Abstract
Age-related macular degeneration (AMD) is the leading cause of irreversible blindness in the elderly of industrialized nations, and there is increasing evidence to support a role for chronic inflammation in its pathogenesis. Mitochondrial DNA (mtDNA) has been recently reported to be pro-inflammatory in various diseases such as Alzheimer’s and heart failure. Here, we report that intracellular mtDNA induces ARPE-19 cells to secrete IL-6 and IL-8, inflammatory cytokines that have consistently been associated with AMD onset and progression. The induction was dependent on the size of mtDNA, but not on specific sequence. Oxidative stress plays a major role in the development of AMD, and our findings indicate that mtDNA induces IL-6 and IL-8 more potently when oxidized. Cytokine induction was mediated by STING (Stimulator of Interferon Genes) and NF-κB as evidenced by abrogation of the cytokine response with use of specific inhibitors (siRNA and Bay 11–7082, respectively). Finally, mtDNA primed the NLRP3 inflammasome and weakly activated it as shown by caspase-1 activation and mature IL-1β secretion. This study contributes to our understanding of the potential pro-inflammatory role of mtDNA in the pathogenesis of AMD.
Keywords: Age-related macular degeneration, Retinal pigment epithelium, Mitochondrial DNA, Inflammation, NLRP3 inflammasome
Graphical abstract
Introduction
Mitochondrial damage and dysfunction are associated with aging and the development of neurodegenerative diseases1,2, including age-related macular degeneration (AMD)3,4. The examination of retinas from patients with AMD has consistently revealed mitochondrial DNA (mtDNA) damage and a decreased capacity for repair3–7. More specifically, ultrastructural studies point to a decline in the number and structural integrity of mitochondria in the RPE of aged and AMD eyes8. Although it remains uncertain whether this finding is a cause or a consequence of disease, mitochondrial damage can be conceptualized to contribute to the development of AMD by way of increased oxidative stress and deregulated apoptosis and necrosis, which culminate in RPE death4.
Inflammation is another plausible link between mitochondrial damage and disease. In diverse pathologies that cause cell injury or death, mitochondria can act as a major source of damage-associated molecular patterns (DAMPs)9. These are danger signals of various origins that act by engaging intracellular or extracellular pattern recognition receptors (PRRs) in order to stimulate an immune response and cytokine production. Among the proposed DAMPs, mtDNA has received considerable attention due to its immunostimulatory potential when uncontained9. In fact, mtDNA release has been reported in various pathologic conditions, including rheumatoid arthritis10, cardiomyopathy11, trauma12, systemic inflammation13–17, and neurodegeneration18,19. Since inflammation is a an emerging major feature of AMD, and given the compromised structural integrity of mitochondria in the RPE of AMD patients, we hypothesized mtDNA release as potentially inflammatogenic in the pathogenesis of AMD.
In particular, mtDNA has been found to induce the pro-inflammatory cytokines IL-6 and IL-8 in vitro and in vivo12,13,17–20. Both of these cytokines have been associated with AMD progression21–25 and the development of choroidal neovascularization (CNV)26–28. In macrophages, mtDNA induces the NLRP3 inflammasome29,30, which has also been recently reported to be involved in the pathogenesis of AMD31–33. NLRP3 activation leads to cleavage of the pro form of caspase-1 into its mature form, with the latter cleaving the pro forms of IL-β and IL-18 into their mature forms, which are subsequently secreted by the cell34. Since the RPE is directly involved in the pathogenesis of AMD35, we thought it meaningful to examine whether mtDNA can induce the pro-inflammatory cytokines IL-6 and IL-8, and the NLRP3 inflammasome in ARPE-19 cells.
In this study, we report that mtDNA stimulates IL-6 and IL-8 secretion as well as priming of the NLRP3 inflammasome in ARPE-19 cells. Those effects were mediated by a common pathway involving Stimulator of Interferon Genes (STING), an adaptor protein that is increasingly being viewed as a central mediator of the DNA sensing pathways36. We also report that mtDNA activates the NLRP3 inflammasome, leading to the secretion of IL-1β from ARPE-19 cells. Together, these findings provide a novel mechanistic link between mitochondrial damage and inflammation in AMD based on the inflammatogenic potential of mtDNA.
Materials and Methods
Cell Lines and Reagents
ARPE-19 cells (CRL-2302), THP-1 cells (TIB-202), and RPMI-1640 medium (30–2001) were purchased from the American Type Culture Collection (ATCC) (Manassas, VA). Human primary retinal pigment epithelial cells (hRPE) (194987) were obtained from Lonza (Walkersville, MD). Dulbecco’s Modified Eagle Medium (DMEM/F-12) (11330-057), heat-inactivated fetal bovine serum (HI-FBS) (10438-026) and Penicillin-Streptomycin (15140122) were purchased from Life Technologies (Grand Island, NY). HP X-tremeGene transfection agent (06366244001) was obtained from Roche (Indianapolis, IN), recombinant human IL-1α (200-LA-010) from R&D Systems (Minneapolis, MN), and Bay 11–7082 inhibitor (196870) from EMD Millipore (Billerica, MA). Anti-IL-18 antibody (ab137664), anti-IL-6 antibody (ab32530), and anti-TATA binding protein (TBP) (51841) were obtained from Abcam (Cambridge, MA). Anti-IL-1β antibody (MAB201), anti-IL-8 antibody (MAB208), and anti-STING (MAB7169) were purchased from R&D Systems. Anti-caspase-1 (sc-515) was purchased from Santa Cruz Biotechnology (Dallas, TX). Anti-β-tubulin antibody (2128), anti-A20 (5630) and HRP-linked secondary antibodies (7074S, 7076S) were obtained from Cell Signaling Technology (Danvers, MA). ELISA kits for IL-1β (DLB50), IL-18 (7620) and IL-8 (D8000C) were purchased from R&D. Taq Polymerase (M0267S) was purchased from New England Biolabs (Ipswich, MA).
Cell Culture
ARPE-19 cells were maintained in DMEM/F-12 medium supplemented with 10% heat-inactivated FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin. THP-1 cells were grown in RPMI-1640 medium supplemented with 10% HI-FBS, 100 U/mL penicillin, 100 µg/mL streptomycin, and 50 µM 2-mercaptoethanol. All cells were grown in humidified 5% CO2 at 37°C and passaged every 2–3 days.
Preparation of mtDNA
Total DNA was extracted from ARPE-19 cells using QIAamp DNA mini kit (51304) from Qiagen (Valencia, CA) according to the manufacturer’s protocol. From the extracted DNA, specific mtDNA fragments were amplified by PCR on Bio-Rad BiCycler (Hercules, CA) using primers and methods from a published protocol37. The PCR products were run on a gel to verify specificity before being purified using a PCR purification kit (28104) from Qiagen. The concentration of DNA was then measured using NanoDrop 2000 (ThermoFisher Scientific, Waltham, MA). Total DNA template was undetectable in the final elute as determined by blank controls. To generate oxidized mtDNA, 8-oxo-dGTP (N-2034) from TriLink Biotechnologies (San Diego, CA) was added to the PCR reaction mixture at the same concentration as the other nucleotides. To generate FAM-tagged mtDNA, FAM-tagged primers were used. All primers were ordered from the Massachusetts General Hospital DNA core facility (Boston, MA).
mtDNA Transfection into Cells
ARPE-19 cells were seeded in 24-well plates at a density of 3x104 cells/well in complete DMEM/F12 medium. The following day, IL-1α was added at 5ng/mL to prime the cells. 24 hours later, culture medium was changed to antibiotic-free DMEM/F12 medium supplemented with 1% FBS, and cells were transfected at 70–80% confluence with 1µg/mL mtDNA using HP X-tremeGene agent at a ratio of 1:1 according to the manufacturer’s protocol. Transfection agent alone was used as a control for all experiments. 48 hours later, culture media were collected and cells lysed with NP40 lysis buffer (Invitrogen) supplemented with protease inhibitors (Complete Mini, Roche). For THP-1 experiments, 1.5 x 106 cells were seeded in each well of a 6-well plate in complete RPMI medium supplemented with 200 nM PMA to allow differentiation into macrophages. 24 hours later, medium was renewed and LPS added at 500ng/mL to prime the cells. 3 hours later, medium was changed to antibiotic-free RPMI medium and transfection of mtDNA was performed as per above. 24 hours later, culture media and cell lysates were collected as per above.
Western Blot
Total protein concentration was determined by DC Protein Assay (Bio-Rad, Philadelphia, PA, US). Equal amounts of total protein (for cell lysates) or culture medium (for secreted cytokines) were loaded on a NUPAGE 4–12% bis-tris polyacrylamide gel (Life Technologies, Grand Island, NJ) and subjected to electrophoresis. The proteins were transferred to a PVDF membrane (Millipore, Billerica, MA), and the membrane was subsequently blocked with non-fat milk and incubated with primary antibodies against IL-8 and capsase-1 at 1:500; IL-18, A20, and TBP at 1:1000; IL-1β STING, IL-6 and β-tubulin at 1:2000. The membrane was then washed and incubated with secondary antibodies conjugated to horseradish peroxidase (Cell Signaling Technology) at 1:10000. The membrane was developed with enhanced chemiluminescence (ECL Select) (GE Healthcare, Wauwatosa, WI). The intensity of protein bands was quantified using the software Image Lab 4.1 (Bio-Rad).
mtDNA Tracking
ARPE-19 cells were seeded on EZ Slides (PEZGS0416, Millipore) at 5x104 cells/well in antibiotic-free medium and allowed to settle overnight. The following day, cells were transfected with 1µg/mL of FAM-tagged mtDNA using HP X-tremeGene agent at a ratio of 1:1 according to the manufacturer’s protocol. 24 hours later, cells were carefully washed with PBS before fixation in 4% PFA for 5 minutes. Afterwards, cells were washed again with PBS and mounted using Vectashield mounting medium with DAPI (H-1200) (Vector Laboratories, Burlingame, CA). Slides were visualized on Zeiss Axio Imager M2.
MTT Viability Assay
ARPE-19 cells were transfected with mtDNA in a 96-well plate under the same conditions outlined above. 48 hours later, cells were washed with PBS and incubated for 3 hours with 0.5 mg/mL MTT (Life Technologies) dissolved in PBS. The formed formazan crystals were dissolved by adding 100µL of acidified (0.04 N HCl) isopropanol (Sigma-Aldrich, St. Louis, MO) to each well. Absorbance was read at 590nm using the SpectraMax190 Microplate Reader (Molecular Devices, Sunnyvale, CA).
siRNA experiments
For STING knockdown verification, ARPE-19 cells in 6-well plates were transfected at 70–80% confluence with 20nM of either STING siRNA (Qiagen) or control siRNA in antibiotic-free DMEM/F12 medium using RNAiMAX (Invitrogen) according to the manufacturer’s protocol. 48 hours later, medium was renewed and the same treatment was repeated for 48h, after which cell lysates were collected and immunoblotted for STING. For STING knockdown experiments, ARPE-19 cells were seeded in 6-well plates at a density of 2x105 cells/well in antibiotic-free medium. The following day, they were primed with 5 ng/mL IL-1α and transfected with 20nM of STING or control siRNA for 48 hours using the RNAiMAX protocol. Medium was then renewed and mtDNA (1µg/mL) and siRNA (20nM) were co-transfected using HP X-tremeGene reagent at a 1:1 ratio as per manufacturer’s instructions. 48 hours later, culture media and cell lysates were collected for analysis.
p65 NF-κB nuclear translocation
ARPE-19 cells were transfected with mtDNA in 100mm dishes under the same conditions outlined above. 24 hours later, nuclei were extracted using a kit (40010) (Active Motif, Carlsbad, CA) and proper nuclear fractionation was verified by western blot with TATA binding protein and b-tubulin. p65 NF-κB was quantified in the nuclear extracts using TransAM NF-κB ELISA (43296) (Active Motif) after compensation for total protein concentration.
Caspase-1 activity
ARPE-19 cells were transfected with mtDNA in glass-bottom 24-well plates under the same conditions outlined above. 24 hours later, FAM-YVAD-FMK (#97) (Immunocytochemistry technologies, Bloomington, MN), a probe specific to caspase-1, was added at the concentration recommended by the manufacturer. After a 2-hour incubation at 37°C and 5% CO2, Hoechst was added to the wells at 1µg/mL and cells were reincubated for 10 minutes. Cells were washed carefully with PBS and visualized using Zeiss AxioObserver 21.
Statistical analysis
All experiments were performed in triplicate unless otherwise noted. Statistical analysis was performed on JMP Pro software version 11.2.0 from SAS (Cary, NC). Results are expressed as mean ± SEM. Statistical significance for differences between two treatment groups was determined with Student’s t-test. One-way ANOVA with Tukey post-hoc correction was used for multiple group comparisons. A p-value of < 0.05 was considered statistically significant.
Results
mtDNA induces IL-6 and IL-8 secretion from ARPE-19
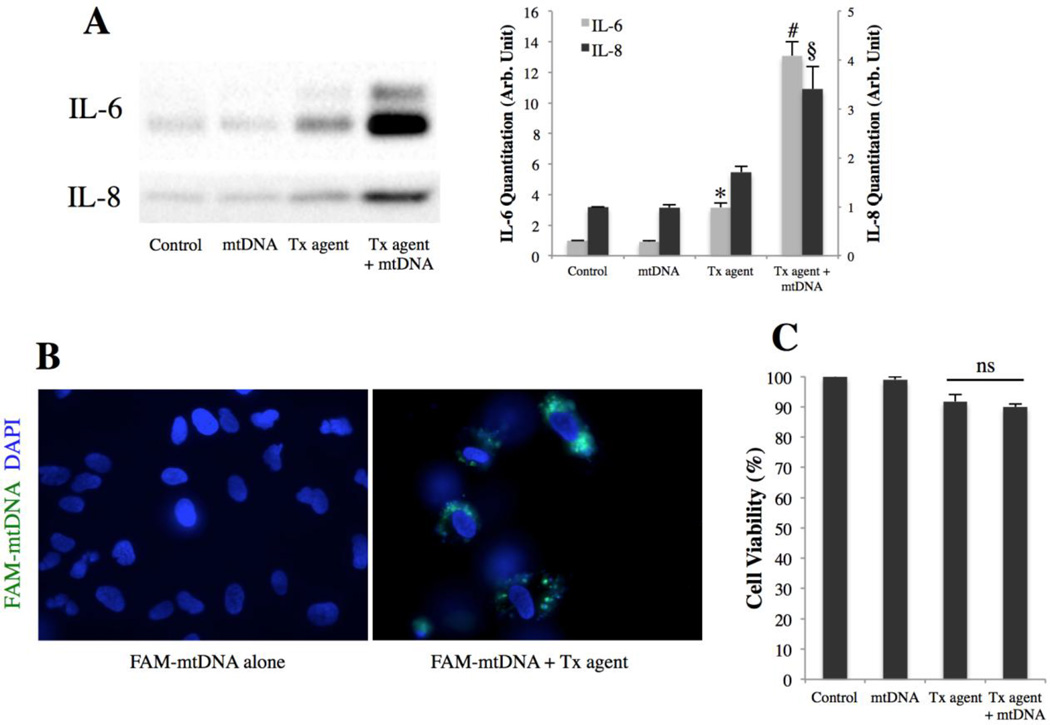
To determine the effect of mtDNA on IL-6 and IL-8 secretion from ARPE-19 cells, a 2kb fragment of mtDNA was amplified by PCR and transfected into ARPE-19 cells. IL-6 and IL-8 were measured in the culture medium 48h post-transfection by western blot. Figure 1A shows that mtDNA induced a 4-fold and a 2-fold increase in the secretion of IL-6 (p<0.001) and IL-8 (p<0.01), respectively, compared to a transfection agent control. In contrast, neither cytokine was induced when mtDNA was used without transfection agent. This suggests that IL-6 and IL-8 are induced by intracellular mtDNA rather than extracellular mtDNA binding to a cell surface receptor. To confirm this, labeled mtDNA was generated by PCR using FAM-tagged primers, and ARPE-19 cells were treated for 24h with labeled mtDNA with or without transfection agent. Figure 1B shows mtDNA being present in an intracellular cytoplasmic location only when combined with a transfection agent. This finding, in conjunction with the previous result (Fig. 1A), indicates that only intracellular mtDNA can induce IL-6 and IL-8 secretion. To assess whether the observed cytokine response was due to cell death, cell viability was determined using an MTT assay at 48h post-transfection with the same mtDNA dose. mtDNA did not cause significant cell death compared to a transfection agent control, indicating that mtDNA induces IL-6 and IL-8 secretion rather than merely causing cell death and release of cytokines.
Figure 1. mtDNA induces IL-6 and IL-8 secretion from ARPE-19 cells.
(A) IL-6 and IL-8 secretion from ARPE-19 as determined by western blot of culture media 48h post-transfection with 1µg/mL mtDNA. Right, quantitation by densitometry. *p < 0.05 versus control and mtDNA groups; #p < 0.001 versus all other groups; §p < 0.01 versus all other groups.
(B) ARPE-19 cells 24h after treatment with FAM-tagged mtDNA with or without transfection agent. mtDNA in green, DAPI in blue.
(C) ARPE-19 viability determined by MTT 48h post-transfection with 1µg/mL mtDNA.
Oxidized and larger mtDNA fragments induce a stronger cytokine response
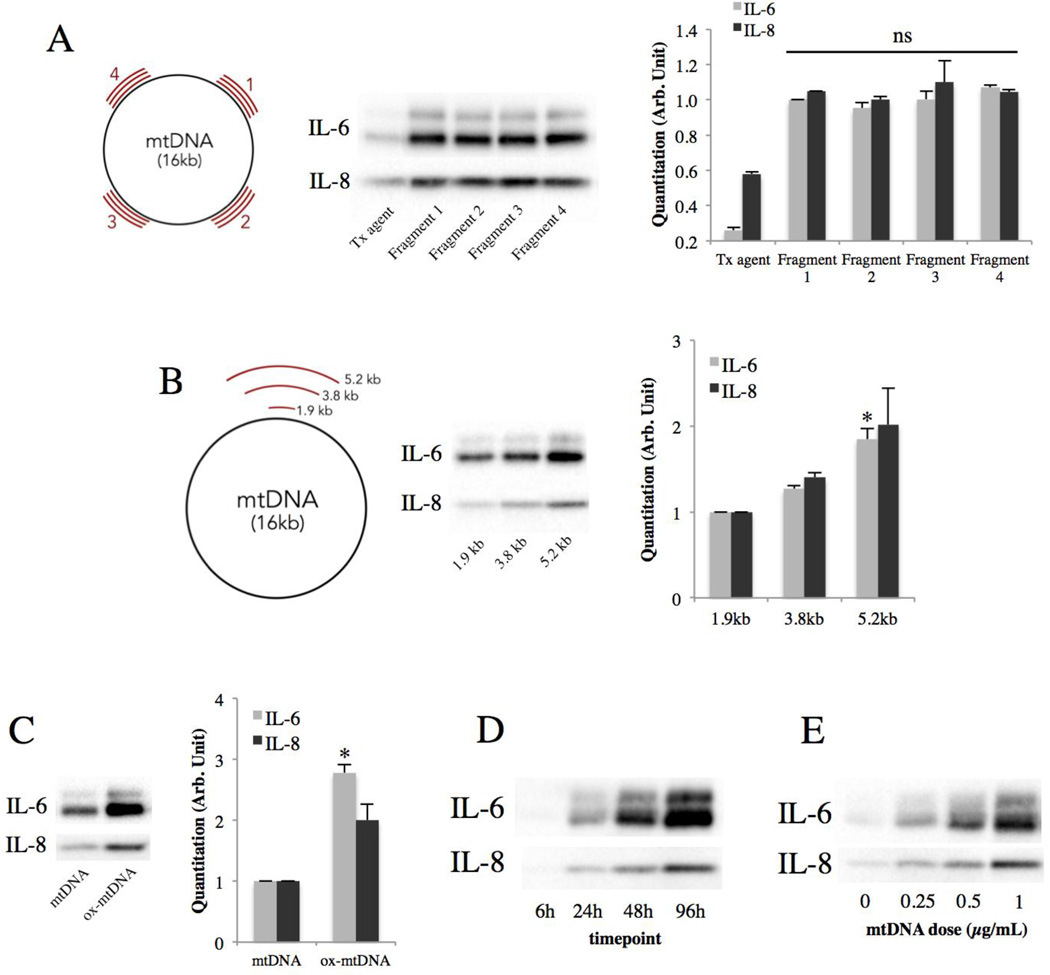
To determine whether the sequence of the mtDNA fragment has any effect on cytokine induction, four 2-kb fragments of mtDNA with different sequences were transfected into ARPE-19 cells. They were found to produce similar inductions of IL-6 and IL-8 (Fig. 2A), indicating that mtDNA induces IL-6 and IL-8 in a sequence-independent fashion. Since AMD has been associated with impaired autophagy38 and RPE death39, mtDNA under such conditions may be fragmented and accumulate in the cell in different sizes40. Therefore, to determine the effect of the size of mtDNA on IL-6 and IL-8 induction, 3 mtDNA fragments of different sizes were generated by PCR and transfected into ARPE-19 cells. The result shows that larger mtDNA fragments induced stronger IL-6 and IL-8 responses (Fig. 2B). Furthermore, mtDNA has been found to be more oxidized in AMD retinas compared to age-matched controls41. To mimic the in vivo situation, oxidized mtDNA was generated by PCR incorporating the modified nucleotide 8-oxoguanine and transfected into ARPE-19 cells. This resulted in a stronger IL-6 and IL-8 secretion compared to non-oxidized mtDNA (Fig. 2C). Finally, the time course and dose response were evaluated, and both IL-6 and IL-8 were found to be induced in a time and dose-dependent fashion (Fig. 2D and 2E).
Figure 2. Oxidized and larger mtDNA fragments induce a stronger cytokine response.
(A) IL-6 and IL-8 secretion from ARPE-19 cells 48h post-transfection with 1µg/mL of 2-kb mtDNA fragments of different sequences but similar sizes (schematic). Right, quantitation by densitometry.
(B) IL-6 and IL-8 secretion from ARPE-19 cells 48h post-transfection with 1µg/mL of mtDNA fragments of different sizes (schematic). Right, quantitation by densitometry. *p < 0.05 versus all other groups.
(C) IL-6 and IL-8 secretion from ARPE-19 cells 48h post-transfection with 1µg/mL of mtDNA or oxidized mtDNA. Right, quantitation by densitometry. *p < 0.01 versus control.
(D) Time course of IL-6 and IL-8 induction by 1µg/mL of mtDNA.
(E) Effect of mtDNA dose on IL-6 and IL-8 secretion at 48h.
mtDNA induces IL-6 and IL-8 secretion through STING
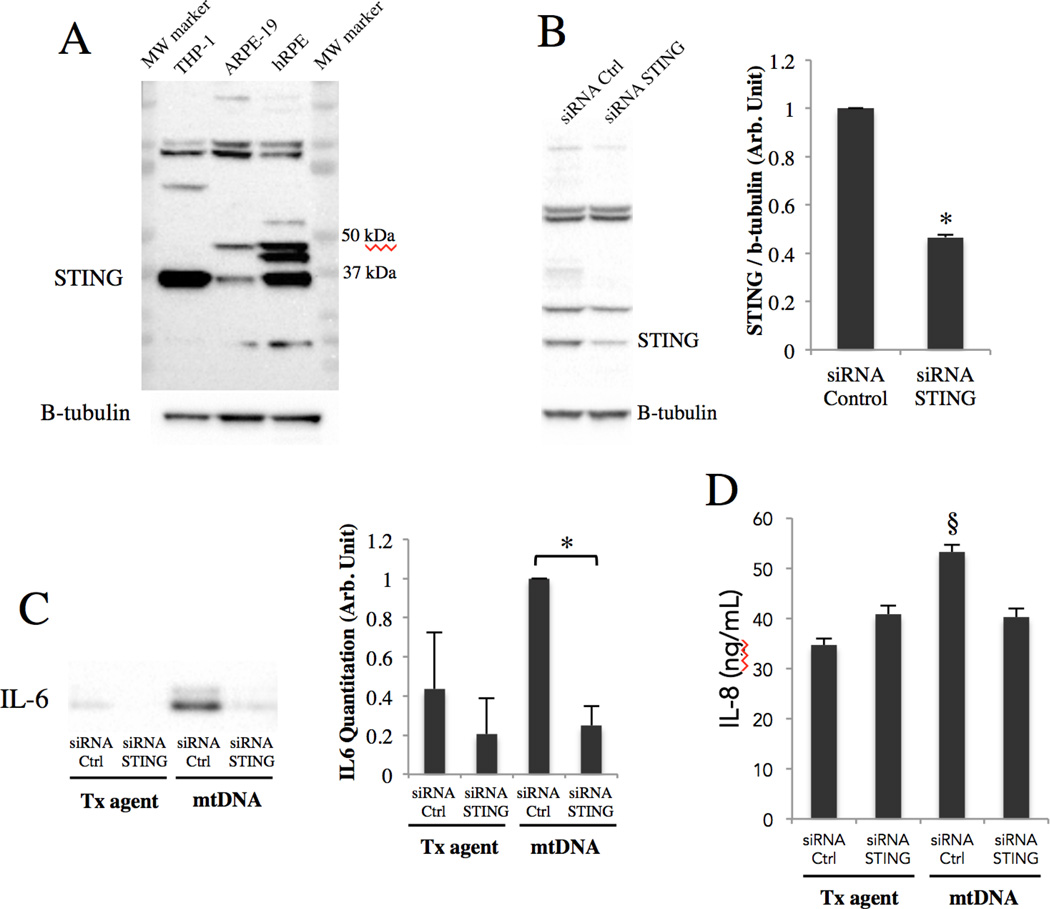
STING, an endoplasmic reticulum transmembrane protein, has been shown to be important in the immune response to foreign DNA in the context of infection, and self DNA in the context of autoimmunity42. Our experiments show that it is expressed in ARPE-19 cells and even more so in primary human RPE cells (Fig. 3A), and this expression can be successfully knocked down in ARPE-19 cells with siRNA specific to STING (Fig. 3B). To explore whether STING is involved in mtDNA-induced IL-6 and IL-8 secretion, mtDNA was transfected into ARPE-19 cells where STING had been knocked down with siRNA. The result shows that STING knockdown partially abrogated mtDNA-induced IL-6 (Fig. 3C) and IL-8 (Fig. 3E) secretion, indicating that STING mediates the induction of IL-6 and IL-8 secretion by mtDNA.
Figure 3. mtDNA induces IL-6 and IL-8 secretion through STING.
(A) STING immunoblotting of untreated ARPE-19 and primary human RPE lysates alongside a THP-1 lysate (positive control for STING). 40ug of total protein loaded per lane. Prominent band seen at STING’s predicted molecular weight (≈40kDa).
(B) ARPE-19 cells were treated for a total of 96h with 20nM of either STING siRNA or control siRNA and lysates immunoblotted with STING monoclonal antibody. Right, quantitation by densitometry. *p < 0.001 versus control siRNA.
(C&D) Along with mtDNA transfection, ARPE-19 cells were pre and co-treated with STING or control siRNA, and culture media were collected at 48h and subjected to western blot for IL-6 (C) and ELISA for IL-8 (D), as western blot did not provide sufficient resolution to study differences in IL-8 levels. *p < 0.05; §p < 0.01 versus all other groups.
mtDNA induces IL-6 and IL-8 secretion via NF-κB
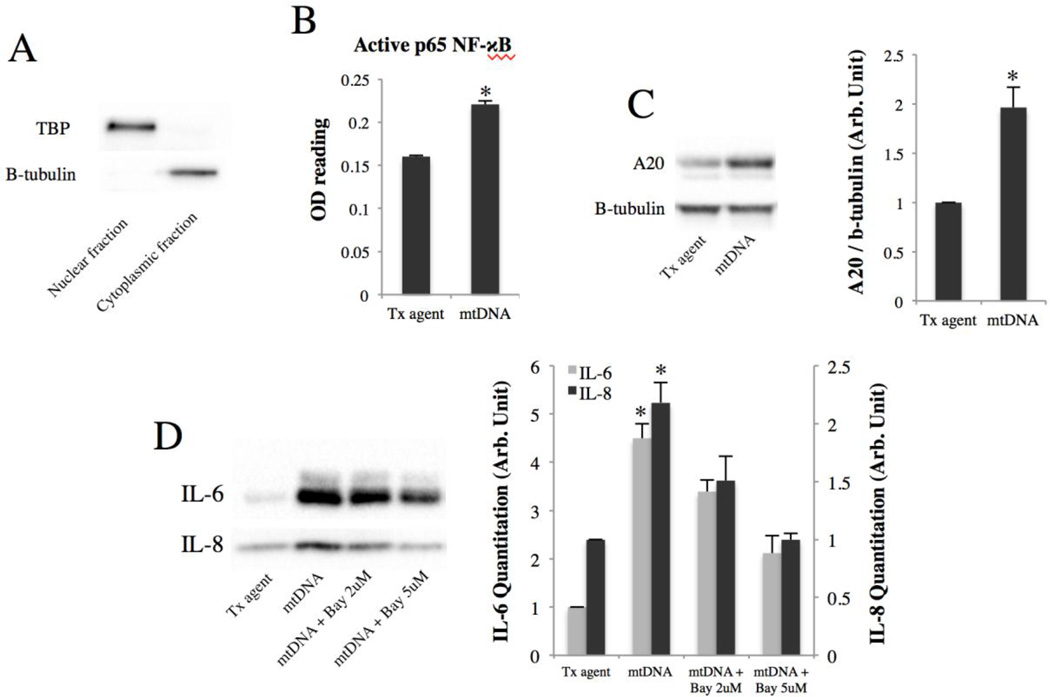
Because the NF-κB pathway has been recently shown to be activated by STING 43,44, we sought to determine whether it is also activated by mtDNA. The levels of NF-κB subunit p65 were measured by ELISA in nuclear isolates of ARPE-19 cells 24h post-transfection with mtDNA. Figure 4B shows that mtDNA induces p65 nuclear translocation 1.4-fold (p<0.01). mtDNA also induces the expression of A20 (Fig. 4C), a marker of NF-κB activation. These results together indicate that mtDNA activates the NF-κB pathway. To test whether NF-κB mediates the IL-6 and IL-8 induction by mtDNA, ARPE-19 cells were briefly pre-incubated with different concentrations of an NF-κB inhibitor (Bay 11–7082) before mtDNA transfection. This led to a dose-dependent abrogation of mtDNA-induced IL-6 and IL-8 secretion (Fig. 4D), indicating that the NF-κB pathway is involved in mediating IL-6 and IL-8 induction by mtDNA.
Figure 4. mtDNA induces IL-6 and IL-8 secretion via NF-κB.
(A) Nuclear fractionation efficiency verified with tata-binding protein (TBP) and b-tubulin.
(B) p65 NF-κB ELISA performed on nuclear isolates of ARPE-19 cells 24h post-transfection with mtDNA. *p < 0.01 versus control.
(C) A20 expression level 48h post-transfection of mtDNA. Right, quantitation by densitometry. *p < 0.05 versus control.
(D) ARPE-19 cells were pre-incubated for 30 minutes with an irreversible NF-κB inhibitor (Bay 11–7082) before mtDNA transfection. After 48h, culture media were collected and immunoblotted for IL-6 and IL-8. Right, quantitation by densitometry. *p < 0.05 versus control and [mtDNA + Bay 5uM] groups.
mtDNA primes the NLRP3 inflammasome through STING and NF-κB
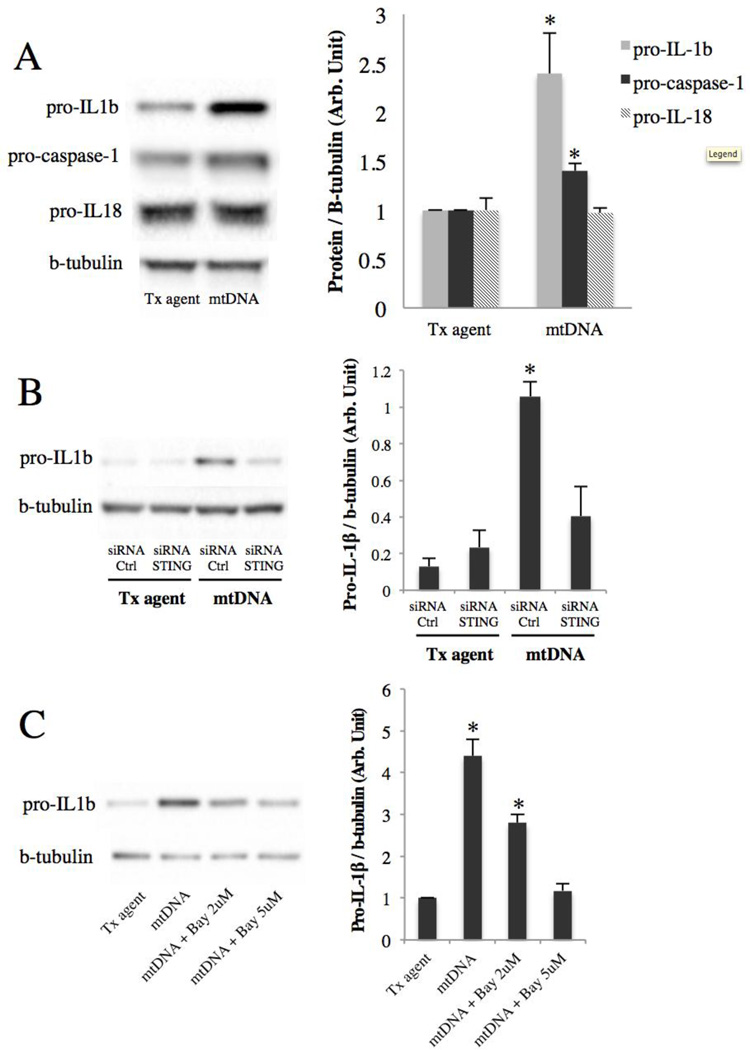
Since the NLRP3 inflammasome is primed by NF-κB34,45,46, we hypothesized that mtDNA could prime the NLRP3 inflammasome. To test this hypothesis, the levels of the pro forms of IL-1β, IL-18, and caspase-1 in ARPE-19 cells transfected with mtDNA were analyzed by western blot. The result shows that mtDNA upregulates pro-IL-1β 2.4-fold (p<0.05) and pro-caspase-1 1.4-fold (p<0.05) compared to control (Fig. 5A), indicating that mtDNA primes the NLRP3 inflammasome. In contrast, mtDNA did not change the level of pro-IL-18 (Fig. 5A). To test whether the pro-IL-1β induction is mediated by STING and NF-κB, the same inhibition strategy used previously was followed. The result shows that mtDNA induces pro-IL-1β via STING, as evidenced by abrogation of the induction with STING knockdown (Fig. 5B). Also, NF-κB inhibition dose-dependently abrogated the induction (Fig. 5C), indicating that mtDNA induces pro-IL-1β through the NF-κB pathway. Together these findings indicate that mtDNA primes the NLRP3 inflammasome through a STING/NF-κB pathway.
Figure 5. mtDNA primes the NLRP3 inflammasome through STING and NF-κB.
(A) Immunoblots of ARPE-19 lysates 48h after mtDNA transfection. Right, quantitation by densitometry. *p < 0.05 versus control.
(B) Along with mtDNA transfection, ARPE-19 cells were pre and co-treated with STING or control siRNA, and lysates were collected at 48h and immunoblotted for pro-IL-1β. Right, quantitation by densitometry. *p < 0.05 versus all other groups.
(C) ARPE-19 cells were pre-incubated for 30 minutes with an irreversible NF-κB inhibitor (Bay 11–7082) before mtDNA transfection. After 48h, lysates were collected and immunoblotted for pro-IL-1β. Right, quantitation by densitometry. *p < 0.05 versus all other groups.
mtDNA and the NLRP3 inflammasome in the RPE
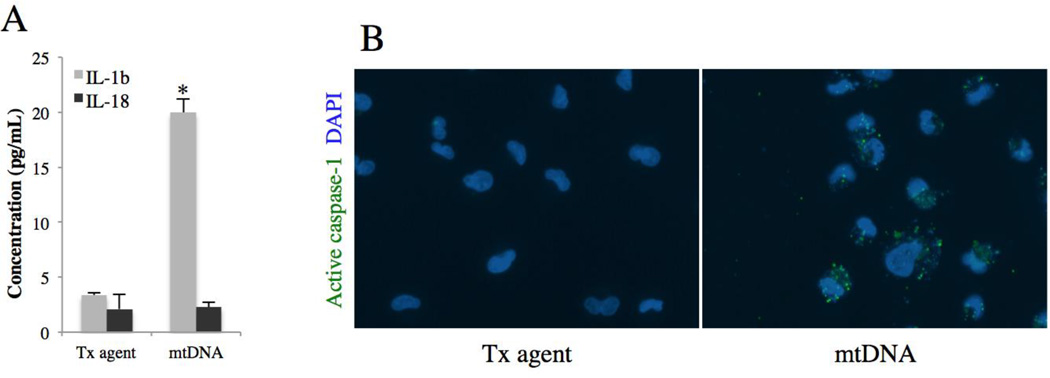
To determine whether mtDNA also activates NLRP3, we performed ELISA for mature IL-1β and IL-18 on the culture medium of ARPE-19 48h post-transfection with mtDNA. Figure 6A shows that, in ARPE-19 cells, mtDNA induces secretion of IL-1β 5.9-fold compared to control (p<0.01, n=4), but not IL-18. Although these ELISA assays are designed to detect the mature forms of IL-1β and IL-18, the pro forms of these cytokines do cross-react if present in high quantities. For this reason, western blotting was performed yet repeatedly failed to detect any mature form in either cell lysates or culture media. This is not due to technical difficulties on our part since we could detect the mature form of IL-1β in THP-1 cells after mtDNA transfection (Suppl. Fig). The major caspase involved in the maturation of IL-1β is caspase 1. Its activity was assessed in ARPE-19 cells 24h after mtDNA transfection using FAM-YVAD-FMK probe, a molecule that fluoresces when bound to active caspase-1. Imaging showed that cells transfected with mtDNA had increased FAM-YVAD-FMK signal (Fig. 6B), suggesting potential activation of caspase-1 by mtDNA in ARPE-19 cells.
Figure 6. mtDNA and the NLRP3 inflammasome in the RPE.
(A) IL-1β and IL-18 ELISA on culture media of ARPE-19 cells 48h post-transfection with mtDNA. Results are mean ± SEM, n=4. *p < 0.01 versus control.
(B) ARPE-19 caspase-1 activity assessed 24h after mtDNA transfection using fluorescent FLICA probe FAM-YVAD-FMK.
Discussion
AMD is associated with a number of risk factors, including age, smoking, obesity and cardiovascular disease. Despite the uncertainty surrounding the pathophysiology of this disease, the last decade has seen increasing evidence to support a role for mitochondrial dysfunction in its development4–7. Compared to age-matched controls, RPE cells of AMD patients have been reported to have a decline in the number and structural integrity of mitochondria8 as well as a decreased content of electron transport chain proteins47, indicating impaired mitochondrial biogenesis even at early stages of AMD. Furthermore, damage to mtDNA has been documented in the RPE of AMD patients even before vision loss occurs5–7, suggesting the potential benefit of early interventions to rescue mitochondrial function in slowing down disease progression. Recent work has also identified a role not only for damaged mtDNA but also for inherited mitochondrial DNA variants48. In that study Kenney et al found that J and H cytoplasmic hybrids have significantly altered expression of several nuclear genes involving alternative complement, inflammation and apoptosis pathways. The same group in a separate study49 has also found that mtDNA variants mediate energy production and expression levels for CFH, C3 and EFEMP1, genes known to be involved in AMD50–57. Since altered mitochondrial function is not only present but also likely contributory to AMD progression, we postulated release of inflammatogenic mtDNA as a potential mechanism for disease perpetration.
In fact, a wealth of evidence supports a role for inflammation in the development and progression of AMD35,58. Specifically, clinical and genetic studies have revealed the importance of IL-6 and IL-8 in the pathogenesis of AMD. IL-6 is a pleiotropic inflammatory cytokine that can exaggerate immune and inflammatory responses, and its level has been associated with AMD onset24 and progression22. IL-8 is a member of the CXC chemokine family and a potent chemoattractant for neutrophils and monocytes, and IL-8 haplotypes have been found to confer an increased risk of AMD21,23,25. In our study, intracellular mtDNA induced IL-6 and IL-8 secretion from ARPE-19 cells, suggesting that mtDNA can potentially contribute to the inflammatory component of AMD. Both interleukins studied have also been associated with wet AMD, as they have been correlated with CNV and macular thickness in exudative AMD27,28. Furthermore, both cytokines are pro-angiogenic and induce VEGF59–62, a crucial factor in CNV development. In aged RPE cells with compromised mitochondrial integrity, mtDNA molecules that leak into the cytosol may therefore potentiate a local inflammatory microenvironment that furthers progression of AMD and CNV. This phenomenon may be even more prominent in situations where mtDNA degradation is impeded, such as impaired autophagy in the RPE. The latter phenomenon has been associated with AMD38, and it could cause accumulation of damaged organelles and molecules in the RPE, including mtDNA. Our experiments show that larger mtDNA fragments and higher mtDNA doses cause more IL-6 and IL-8 secretion. These findings suggest that RPE cells in AMD may be even more susceptible to mtDNA-induced damage due to their declining autophagic function and an inability to break down mtDNA into its basic nucleotide constituents.
Oxidative stress has also been implicated in aging and AMD63, and part of the evidence comes from genetic studies that showed an association between antioxidant enzyme gene polymorphisms and AMD64,65. Furthermore, mitochondria are the main cellular source of oxidative stress66, and mtDNA has been found to be oxidized in AMD retinas41. In our study, oxidized mtDNA induced a stronger IL-6 and IL-8 response compared to non-oxidized mtDNA. This suggests that mtDNA may be even more inflammatogenic in AMD patients by virtue of its oxidized status, worsening a vicious cycle of inflammation and damage that progressively leads to mitochondrial failure and death of the RPE. This phenomenon may be particularly pronounced in AMD variants with compromised anti-oxidative defense mechanisms64,65. Although our findings indicate that induction of IL-6 and IL-8 was sequence-independent, we cannot exclude the importance of nucleotide composition, especially in oxidative situations where differential nucleotide susceptibility to oxidation will translate into different overall oxidative charges across fragments with different nucleotide constitutions. Studying this phenomenon would also require the use of smaller mtDNA fragments where dissimilar nucleotide compositions are more likely to be found.
Extracellular mtDNA had no effect on either IL-6 or IL-8 secretion, at least within the resolving capacity of western blot. This could be explained by a lack of surface receptors for mtDNA on ARPE-19 cells. Practically all the DNA sensors described to date are intracellular36, including Toll-like receptor 9 (TLR9), a transmembrane receptor of unmethylated CpG motifs that has been confirmed to be predominantly intracellular rather than on the cell membrane67,68, including in ARPE-1969. Our findings are also in concordance with previous evidence that extracellular mtDNA has no immunostimulatory effect unless used with a molecule that aids its uptake into the cell20,70. In other studies, extracellular mtDNA was mildly immunostimulatory in certain cell types, but only at very high concentrations (>10µg/mL)13,20. In ARPE-19 cells, extracellular CpG DNA has been shown to induce IL-8 secretion only at the dose of 84µg/mL, almost 2 orders of magnitude higher than the concentration we used, and the authors attributed the effect to CpG DNA that had been internalized by phagocytosis69.
Although it remains unclear which DNA sensors are involved in sensing intracellular mtDNA, we have identified STING as an important downstream mediator of the inflammatory pathways triggered by mtDNA in ARPE-19 cells as evidenced by STING knockdown experiments. This transmembrane endoplasmic reticulum protein has been clearly demonstrated to take center stage in the response to intracellular foreign and self-DNA36,42. Unlike DNA sensors, which exhibit overlapping and redundant functions, STING seems to constitute a common downstream node to most DNA-sensing pathways, a property that might be advantageous in therapeutic efforts to modulate the innate immune defenses of the cell. Our study extends the importance of STING to RPE cells in their inflammatory response to mtDNA. To our knowledge, this is the first report of STING or any DNA sensing function in RPE cells. Such knowledge may prove valuable in understanding the consequences of insufficient autophagy in the face of the high metabolic demands that constantly weigh on the aging RPE. Once activated, STING can in turn stimulate NF-κB43,44. Our findings indicate that mtDNA can indeed activate this latter pathway. NF-κB blockade has been shown to inhibit choroidal neovascularization71,72, highlighting the importance of NF-κB in the progression of AMD. NF-κB also regulates the expression of many genes in RPE cells, including IL-6 and IL-873,74, and this is corroborated by our experiments of NF-κB inhibition abrogating the induction of cytokines.
Recent studies have shown NF-κB to be critical for priming of the NLRP3 inflammasome in RPE cells75. The NLRP3 inflammasome has recently received widespread attention for its possible association with both dry and wet AMD31–33, and several stimuli have been shown to trigger its activation in ARPE-19 cells76–80. We found that mtDNA primes but does not conclusively activate the NLRP3 inflammasome through a STING/NF-κB pathway as evidenced by upregulation of the pro forms of IL-1β and caspase-1 but minimal upregualtion of the mature form. Pro-IL-18 appeared to be constitutively expressed in our cells, which is in accordance with previous findings in ARPE-1976,81 and other cell types34. Our data suggests a potential low-level activation of the NLRP3 inflammasome by mtDNA in ARPE-19 cells as evidenced by secretion of low levels of mature IL-1β to the culture medium and increased FAM-YVAD-FMK signal. It should be noted however that concerns about the specificity of the latter probe have been raised82. More importantly, because of differences and serious deficiencies in AMD experimental models, there is still no consensus as to whether NLRP3 activity is detrimental or protective in the first place. One study suggested a protective effect on CNV through the anti-angiogenic effect of IL-18 using an acute laser-injury CNV model33, whereas another study suggested a detrimental effect through IL-1β using a VEGF-A-overexpressing mouse31. The role of these cytokines in the pathogenesis of AMD remains unclear. Our data unfortunately cannot argue one way or another. The low-level inflammasome activation that we observed could be compensatory and protective, or detrimental, or even just a “spillover” of overall inflammation. In addition, NLRP3 activation in RPE cells is orders of magnitude less than that of inflammatory cells, and it still remains to be answered if a chronic low-level activation of the inflammasome in RPE cells could play a role in the evolution of AMD. One can argue that in contrast to acute inflammatory processes where highly potent inflammatory cells may be needed, AMD and other chronic diseases such as atherosclerosis are characterized by a state of para-inflammation83,84, a term used to describe a response to stress that is intermediate between the basal state and inflammation. In our study, we found that mtDNA could induce secretion of the mature form of IL-1β in THP-1 cells85, a macrophage cell line, indicating that mtDNA can activate NLRP3 in macrophages, consistent with previous reports in bone-marrow-derived macrophages29,30. This suggests a role for aged macrophages in licensing NLRP3 activation within the retinal microenvironment when exposed to their own accumulated mtDNA or mtDNA that has been phagocytized from dying retinal cells such as the RPE. Moreover, previous evidence of a direct physical interaction between mtDNA and NLRP330 makes the applicabiliy of NLRP3 activation by mtDNA in other cell types more plausible, at least conceptually.
In summary, this study elucidates the pro-inflammatory properties of mtDNA in ARPE-19 cells and suggests inflammation as a plausible mechanistic link between mitochondrial damage and AMD, one in which accumulating inflammatogenic mtDNA in aged RPE cells can create an inflammatory microenvironment that aids in the development and progression of AMD. Our findings also suggest STING and NF-κB as interesting therapeutic targets to explore in the future for the purpose of modulating innate immune defenses in the aging RPE.
Highlights.
-
-
mtDNA induces IL-6 and IL-8 secretion from ARPE-19 cells via a STING/NF-kB pathway
-
-
Oxidized and larger mtDNA is more immunostimulatory
-
-
mtDNA primes and potentially activates the NLRP3 inflammasome in ARPE-19 cells
Acknowledgments
The authors would like to thank Mien Hoang and Keiko Kataoka for their technical support. This work was supported by: NEI R21EY023079-01A1 (DGV); The Yeatts Family Foundation (DGV, JWM); The Loefflers Family Fund (DGV, JWM); the 2013 Macula Society Research Grant award (DGV); a Physician Scientist Award (DGV), an unrestricted grant (JWM) from the Research to Prevent Blindness Foundation; NEI grant EY014104 (MEEI Core Grant); and Bayer Healthcare 2013 Global Ophthalmology Award (DEM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosure(s): The author(s) have no proprietary or commercial interest in any materials discussed in this article.
References
- 1.Reeve AK, Krishnan KJ, Turnbull D. Mitochondrial DNA mutations in disease, aging, and neurodegeneration. Ann N Y Acad Sci. 2008;1147:21–29. doi: 10.1196/annals.1427.016. [DOI] [PubMed] [Google Scholar]
- 2.Currais A. Ageing and inflammation – A central role for mitochondria in brain health and disease. Ageing Res Rev. 2015:1–13. doi: 10.1016/j.arr.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Blasiak J, Glowacki S, Kauppinen A, Kaarniranta K. Mitochondrial and nuclear DNA damage and repair in age-related macular degeneration. Int J Mol Sci. 2013;14:2996–3010. doi: 10.3390/ijms14022996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarrett SG, Lewin AS, Boulton ME. The importance of Mitochondria in age-related and inherited eye disorders. Ophthalmic Res. 2010;44:179–190. doi: 10.1159/000316480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karunadharma PP, Nordgaard CL, Olsen TW, Ferrington Da. Mitochondrial DNA damage as a potential mechanism for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2010;51(11):5470–5479. doi: 10.1167/iovs.10-5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin H, Xu H, Liang F-Q, et al. Mitochondrial DNA damage and repair in RPE associated with aging and age-related macular degeneration. Invest Ophthalmol Vis Sci. 2011;52(6):3521–3529. doi: 10.1167/iovs.10-6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terluk MR, Kapphahn RJ, Soukup LM, et al. Investigating mitochondria as a target for treating age-related macular degeneration. J Neurosci. 2015;35(18):7304–7311. doi: 10.1523/JNEUROSCI.0190-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feher J, Kovacs I, Artico M, Cavallotti C, Papale A, Balacco Gabrieli C. Mitochondrial alterations of retinal pigment epithelium in age-related macular degeneration. Neurobiol Aging. 2006;27:983–993. doi: 10.1016/j.neurobiolaging.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 9.Krysko DV, Agostinis P, Krysko O, et al. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol. 2011;32(4):157–164. doi: 10.1016/j.it.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Collins LV, Hajizadeh S, Holme E, Jonsson I-M, Tarkowski A. Endogenously oxidized mitochondrial DNA induces in vivo and in vitro inflammatory responses. J Leukoc Biol. 2004;75(6):995–1000. doi: 10.1189/jlb.0703328. [DOI] [PubMed] [Google Scholar]
- 11.Oka T, Hikoso S, Yamaguchi O, et al. Corrigendum: Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;490(7397):292–292. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Raoof M, Chen Y, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464(7285):104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang JZ, Liu Z, Liu J, Ren JX, Sun TS. Mitochondrial DNA induces inflammation and increases TLR9/NF-κB expression in lung tissue. Int J Mol Med. 2014;33:817–824. doi: 10.3892/ijmm.2014.1650. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Puskarich MA, Shapiro NI, Trzeciak S, Kline JA, Jones AE. Plasma levels of mitochondrial DNA in patients presenting to the emergency department with sepsis. Shock. 2012;38(4):337–340. doi: 10.1097/SHK.0b013e318266a169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinti M, Cevenini E, Nasi M, et al. Circulating mitochondrial DNA increases with age and is a familiar trait: Implications for “inflamm-aging”. Eur J Immunol. 2014;44(5):1552–1562. doi: 10.1002/eji.201343921. [DOI] [PubMed] [Google Scholar]
- 16.Marques PE, Amaral SS, Pires DA, et al. Chemokines and mitochondrial products activate neutrophils to amplify organ injury during mouse acute liver failure. Hepatology. 2012;56(5):1971–1982. doi: 10.1002/hep.25801. [DOI] [PubMed] [Google Scholar]
- 17.Cao H, Ye H, Sun Z, et al. Circulatory Mitochondrial DNA Is a Pro-Inflammatory Agent in Maintenance Hemodialysis Patients. PLoS One. 2014;9:e113179. doi: 10.1371/journal.pone.0113179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathew A, Lindsley Ta, Sheridan A, et al. Degraded mitochondrial dna is a newly identified subtype of the damage associated molecular pattern (DAMP) family and possible trigger of neurodegeneration. J Alzheimer’s Dis. 2012;30:617–627. doi: 10.3233/JAD-2012-120145. [DOI] [PubMed] [Google Scholar]
- 19.Wilkins HM, Carl SM, Weber SG, et al. Mitochondrial Lysates Induce Inflammation and Alzheimer’s Disease-Relevant Changes in Microglial and Neuronal Cells. J Alzheimers Dis. 2014 doi: 10.3233/JAD-142334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pazmandi K, Agod Z, Kumar BV, et al. Oxidative modification enhances the immunostimulatory effects of extracellular mitochondrial DNA on plasmacytoid dendritic cells. Free Radic Biol Med. 2014;77:281–290. doi: 10.1016/j.freeradbiomed.2014.09.028. [DOI] [PubMed] [Google Scholar]
- 21.Tsai Y-Y, Lin J-M, Wan L, et al. Interleukin gene polymorphisms in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49(2):693–698. doi: 10.1167/iovs.07-0125. [DOI] [PubMed] [Google Scholar]
- 22.Seddon JM, George S, Rosner B, Rifai N. Progression of age-related macular degeneration: prospective assessment of C-reactive protein, interleukin 6, and other cardiovascular biomarkers. Arch Ophthalmol. 2005;123(6):774–782. doi: 10.1001/archopht.123.6.774. [DOI] [PubMed] [Google Scholar]
- 23.Ricci F, Staurenghi G, Lepre T, et al. Haplotypes in IL-8 Gene Are Associated to Age-Related Macular Degeneration: A Case-Control Study. PLoS One. 2013;8(6):e66978. doi: 10.1371/journal.pone.0066978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein R, Myers CE, Cruickshanks KJ, et al. Markers of inflammation, oxidative stress, and endothelial dysfunction and the 20-year cumulative incidence of early age-related macular degeneration: the Beaver Dam Eye Study. JAMA Ophthalmol. 2014;132(4):446–455. doi: 10.1001/jamaophthalmol.2013.7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goverdhan SV, Ennis S, Hannan SR, et al. Interleukin-8 promoter polymorphism −251A/T is a risk factor for age-related macular degeneration. Br J Ophthalmol. 2008;92(4):537–540. doi: 10.1136/bjo.2007.123190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izumi-Nagai K, Nagai N, Ozawa Y, et al. Interleukin-6 receptor-mediated activation of signal transducer and activator of transcription-3 (STAT3) promotes choroidal neovascularization. Am J Pathol. 2007;170(6):2149–2158. doi: 10.2353/ajpath.2007.061018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miao H, Tao Y, Li X. Inflammatory cytokines in aqueous humor of patients with choroidal neovascularization. [Accessed February 18, 2015];Mol Vis. 2012 18:574–580. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3298419&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]
- 28.Roh MI, Kim HS, Song JH, Lim JB, Koh HJ, Kwon OW. Concentration of cytokines in the aqueous humor of patients with naive, recurrent and regressed CNV associated with amd after bevacizumab treatment. Retina. 2009;29(4):523–529. doi: 10.1097/IAE.0b013e318195cb15. [DOI] [PubMed] [Google Scholar]
- 29.Nakahira K, Haspel JA, Rathinam VaK, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12(3):222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimada K, Crother TR, Karlin J, et al. Oxidized Mitochondrial DNA Activates the NLRP3 Inflammasome during Apoptosis. Immunity. 2012;36(3):401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marneros A. NLRP3 inflammasome blockade inhibits VEGF-A-induced age-related macular degeneration. Cell Rep. 2013;4(5):945–958. doi: 10.1016/j.celrep.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarallo V, Hirano Y, Gelfand BD, et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149:847–859. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doyle SL, Campbell M, Ozaki E, et al. NLRP3 has a protective role in age-related macular degeneration through the induction of IL-18 by drusen components. Nat Med. 2012;18(5):791–798. doi: 10.1038/nm.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157(5):1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Ambati J, Fowler BJ. Mechanisms of age-related macular degeneration. Neuron. 2012;75:26–39. doi: 10.1016/j.neuron.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paludan S, Bowie A. Immune Sensing of DNA. Immunity. 2013;38(5):870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bannwarth S, Procaccio V, Paquis-Flucklinger V. Rapid identification of unknown heteroplasmic mutations across the entire human mitochondrial genome with mismatch-specific Surveyor Nuclease. Nat Protoc. 2006;1(4):2037–2047. doi: 10.1038/nprot.2006.318. [DOI] [PubMed] [Google Scholar]
- 38.Mitter SK, Song C, Qi X, et al. Dysregulated autophagy in the RPE is associated with increased susceptibility to oxidative stress and AMD. Autophagy. 2014 February 2015;10:1989–2005. doi: 10.4161/auto.36184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarks SH. Ageing and degeneration in the macular region: a clinico-pathological study. Br J Ophthalmol. 1976;60(5):324–341. doi: 10.1136/bjo.60.5.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garg AD, Nowis D, Golab J, Vandenabeele P, Krysko DV, Agostinis P. Immunogenic cell death, DAMPs and anticancer therapeutics: An emerging amalgamation. Biochim Biophys Acta - Rev Cancer. 2010;1805(1):53–71. doi: 10.1016/j.bbcan.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Udar N, Atilano SR, Memarzadeh M, et al. Mitochondrial DNA haplogroups associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2009;50(6):2966–2974. doi: 10.1167/iovs.08-2646. [DOI] [PubMed] [Google Scholar]
- 42.Burdette DL, Vance RE. STING and the innate immune response to nucleic acids in the cytosol. Nat Immunol. 2013;14(1):19–26. doi: 10.1038/ni.2491. [DOI] [PubMed] [Google Scholar]
- 43.Abe T, Barber GN. Cytosolic-DNA-Mediated, STING-Dependent Proinflammatory Gene Induction Necessitates Canonical NF-κB Activation through TBK1. J Virol. 2014;88(10):5328–5341. doi: 10.1128/JVI.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blaauboer SM, Gabrielle VD, Jin L. MPYS/STING-Mediated TNF-, Not Type I IFN, Is Essential for the Mucosal Adjuvant Activity of (3'-5')-Cyclic-Di-Guanosine-Monophosphate In Vivo. J Immunol. 2013;192:492–502. doi: 10.4049/jimmunol.1301812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bauernfeind FG, Horvath G, Stutz A, et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183(2):787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiao Y, Wang P, Qi J, Zhang L, Gao C. TLR-induced NF-kB activation regulates NLRP3 expression in murine macrophages. FEBS Lett. 2012;586(7):1022–1026. doi: 10.1016/j.febslet.2012.02.045. [DOI] [PubMed] [Google Scholar]
- 47.Nordgaard CL, Berg KM, Kapphahn RJ, et al. Proteomics of the retinal pigment epithelium reveals altered protein expression at progressive stages of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2006;47(3):815–822. doi: 10.1167/iovs.05-0976. [DOI] [PubMed] [Google Scholar]
- 48.Kenney MC, Chwa M, Atilano SR, et al. Inherited mitochondrial DNA variants can affect complement, inflammation and apoptosis pathways: insights into mitochondrial-nuclear interactions. Hum Mol Genet. 2014;23(13):3537–3551. doi: 10.1093/hmg/ddu065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kenney MC, Chwa M, Atilano SR, et al. Mitochondrial DNA variants mediate energy production and expression levels for CFH, C3 and EFEMP1 genes: implications for age-related macular degeneration. PLoS One. 2013;8(1):e54339. doi: 10.1371/journal.pone.0054339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okamoto H, Umeda S, Obazawa M, et al. Complement factor H polymorphisms in Japanese population with age-related macular degeneration. [Accessed August 8, 2015];Mol Vis. 2006 12:156–158. http://www.ncbi.nlm.nih.gov/pubmed/16541016. [PubMed] [Google Scholar]
- 52.Yanagisawa S, Kondo N, Miki A, et al. A common complement C3 variant is associated with protection against wet age-related macular degeneration in a Japanese population. PLoS One. 2011;6(12):e28847. doi: 10.1371/journal.pone.0028847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida T, DeWan A, Zhang H, et al. HTRA1 promoter polymorphism predisposes Japanese to age-related macular degeneration. [Accessed August 8, 2015];Mol Vis. 2007 13:545–548. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2652018&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]
- 54.Fu L, Garland D, Yang Z, et al. The R345W mutation in EFEMP1 is pathogenic and causes AMD-like deposits in mice. Hum Mol Genet. 2007;16(20):2411–2422. doi: 10.1093/hmg/ddm198. [DOI] [PubMed] [Google Scholar]
- 55.Marmorstein LY, Munier FL, Arsenijevic Y, et al. Aberrant accumulation of EFEMP1 underlies drusen formation in Malattia Leventinese and age-related macular degeneration. Proc Natl Acad Sci U S A. 2002;99(20):13067–13072. doi: 10.1073/pnas.202491599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stone EM, Lotery AJ, Munier FL, et al. A single EFEMP1 mutation associated with both Malattia Leventinese and Doyne honeycomb retinal dystrophy. Nat Genet. 1999;22(2):199–202. doi: 10.1038/9722. [DOI] [PubMed] [Google Scholar]
- 57.Hageman GS, Anderson DH, Johnson LV, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102(20):7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patel M, Chan C-C. Immunopathological aspects of age-related macular degeneration. Semin Immunopathol. 2008;30(2):97–110. doi: 10.1007/s00281-008-0112-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cohen T, Nahari D, Cerem LW, Neufeld G, Levi BZ. Interleukin 6 induces the expression of vascular endothelial growth factor. [Accessed February 26, 2015];J Biol Chem. 1996 271(2):736–741. doi: 10.1074/jbc.271.2.736. http://www.ncbi.nlm.nih.gov/pubmed/8557680. [DOI] [PubMed] [Google Scholar]
- 60.Heidemann J, Ogawa H, Dwinell MB, et al. Angiogenic effects of interleukin 8 (CXCL8) in human intestinal microvascular endothelial cells are mediated by CXCR2. J Biol Chem. 2003;278(10):8508–8515. doi: 10.1074/jbc.M208231200. [DOI] [PubMed] [Google Scholar]
- 61.Martin D, Galisteo R, Gutkind JS. CXCL8/IL8 stimulates vascular endothelial growth factor (VEGF) expression and the autocrine activation of VEGFR2 in endothelial cells by activating NFkappaB through the CBM (Carma3/Bcl10/Malt1) complex. J Biol Chem. 2009;284(10):6038–6042. doi: 10.1074/jbc.C800207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tzeng H-E, Tsai C-H, Chang Z-L, et al. Interleukin-6 induces vascular endothelial growth factor expression and promotes angiogenesis through apoptosis signal-regulating kinase 1 in human osteosarcoma. Biochem Pharmacol. 2013;85(4):531–540. doi: 10.1016/j.bcp.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 63.Beatty S, Koh H, Phil M, Henson D, Boulton M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. [Accessed February 24, 2015];Surv Ophthalmol. 45(2):115–134. doi: 10.1016/s0039-6257(00)00140-5. http://www.ncbi.nlm.nih.gov/pubmed/11033038. [DOI] [PubMed] [Google Scholar]
- 64.Kimura K, Isashiki Y, Sonoda S, Kakiuchi-Matsumoto T, Ohba N. Genetic association of manganese superoxide dismutase with exudative age-related macular degeneration. [Accessed July 22, 2015];Am J Ophthalmol. 2000 130(6):769–773. doi: 10.1016/s0002-9394(00)00552-3. http://www.ncbi.nlm.nih.gov/pubmed/11124296. [DOI] [PubMed] [Google Scholar]
- 65.Kan M, Liu F, Weng X, et al. Association study of newly identified age-related macular degeneration susceptible loci SOD2, MBP, and C8orf42 in Han Chinese population. Diagn Pathol. 2014;9:73. doi: 10.1186/1746-1596-9-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muller F. The nature and mechanism of superoxide production by the electron transport chain: Its relevance to aging. J Am Aging Assoc. 2000;23(4):227–253. doi: 10.1007/s11357-000-0022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leifer CA, Kennedy MN, Mazzoni A, Lee C, Kruhlak MJ, Segal DM. TLR9 is localized in the endoplasmic reticulum prior to stimulation. [Accessed February 26, 2015];J Immunol. 2004 173(2):1179–1183. doi: 10.4049/jimmunol.173.2.1179. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2757936&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Latz E, Schoenemeyer A, Visintin A, et al. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5(2):190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 69.Ebihara N, Chen L, Tokura T, Ushio H, Iwatsu M, Murakami A. Distinct functions between toll-like receptors 3 and 9 in retinal pigment epithelial cells. Ophthalmic Res. 2007;39:155–163. doi: 10.1159/000103235. [DOI] [PubMed] [Google Scholar]
- 70.Ries M, Schuster P, Thomann S, Donhauser N, Vollmer J, Schmidt B. Identification of novel oligonucleotides from mitochondrial DNA that spontaneously induce plasmacytoid dendritic cell activation. J Leukoc Biol. 2013 Jul;94:123–135. doi: 10.1189/jlb.0612278. [DOI] [PubMed] [Google Scholar]
- 71.Izumi-Nagai K, Nagai N, Ohgami K, et al. Macular pigment lutein is antiinflammatory in preventing choroidal neovascularization. Arterioscler Thromb Vasc Biol. 2007;27(12):2555–2562. doi: 10.1161/ATVBAHA.107.151431. [DOI] [PubMed] [Google Scholar]
- 72.Izumi-Nagai K, Nagai N, Ohgami K, et al. Inhibition of choroidal neovascularization with an anti-inflammatory carotenoid astaxanthin. Invest Ophthalmol Vis Sci. 2008;49(4):1679–1685. doi: 10.1167/iovs.07-1426. [DOI] [PubMed] [Google Scholar]
- 73.Liu X-C, Liu X-F, Jian C-X, Li C-J, He S-Z. IL-33 is induced by amyloid-β stimulation and regulates inflammatory cytokine production in retinal pigment epithelium cells. Inflammation. 2012;35(2):776–784. doi: 10.1007/s10753-011-9379-4. [DOI] [PubMed] [Google Scholar]
- 74.Chen Y, Kijlstra A, Chen Y, Yang P. IL-17A stimulates the production of inflammatory mediators via Erk1/2, p38 MAPK, PI3K/Akt, and NF-κB pathways in ARPE-19 cells. [Accessed February 26, 2015];Mol Vis. 2011 17:3072–3077. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3233389&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]
- 75.Kerur N, Hirano Y, Tarallo V, et al. TLR-independent and P2X7-dependent signaling mediate Alu RNA-induced NLRP3 inflammasome activation in geographic atrophy. Investig Ophthalmol Vis Sci. 2013;54:7395–7401. doi: 10.1167/iovs.13-12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi G, Chen S, Wandu WS, et al. Inflammasomes induced by 7-ketocholesterol and other stimuli in RPE and in bone marrow-derived cells differ markedly in production of IL-1β and IL-18. Invest Ophthalmol Vis Sci. 2015 doi: 10.1167/iovs.14-14557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kauppinen A, Niskanen H, Suuronen T, Kinnunen K, Salminen A, Kaarniranta K. Oxidative stress activates NLRP3 inflammasomes in ARPE-19 cells-Implications for age-related macular degeneration (AMD) Immunol Lett. 2012;147(1–2):29–33. doi: 10.1016/j.imlet.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 78.Tseng WA, Thein T, Kinnunen K, et al. NLRP3 inflammasome activation in retinal pigment epithelial cells by lysosomal destabilization: implications for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2013;54:110–120. doi: 10.1167/iovs.12-10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anderson Oa, Finkelstein A, Shima DT. A2E Induces IL-1β Production in Retinal Pigment Epithelial Cells via the NLRP3 Inflammasome. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0067263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Piippo N, Korkmaz A, Hytti M, et al. Decline in cellular clearance systems induces inflammasome signaling in human ARPE-19 cells. Biochim Biophys Acta. 2014;1843(12):3038–3046. doi: 10.1016/j.bbamcr.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 81.Hu Y, Lin H, Dib B, et al. Cholesterol Crystals Induce Inflammatory Cytokines Expression in a Human Retinal Pigment Epithelium Cell Line by Activating the NF-kappaB Pathway. [Accessed February 26, 2015];Discov Med. 18(97):7–14. http://www.discoverymedicine.com/Yijun-Hu/2014/07/cholesterol-crystals-induce-inflammatory-cytokines-expression-in-arpe-19-cells-by-activating-the-nf-kappab-pathway/ [PMC free article] [PubMed] [Google Scholar]
- 82.Darzynkiewicz Z, Pozarowski P. All that glitters is not gold: all that FLICA binds is not caspase. A caution in data interpretation--and new opportunities. Cytometry A. 2007;71(8):536–537. doi: 10.1002/cyto.a.20425. [DOI] [PubMed] [Google Scholar]
- 83.Xu H, Chen M, Forrester JV. Para-inflammation in the aging retina. Prog Retin Eye Res. 2009;28(5):348–368. doi: 10.1016/j.preteyeres.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 84.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454(7203):428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 85.Dib B, Lin H, Maidana DE, et al. Mitochondrial DNA activates the NLRP3 inflammasome in THP-1 macrophages. Data in Brief. Submitted. [Google Scholar]