Abstract
Introduction:
Cryptococcal meningitis (CM) may be caused by several species of Cryptococcus.
Case presentation:
We describe a fatal case of CM in a HIV-positive patient from Ivory Coast infected by Cryptococcus neoformans VNI and Cryptococcus deuterogattii. Isolates were recovered from cerebrospinal fluid (CSF) prior to systemic antifungal treatment. Six isolates were studied (the entire culture plus five isolated colonies from it). Serotyping was performed via LAC 1 and CAP 64 gene amplification. Genotyping was performed using restriction fragment length polymorphism (RFLP) analysis of the URA5 gene, (GACA)4, (GTG)5 and M13 PCR fingerprinting. URA5-RFLP analysis identified the original culture with two different molecular type combinations. However, URA5-RFLP profiles of the five colonies isolated from the original sample revealed two different species. Four colonies were identified as C. deuterogattii and the last isolate as C. neoformans VNI. The in vitro susceptibility profile was determined using the standard method according to the CLSI M27-A3 protocol. The isolates were susceptible to the tested antifungals (fluconazole, flucytosine and amphotericin B). Treatment with fluconazole (1200 mg day−1) was initiated; however, the patient died 17 days after the onset of antifungal therapy.
Conclusion:
This is the first reported case of mixed infection with C. neoformans and C. deuterogattii in a HIV-positive patient.
Keywords: AIDS, Cryptococcus deuterogattii, Cryptococcus neoformans, genotyping, mixed infection, antifungal susceptibility
Introduction
Cryptococcus complex species is a major cause of fungal meningoencephalitis in immunocompromised patients in Africa (Aoussi et al., 2012; Bertout et al., 2013). This encapsulated basidiomycetous yeast is widespread in the environment, where it is associated most commonly with avian excreta and tree hollows (Connolly et al., 1999). The initial infection is acquired by inhalation of fungal cells from an environmental source where mixed yeast populations may co-exist. The taxonomy of the Cryptococcus neoformans/Cryptococcus gattii complex species has recently been revised and contains seven proposed species and thirteen genotypes (Hagen et al., 2015). A single isolate of Cryptococcus complex species has been thought to be responsible for the disease (Desnos-Ollivier et al., 2010). However, the isolation of strains with different genotypes, serotypes or species during the same episode in a unique sample makes infestation by multiple strains likely (Aminnejad et al., 2012; Barchiesi et al., 1995; Bertout et al., 2013; Bovers et al., 2006, 2008; Igreja et al., 2004; Illnait-Zaragozí et al., 2010; Mlinaric-Missoni et al., 2011). Isolates are usually susceptible to the standard clinically used antifungals , including fluconazole, 5-fluorocytosine and amphotericin B (Mlinaric-Missoni et al., 2011; Favalessa et al., 2014). The use of antifungal agents, particularly in long-term suppressive regimens, has raised concern about the development of drug resistance in C. neoformans. Resistance to antifungal drugs is scarce among clinical isolates of C. neoformans but has been reported. Nevertheless, reduced susceptibility between different species of Cryptococcus has been demonstrated worldwide (Chong et al., 2010; Chowdhary et al., 2011; Trilles et al., 2012).
We describe the first report of mixed infection with C. neoformans VNI and C. deuterogattii in a HIV-positive patient. The possibility of mixed infection must be considered for the management of cryptococcosis.
Case report
In January 2014, a 41-year-old male was hospitalized in the Infectious and Tropical Diseases Unit of Treichville (Abidjan, Ivory Coast) because of complaints of high-grade fever, and severe headache lasting for three weeks associated with depressed level of consciousness and multiple episodes of vomiting.
Known to be HIV-1 positive since 2012, this patient was under second line of antiretroviral treatment combining tenofovir (245 mg day−1), emtricitabine (200 mg day−1) and lopinavir/ritonavir (400/100 mg twice daily).
Investigations
On physical examination, the patient was confused and his speech was incoherent. No meningismus or focal neurological signs were found. Full blood count revealed a decrease of haemoglobin down to 7.6 g dl−1, CD4 count was 75 cells mm−3 and the plasma HIV viral load was 704 275 copies ml−1. Lumbar puncture revealed an elevated opening pressure (40 cm H2O) of the cerebrospinal fluid (CSF) containing a low glucose concentration (0.17 g l−1) and elevated protein level (1.35 g l−1). CSF bacterial cultures were sterile. However, direct microscopic examination of the CSF using India ink detected numerous encapsulated yeasts suggesting Cryptococcus spp. and a diagnosis of cryptococcal meningitis (CM) was given by the physicians.
Diagnosis
Culture of the CSF on Sabouraud dextrose agar medium (Biomerieux) at 37 °C for 3 days allowed isolation of smooth yellowish colonies that subsequently showed to be urease-positive. A positive test was observed by a colour change from orange to pink, due to the production of urease by Cryptococcus complex species in the medium. For urease-positive cultures, culture on Niger seeds agar, as previously described, was carried out to certify that the strains present are effectively of Cryptococcus species (Staib et al., 1987). Colonies of Cryptococcus complex species were identified by production of brown melanin pigment. Finally, the genus and the species were confirmed by using the API 20C test (Biomerieux).
Phenotypic characterization of the Cryptococcus species was achieved by chemotyping in l-canavanine-glycine-bromothymol blue (CGB) agar. CGB agar was used to differentiate C. neoformans complex species and C. gattii complex species as described previously (Klein et al., 2009). Growth of C. gattii complex species on CGB agar produced a blue color, indicating the assimilation of glycine, while C. neoformans complex species failed to cause a colour change. Six isolates (entire culture and five isolated colonies) were separated for further investigations. A blue-colour change on CGB agar suggesting C. gattii was observed for four out of five colonies. Serotyping was performed via LAC 1 and CAP 64 gene amplification. To gain more interpretation of the isolates profiles, genotyping was performed using M13, (GACA)4 and (GTG)5 primers and RFLP analysis of the URA5 gene. Mixed patterns of both C. neoformans VNI and Cryptococcus deuterogattii species were observed in the entire culture. Concerning the isolated colonies, one was identified as C. neoformans VNI and four as C. deuterogattii.
Susceptibilities to amphotericin B, flucytosine and fluconazole were tested by the broth microdilution method according to the M27-A3 CLSI protocol, 2008 (CLSI, 2008). For Cryptococcus species, clinical break-points have not been established by the CLSI, so we used epidemiological cut-off values as described previously (Espinel-Ingroff et al., 2012a, b; Pfaller et al., 2005): fluconazole and flucytosine, susceptible ≤8 µg ml−1; amphotericin B, susceptible ≤1 µg ml−1. All isolates exhibited a low MIC value to fluconazole (4 µg ml−1), flucytosine (2 µg ml−1) and amphotericin B (0.5 µg ml−1).
Treatment
Patient received high oral dose of fluconazole (1200 mg day−1).
Outcome and follow-up
On day 12 of admission, the patient continued to display altered mental status (GCS 6/15) with unresolved pyrexia, headaches and persistently low haemoglobin unresponsive to transfusion. He died at day 17 after the initiation of antifungal therapy.
Discussion
Due to the importance of the C. neoformans/C. gattii species complex as human fungal pathogens, several research groups are currently focusing on the molecular determination of the number of genetically divergent subgroups within each species. The application of molecular methods, such as PCR fingerprinting, amplified fragment length polymorphism (AFLP) and, more recently, multi-locus sequence typing (MLST) has led to a better insight into the taxonomy and identification of the causative agents of cryptococcosis (Bovers et al., 2006, 2008). Advances in phylogenetic and genotypic studies now suggest seven species in the C. gattii/C. neoformans species complex. Interspecies hybrids between C. gattii and C. neoformans were also considered: C. neoformans var. neoformans AFLP2/VNIV × C. gattii AFLP4/VGI (serotype BD, genotype AFLP8), C. neoformans var. grubii AFLP1/VNI × C. gattii AFLP4/VGI (serotype AB, genotype AFLP9) and C. neoformans var. grubii AFLP1/VNI × C. deuterogattii AFLP6/VGII (serotype AB, genotype AFLP11) (Aminnejad et al., 2012; Bovers et al., 2006, 2008).
Previous worldwide studies reported a greater genetic diversity among cryptococcal isolates (Aminnejad et al., 2012; Bertout et al., 2013; Desnos-Ollivier et al., 2010). In the past, a single isolate of Cryptococcus complex species has been thought to be responsible for the disease (Desnos-Ollivier et al., 2010). Various combinations of strains with different genotypes of the same species were found during the same episode of cryptococcosis, sometimes in the same sample (Barchiesi et al., 1995; Bertout et al., 2013; Haynes et al., 1995; Igreja et al., 2004; Illnait -Zaragozí et al., 2010; Mlinaric-Missoni et al., 2011). These results provided evidence for mixed infection with genetically distinct strains acquired from the environment. Infrequently, different serotype or different Cryptococcus species have also been identified during the same episode of CM (Aminnejad et al., 2012; Bovers et al., 2006, 2008; Desnos-Ollivier et al., 2010).
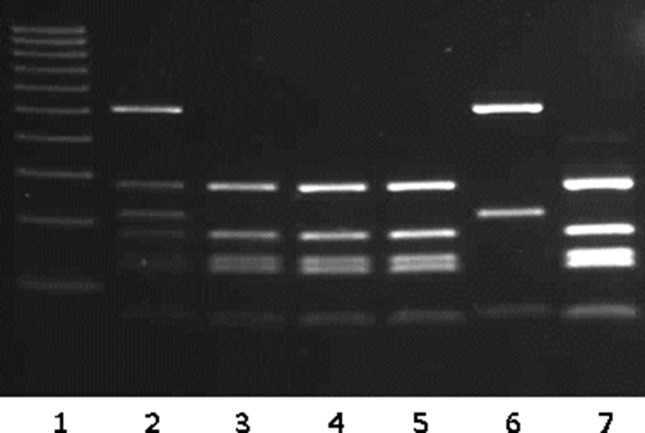
In the present case, serotyping performed on the culture was unable to identify a causative organism. URA5-RFLP analysis identified the original culture with two different molecular type combinations. The M13 PCR fingerprinting profile of this strain confirmed that the original sample contained fragments of both proposed parental groups (VNI and VGII). However, URA5-RFLP profiles of the five colonies isolated from the original sample revealed two different species (Fig. 1). Four colonies were identified as C. deuterogattii and the last isolate as C. neoformans VNI. To the best of our knowledge, our study is the first report in the literature documenting the occurrence of a mixed infection of both C. deuterogattii and C. neoformans VNI in a HIV- positive patient. In contrast, by using the same molecular typing technique, a previous study reported the occurrence of hybrids between C. deuterogattii and C. neoformans (Aminnejad et al., 2012). Until recently, C. gattii species complex was believed to primarily infect immunocompetent individuals, but our results and other reports demonstrated that immunocompromised patients could also be infected by these fungal species (Desnos-Ollivier et al., 2010; Illnait-Zaragozí et al., 2010; Mlinaric-Missoni et al., 2011).
Fig. 1.
URA5 gene restriction fragment length polymorphism (RFLP) of the isolates.
Lanes: 1, Ladder 100 pb (Sigma Aldrich); 2, entire culture; 3–7, the five isolated colonies with lanes 3, 4, 5 and 7 corresponding to C. deuterogattii profiles and lane 6 corresponding to C. neoformans VNI profile.
It is known that exposure to environmental sources, such as contaminated trees and soil can lead to cryptococcosis infection in humans and animals. Recently, a new natural habitat for C. neoformans strains was described in Brazil, consisting of hollow trees in which both varieties of the yeast may be found simultaneously (Lazera et al., 2000). An in vivo study described the isolation of two varieties of C. neoformans from the upper respiratory tract of a koala, indicating a mixed colonization (Connolly et al., 1999). Our finding suggested the existence of environmental C. deuterogattii and C. neoformans species in Ivory Coast. Future studies will focus on recovery of Cryptococcus from soil, eucalyptus trees, flowers and pigeon droppings near human habitats. Understanding the environmental source of infection is critical for understanding the ecology and pathogenesis of this fungus.
Concerning the antifungal susceptibility, isolates of C. neoformans AFLP1/VNI were described in literature to have a significantly higher geometric mean MIC for fluconazole than isolates of the two subgenotypes AFLP1A/VNB/VNII and AFLP1B/VNII (Chong et al., 2010). C. neoformans has been observed to be more susceptible to several antifungals when compared to C. gattii (Chowdhary et al., 2011) and C. deuterogattii (Trilles et al., 2012), but less susceptible for 5-fluorocytosine and amphotericin B when compared to C. gattii (Chau et al., 2010). In our study, results showed that the two isolated species were susceptible to fluconazole, flucytosine and amphotericin B.
In Ivory Coast, despite the availability of antiretroviral treatment, most patients admitted for CM consult at late stages of the disease (Aoussi et al., 2012). The delay for consultation is even longer in cases of CM in HIV-negative patients because of the insidious evolution of symptoms reported in most series (Favalessa et al., 2014; Mlinaric-Missoni et al., 2011). At the time of admission, the patient presented severe headache, fever and vomiting. Previous studies have shown that headache is the predominant clinical sign of patients affected by C. gattii (Cookman et al., 2013; Favalessa et al., 2014, Steele et al., 2010).
Nevertheless, patients with mixed infections did not differ significantly from those with single infections in terms of underlying disease and clinical presentation (Desnos-Ollivier et al., 2010). For this case, the consultation at late stage, the severe immunodepression and the elevated CSF intracranial pressure could explain the early death of the patient.
This study is the first report on the presence of C. neoformans VNI and C. deuterogattii in a HIV-positive patient. It also highlights the first clinical isolation of C. deuterogattii from Ivory Coast.
Acknowledgements
The authors thank the Infectious and Tropical Diseases Unit study of the teaching hospital of Treichville. We thank D. Castel and R. N’cho for technical help. We thank also D T. Ngouana for critically reviewing the manuscript. The present study received the approval of the Sciences Ethical committees of life and health of Ivory Coast.
Abbreviations:
- CM
Cryptococcal meningitis
- CSF
cerebrospinal fluid
References
- Aminnejad M., Diaz M., Arabatzis M., Castañeda E., Lazera M., Velegraki A., Marriott D., Sorrell T. C., Meyer W. (2012). Identification of novel hybrids between Cryptococcus neoformans var. grubii VNI and Cryptococcus gattii VGII. Mycopathologia 173337–346. 10.1007/s11046-011-9491-x [DOI] [PubMed] [Google Scholar]
- Aoussi E. F., Ehui E., Dembélé J. P., Kolia-Diafouka P., Elloh N. F., Ouattara S. I., Tanon K. A., Doumbia A., Adou-Bryn K. D. (2012). Cryptoccocal meningitis and HIV in the era of HAART in Côte d'Ivoire. Med Mal Infect 42349–354. 10.1016/j.medmal.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Barchiesi F., Hollis R. J., Messer S. A., Scalise G., Rinaldi M. G., Pfaller M. A. (1995). Electrophoretic karyotype and in vitro antifungal susceptibility of Cryptococcus neoformans isolates from AIDS patients. Diagn Microbiol Infect Dis 2399–103. 10.1016/0732-8893(95)00169-7 [DOI] [PubMed] [Google Scholar]
- Bertout S., Drakulovski P., Kouanfack C., Krasteva D., Ngouana T., Dunyach-Rémy C., Dongtsa J., Aghokeng A., Delaporte E., other authors (2013). Genotyping and antifungal susceptibility testing of Cryptococcus neoformans isolates from Cameroonian HIV-positive adult patients. Clin Microbiol Infect 19763–769. 10.1111/1469-0691.12019 [DOI] [PubMed] [Google Scholar]
- Bovers M., Hagen F., Kuramae E. E., Diaz M. R., Spanjaard L., Dromer F., Hoogveld H. L., Boekhout T. (2006). Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res 6599–607. 10.1111/j.1567-1364.2006.00082.x [DOI] [PubMed] [Google Scholar]
- Bovers M., Hagen F., Kuramae E. E., Hoogveld H. L., Dromer F., St-Germain G., Boekhout T. (2008). AIDS patient death caused bynovel Cryptococcus neoformans × C. gattii hybrid. Emerg Infect Dis 141105–1108. 10.3201/eid1407.080122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau T. T., Mai N. H., Phu N. H., Nghia H. D., Chuong L. V., Sinh D. X., Duong V. A., Diep P. T., Campbell J. I. (2010). A prospective descriptive study of cryptococcal meningitis in HIV uninfected patients in Vietnam – high prevalence of Cryptococcus neoformans var grubii in the absence of underlying disease. BMC Infect Dis 10199. 10.1186/1471-2334-10-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong H. S., Dagg R., Malik R., Chen S., Carter D. (2010). In vitro susceptibility of the yeast pathogen Cryptococcus to fluconazole and other azoles varies with molecular genotype. J Clin Microbiol 484115–4120. 10.1128/JCM.01271-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary A., Randhawa H. S., Sundar G., Kathuria S., Prakash A., Khan Z., Sun S., Xu J. (2011). In vitro antifungal susceptibility profiles and genotypes of 308 clinical and environmental isolates of Cryptococcus neoformans var. grubii and Cryptococcus gattii serotype B from north-western India. J Med Microbiol 60 961– 967. 10.1099/jmm.0.029025-0 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (2008). Reference Methods for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi, Approved Standard. 2nd edn, Wayne, PA: Clinical Laboratory Standards Institute. [Google Scholar]
- Connolly J. H., Krockenberger M. B., Malik R., Canfield P. J., Wigney D. I., Muir D. B. (1999). Asymptomatic carriage of Cryptococcus neoformans in the nasal cavity of the koala (Phascolarctos cinereus). Med Mycol 37331–338. 10.1046/j.1365-280X.1999.00236.x [DOI] [PubMed] [Google Scholar]
- Cookman L., Hugi M. (2013). Meningitis secondary to Cryptococcus gattii, an emerging pathogen affecting immunocompetent hosts. World J Emerg Med 4151–153. 10.5847/wjem.j.issn.1920-8642.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnos-Ollivier M., Patel S., Spaulding A. R., Charlier C., Garcia-Hermoso D., Nielsen K., Dromer F. (2010). Mixed infections and in vivo evolution in the human fungal pathogen Cryptococcus neoformans. MBio 1e00091-10. 10.1128/mBio.00091-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinel-Ingroff A., Aller A. I., Canton E., Castanon-Olivares L. R., Chowdhary A., Cordoba S., Cuenca-Estrella M., Fothergill A., Fuller J. (2012a). Cryptococcus neoformans-Cryptococcus gattii species complex: an international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for fluconazole, itraconazole, posaconazole, and voriconazole. Antimicrob Agents Chemother 565898–5906. 10.1128/AAC.01115-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinel-Ingroff A., Chowdhary A., Cuenca-Estrella M., Fothergill A., Fuller J., Hagen F., Govender N., Guarro J, Johnson E. (2012b). Cryptococcus neoformans-cryptococcus gattii species complex: an international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for amphotericin B and flucytosine. Antimicrob Agents Chemother 563107–3113. 10.1128/AAC.06252-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favalessa O. C., Lazera M. S., Wanke B., Vieira A. C., Trilles L., Takahara D. T., Tadano T., Dias L. B., Vieira A. C., Novack G. V. (2014). Fatal Cryptococcus gattii genotype AFLP6/VGII infection in a HIV-negative patient: case report and a literature review. Mycoses 57639––643.. [DOI] [PubMed] [Google Scholar]
- Hagen F., Khayhan K., Theelen B., Kolecka A., Polacheck I., Sionov E., Falk R., Parnmen S., Lumbsch H. T. (2015). Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol 7816–48. 10.1016/j.fgb.2015.02.009 [DOI] [PubMed] [Google Scholar]
- Haynes K. A., Sullivan D. J., Coleman D. C., Clarke J. C., Emilianus R., Atkinson C., Cann K. J. (1995). Involvement of multiple Cryptococcus neoformans strains in a single episode of cryptococcosis and reinfection with novel strains in recurrent infection demonstrated by random amplification of polymorphic DNA and DNA fingerprinting. J Clin Microbiol 3399–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igreja R. P., Mdos S. L., Wanke B., Galhardo M. C., Kidd S. E., Meyer W. (2004). Molecular epidemiology of Cryptococcus neoformans isolates from AIDS patients of the brazilian city, Rio de Janeiro. Med Mycol 42229–238. 10.1080/13693780310001644743 [DOI] [PubMed] [Google Scholar]
- Illnait-Zaragozí M. T., Martínez-Machín G. F., Fernández-Andreu C. M., Hagen F., Boekhout T., Klaassen C. H., Meis J. F. (2010). Microsatellite typing and susceptibilities of serial Cryptococcus neoformans isolates from Cuban patients with recurrent cryptococcal meningitis. BMC Infect Dis 10289. 10.1186/1471-2334-10-289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein K. R., Hall L., Deml S. M., Rysavy J. M., Wohlfiel S. L., Wengenack N. L. (2009). Identification of Cryptococcus gattii by use of L-canavanine glycine bromothymol blue medium and DNA sequencing. J Clin Microbiol 473669–3672. 10.1128/JCM.01072-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazera M. S., Salmito Cavalcanti M. A., Londero A. T., Trilles L., Nishikawa M. M., Wanke B. (2000). Possible primary ecological niche of Cryptococcus neoformans. Med Mycol 38379–383. 10.1080/714030957 [DOI] [PubMed] [Google Scholar]
- Mlinaric-Missoni E., Hagen F., Chew W. H., Vazic-Babic V., Boekhout T., Begovac J. (2011). In vitro antifungal susceptibilities and molecular typing of sequentially isolated clinical Cryptococcus neoformans strains from Croatia. J Med Microbiol 601487–1495. 10.1099/jmm.0.031344-0 [DOI] [PubMed] [Google Scholar]
- Pfaller M. A., Messer S. A., Boyken L., Rice C., Tendolkar S., Hollis R. J., Doern G. V., Diekema D. J. (2005). Global trends in the antifungal susceptibility of Cryptococcus neoformans (1990–2004). J Clin Microbiol 432163–2167. 10.1128/JCM.43.5.2163-2167.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staib F., Seibold M., Antweiler E., Fröhlich B., Weber S., Blisse A. (1987). The brown colour effect (BCE) of Cryptococcus neoformans in the diagnosis, control and epidemiology of C. neoformans infections in AIDS patients. Zentralbl Bakteriol Mikrobiol Hyg A 266167–177. 10.1016/S0176-6724(87)80030-5 [DOI] [PubMed] [Google Scholar]
- Steele K. T., Thakur R., Nthobatsang R., Steenhoff A. P., Bisson G. P. (2010). In-hospital mortality of HIV-infected cryptococcal meningitis patients with C. Gattii and C. neoformans infection in Gaborone, Botswana. Med Mycol 481112–1115. 10.3109/13693781003774689 [DOI] [PubMed] [Google Scholar]
- Trilles L., Meyer W., Wanke B., Guarro J., Lazéra M. (2012). Correlation of antifungal susceptibility and molecular type within the Cryptococcus neoformans/C. gattii species complex. Med Mycol 50328–332. 10.3109/13693786.2011.602126 [DOI] [PubMed] [Google Scholar]