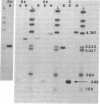
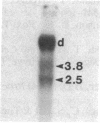
Abstract
We have determined from nucleotide sequence analysis that the subterminal and terminal exons of a respiratory chain NADH dehydrogenase subunit I gene in broad bean mitochondrial DNA (mtDNA) are separated by a group II intron. Within this intron is a 687-codon open reading frame that, from considerations of similarity between amino acid sequences predicted from this open reading frame and maturase-coding sequences in group II introns of certain fungal mitochondrial genes, appears to encode a maturase-related protein. Transcripts complementary to this broad bean sequence (designated a mat-r gene) were detected among RNAs isolated from broad bean mitochondria. Data obtained from DNA-DNA hybridizations indicated that soybean and corn mtDNAs also contain a mat-r gene and suggested that only one copy of this gene occurs in each plant mtDNA. The putative protein specified by the broad bean mat-r gene contains amino acid sequences characteristic of reverse transcriptases. Because of this, consideration is given to the possibility that the maturase-related protein may be functional in the mechanisms by which plant mtDNA sequences are rearranged and foreign sequences are incorporated into plant mtDNAs.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailey-Serres J., Hanson D. K., Fox T. D., Leaver C. J. Mitochondrial genome rearrangement leads to extension and relocation of the cytochrome c oxidase subunit I gene in sorghum. Cell. 1986 Nov 21;47(4):567–576. doi: 10.1016/0092-8674(86)90621-5. [DOI] [PubMed] [Google Scholar]
- Benne R., Van den Burg J., Brakenhoff J. P., Sloof P., Van Boom J. H., Tromp M. C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986 Sep 12;46(6):819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Bland M. M., Levings C. S., 3rd, Matzinger D. F. The tobacco mitochondrial ATPase subunit 9 gene is closely linked to an open reading frame for a ribosomal protein. Mol Gen Genet. 1986 Jul;204(1):8–16. doi: 10.1007/BF00330180. [DOI] [PubMed] [Google Scholar]
- Bonitz S. G., Coruzzi G., Thalenfeld B. E., Tzagoloff A., Macino G. Assembly of the mitochondrial membrane system. Structure and nucleotide sequence of the gene coding for subunit 1 of yeast cytochrme oxidase. J Biol Chem. 1980 Dec 25;255(24):11927–11941. [PubMed] [Google Scholar]
- Carignani G., Groudinsky O., Frezza D., Schiavon E., Bergantino E., Slonimski P. P. An mRNA maturase is encoded by the first intron of the mitochondrial gene for the subunit I of cytochrome oxidase in S. cerevisiae. Cell. 1983 Dec;35(3 Pt 2):733–742. doi: 10.1016/0092-8674(83)90106-x. [DOI] [PubMed] [Google Scholar]
- Carignani G., Netter P., Bergantino E., Robineau S. Expression of the mitochondrial split gene coding for cytochrome oxidase subunit I in S. cerevisiae: RNA splicing pathway. Curr Genet. 1986;11(1):55–63. doi: 10.1007/BF00389426. [DOI] [PubMed] [Google Scholar]
- Clary D. O., Wahleithner J. A., Wolstenholme D. R. Sequence and arrangement of the genes for cytochrome b, URF1, URF4L, URF4, URF5, URF6 and five tRNAs in Drosophila mitochondrial DNA. Nucleic Acids Res. 1984 May 11;12(9):3747–3762. doi: 10.1093/nar/12.9.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey R. E., Levings C. S., 3rd, Timothy D. H. Novel recombinations in the maize mitochondrial genome produce a unique transcriptional unit in the Texas male-sterile cytoplasm. Cell. 1986 Feb 14;44(3):439–449. doi: 10.1016/0092-8674(86)90465-4. [DOI] [PubMed] [Google Scholar]
- Fox T. D., Leaver C. J. The Zea mays mitochondrial gene coding cytochrome oxidase subunit II has an intervening sequence and does not contain TGA codons. Cell. 1981 Nov;26(3 Pt 1):315–323. doi: 10.1016/0092-8674(81)90200-2. [DOI] [PubMed] [Google Scholar]
- Gualberto J. M., Wintz H., Weil J. H., Grienenberger J. M. The genes coding for subunit 3 of NADH dehydrogenase and for ribosomal protein S12 are present in the wheat and maize mitochondrial genomes and are co-transcribed. Mol Gen Genet. 1988 Dec;215(1):118–127. doi: 10.1007/BF00331312. [DOI] [PubMed] [Google Scholar]
- Hiesel R., Brennicke A. Cytochrome oxidase subunit II gene in mitochondria of Oenothera has no intron. EMBO J. 1983;2(12):2173–2178. doi: 10.1002/j.1460-2075.1983.tb01719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper M. T., Lambowitz A. M. A novel reverse transcriptase activity associated with mitochondrial plasmids of Neurospora. Cell. 1988 Nov 18;55(4):693–704. doi: 10.1016/0092-8674(88)90228-0. [DOI] [PubMed] [Google Scholar]
- Lambowitz A. M. Infectious introns. Cell. 1989 Feb 10;56(3):323–326. doi: 10.1016/0092-8674(89)90232-8. [DOI] [PubMed] [Google Scholar]
- Lang B. F., Ahne F., Bonen L. The mitochondrial genome of the fission yeast Schizosaccharomyces pombe. The cytochrome b gene has an intron closely related to the first two introns in the Saccharomyces cerevisiae cox1 gene. J Mol Biol. 1985 Aug 5;184(3):353–366. doi: 10.1016/0022-2836(85)90286-4. [DOI] [PubMed] [Google Scholar]
- Lonsdale D. M., Hodge T. P., Howe C. J., Stern D. B. Maize mitochondrial DNA contains a sequence homologous to the ribulose-1,5-bisphosphate carboxylase large subunit gene of chloroplast DNA. Cell. 1983 Oct;34(3):1007–1014. doi: 10.1016/0092-8674(83)90558-5. [DOI] [PubMed] [Google Scholar]
- Matsuura E. T., Domenico J. M., Cummings D. J. An additional class II intron with homology to reverse transcriptase in rapidly senescing Podospora anserina. Curr Genet. 1986;10(12):915–922. doi: 10.1007/BF00398289. [DOI] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Jacquier A. Long-range intron-exon and intron-intron pairings involved in self-splicing of class II catalytic introns. Cold Spring Harb Symp Quant Biol. 1987;52:201–212. doi: 10.1101/sqb.1987.052.01.025. [DOI] [PubMed] [Google Scholar]
- Michel F., Lang B. F. Mitochondrial class II introns encode proteins related to the reverse transcriptases of retroviruses. Nature. 1985 Aug 15;316(6029):641–643. doi: 10.1038/316641a0. [DOI] [PubMed] [Google Scholar]
- Rottmann W. H., Brears T., Hodge T. P., Lonsdale D. M. A mitochondrial gene is lost via homologous recombination during reversion of CMS T maize to fertility. EMBO J. 1987 Jun;6(6):1541–1546. doi: 10.1002/j.1460-2075.1987.tb02398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster W., Brennicke A. Plastid, nuclear and reverse transcriptase sequences in the mitochondrial genome of Oenothera: is genetic information transferred between organelles via RNA? EMBO J. 1987 Oct;6(10):2857–2863. doi: 10.1002/j.1460-2075.1987.tb02587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Bang A. G., Thompson W. F. The watermelon mitochondrial URF-1 gene: evidence for a complex structure. Curr Genet. 1986;10(11):857–869. doi: 10.1007/BF00418532. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Palmer J. D. Extensive and widespread homologies between mitochondrial DNA and chloroplast DNA in plants. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1946–1950. doi: 10.1073/pnas.81.7.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H., Hayashida H., Miyata T. Sequence homology between retroviral reverse transcriptase and putative polymerases of hepatitis B virus and cauliflower mosaic virus. 1983 Oct 27-Nov 2Nature. 305(5937):827–829. doi: 10.1038/305827a0. [DOI] [PubMed] [Google Scholar]
- Wahleithner J. A., Wolstenholme D. R. Mitochondrial plasmid DNAs of broad bean: nucleotide sequences, complex secondary structures, and transcription. Curr Genet. 1987;12(1):55–67. doi: 10.1007/BF00420728. [DOI] [PubMed] [Google Scholar]
- Wahleithner J. A., Wolstenholme D. R. Ribosomal protein S14 genes in broad bean mitochondrial DNA. Nucleic Acids Res. 1988 Jul 25;16(14B):6897–6913. doi: 10.1093/nar/16.14.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissinger B., Hiesel R., Schuster W., Brennicke A. The NADH-dehydrogenase subunit 5 gene in Oenothera mitochondria contains two introns and is co-transcribed with the 5 S rRNA gene. Mol Gen Genet. 1988 Apr;212(1):56–65. doi: 10.1007/BF00322444. [DOI] [PubMed] [Google Scholar]
- Wolstenholme D. R., Macfarlane J. L., Okimoto R., Clary D. O., Wahleithner J. A. Bizarre tRNAs inferred from DNA sequences of mitochondrial genomes of nematode worms. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1324–1328. doi: 10.1073/pnas.84.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y. E., Eickbush T. H. Functional expression of a sequence-specific endonuclease encoded by the retrotransposon R2Bm. Cell. 1988 Oct 21;55(2):235–246. doi: 10.1016/0092-8674(88)90046-3. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. Similarity of reverse transcriptase-like sequences of viruses, transposable elements, and mitochondrial introns. Mol Biol Evol. 1988 Nov;5(6):675–690. doi: 10.1093/oxfordjournals.molbev.a040521. [DOI] [PubMed] [Google Scholar]