabolic engineering of mammalian cells. Methods Cell Biol. 1994;43(Pt A):161–189. doi: 10.1016/s0091-679x(08)60603-2. [DOI] [PubMed] [Google Scholar]
- 19.Peruzzi D, Dharmapuri S, Cirillo A, Bruni BE, Nicosia A, Cortese R, et al. A novel chimpanzee serotype-based adenoviral vector as delivery tool for cancer vaccines. Vaccine. 2009;27:1293–1300. doi: 10.1016/j.vaccine.2008.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai L, Vodros D, Kozlowski PA, Montefiori DC, Wilson RL, Akerstrom VL, et al. GM-CSF DNA: an adjuvant for higher avidity IgG, rectal IgA, and increased protection against the acute phase of a SHIV-89.6P challenge by a DNA/MVA immunodeficiency virus vaccine. Virology. 2007;369:153–167. doi: 10.1016/j.virol.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgins JR, Sutjipto S, Marx PA, Pedersen NC. Shared antigenic epitopes of the major core proteins of human and simian immunodeficiency virus isolates. J Med Primatol. 1992;21:265–269. [PubMed] [Google Scholar]
- 23.Seaton KE, Ballweber L, Lan A, Donathan M, Hughes S, Vojtech L, et al. HIV-1 specific IgA detected in vaginal secretions of HIV uninfected women participating in a microbicide trial in Southern Africa are primarily directed toward gp120 and gp140 specificities. PLoS One. 2014;9:e101863. doi: 10.1371/journal.pone.0101863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, Chavez LL, et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol. 2008;82:12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yates NL, Liao HX, Fong Y, deCamp A, Vandergrift NA, Williams WT, et al. Vaccine-induced Env V1-V2 IgG3 correlates with lower HIV-1 infection risk and declines soon after vaccination. Sci Transl Med. 2014;6:228ra39. doi: 10.1126/scitranslmed.3007730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yates NL, Stacey AR, Nolen TL, Vandergrift NA, Moody MA, Montefiori DC, et al. HIV-1 gp41 envelope IgA is frequently elicited after transmission but has an initial short response half-life. Mucosal Immunol. 2013;6:692–703. doi: 10.1038/mi.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Pena AT, et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, et al. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montefiori DC. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol. 2009;485:395–405. doi: 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
- 32.Ragonnaud E, Andersson AM, Pedersen AE, Laursen H, Holst PJ. An adenoviral cancer vaccine co-encoding a tumor associated antigen together with secreted 4-1BBL leads to delayed tumor progression. Vaccine. 2016;34:2147–2156. doi: 10.1016/j.vaccine.2015.06.087. [DOI] [PubMed] [Google Scholar]
- 33.Williams K, Westmoreland S, Greco J, Ratai E, Lentz M, Kim WK, et al. Magnetic resonance spectroscopy reveals that activated monocytes contribute to neuronal injury in SIV neuroAIDS. J Clin Invest. 2005;115:2534–2545. doi: 10.1172/JCI22953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reimann KA, Tenner-Racz K, Racz P, Montefiori DC, Yasutomi Y, Lin W, et al. Immunopathogenic events in acute infection of rhesus monkeys with simian immunodeficiency virus of macaques. J Virol. 1994;68:2362–2370. doi: 10.1128/jvi.68.4.2362-2370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 36.Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, et al. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol. 2002;76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loeschner K, Hadrup N, Qvortrup K, Larsen A, Gao X, Vogel U, et al. Distribution of silver in rats following 28 days of repeated oral exposure to silver nanoparticles or silver acetate. Part Fibre Toxicol. 2011;8:18. doi: 10.1186/1743-8977-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 40.Hulot SL, Korber B, Giorgi EE, Vandergrift N, Saunders KO, Balachandran H, et al. Comparison of Immunogenicity in Rhesus Macaques of Transmitted-Founder, HIV-1 Group M Consensus, and Trivalent Mosaic Envelope Vaccines Formulated as a DNA Prime, NYVAC, and Envelope Protein Boost. J Virol. 2015;89:6462–6480. doi: 10.1128/JVI.00383-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao HX, Tsao CY, Alam SM, Muldoon M, Vandergrift N, Ma BJ, et al. Antigenicity and immunogenicity of transmitted/founder, consensus, and chronic envelope glycoproteins of human immunodeficiency virus type 1. J Virol. 2013;87:4185–4201. doi: 10.1128/JVI.02297-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuller SD, Wilk T, Gowen BE, Krausslich HG, Vogt VM. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 43.Wyatt LS, Earl PL, Liu JY, Smith JM, Montefiori DC, Robinson HL, et al. Multiprotein HIV type 1 clade B DNA and MVA vaccines: construction, expression, and immunogenicity in rodents of the MVA component. AIDS Res Hum Retroviruses. 2004;20:645–653. doi: 10.1089/0889222041217428. [DOI] [PubMed] [Google Scholar]
- 44.Edwards TG, Wyss S, Reeves JD, Zolla-Pazner S, Hoxie JA, Doms RW, et al. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J Virol. 2002;76:2683–2691. doi: 10.1128/JVI.76.6.2683-2691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sliepen K, Ozorowski G, Burger JA, van Montfort T, Stunnenberg M, LaBranche C, et al. Presenting native-like HIV-1 envelope trimers on ferritin nanoparticles improves their immunogenicity. Retrovirology. 2015;12:82. doi: 10.1186/s12977-015-0210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, et al. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]