Abstract
Living tissues rely heavily on vascular networks to transport nutrients, oxygen and metabolic waste. However, there still remains a need for a simple and efficient approach to engineer vascularized tissues. Here, we created prevascularized tissues with complex three-dimensional (3D) microarchitectures using a rapid bioprinting method – microscale continuous optical bioprinting (μCOB). Multiple cell types mimicking the native vascular cell composition were encapsulated directly into hydrogels with precisely controlled distribution without the need of sacrificial materials or perfusion. With regionally controlled biomaterial properties the endothelial cells formed lumen-like structures spontaneously in vitro. In vivo implantation demonstrated the survival and progressive formation of the endothelial network in the prevascularized tissue. Anastomosis between the bioprinted endothelial network and host circulation was observed with functional blood vessels featuring red blood cells. With the superior bioprinting speed, flexibility and scalability, this new prevascularization approach can be broadly applicable to the engineering and translation of various functional tissues.
Keywords: 3D bioprinting, Vasculature, Tissue engineering, Complex microarchitecture, Biomaterials
1. Introduction
Tissue engineering is an emerging field that develops artificial biological organ substitutes to address the shortage of donor organs for transplantations and provide tissue models for drug testing. One of the most fundamental challenges in the tissue engineering arena is to create functional vasculature that provides vital nutrition, oxygen and waste transport to the cells within the engineered tissue [1–3]. Without proximity (~150–200 μm) to capillary network, cellular viability and function will be compromised within a very short time, especially in highly metabolic and large-scale tissue constructs [2,3]. To induce vascularization in implanted tissue substitutes, one strategy is to recruit the host vasculature by incorporating pro-angiogenic growth factors [4–6]. However, the use of growth factor in large scale is cost-prohibitive and not efficacious in vivo due to the relatively slow ingrowth process, which is unfavorable to cell viability during the first days post implantation [7,8]. Prevascularization of the engineered tissues, by encapsulation of endothelial cells and supportive cells in vitro before implantation, has showed promising results in enhancing the vascularization, blood perfusion and cellular activity of the tissue graft in vivo [7,9–11]. Further studies have demonstrated that compared to a randomly seeded endothelial network, spatially defined endothelial cords prepatterned with a polydimethylsiloxane (PDMS) mold can significantly improve the speed and extent of the vascularization of the engineered tissue after implantation [2]. The PDMS molding approach employed the self-assembly of the cells in collagen followed by the encasement of two fibrin layers, which is relatively time-consuming (over 4 hours) and is limited to simple geometric designs of the vascular network. Recently, 3D stamping based on PDMS molding was used to build 3D scaffolds with different sizes of branched channels and micro-holes[12]. These scaffolds can be further perfused with endothelial cells to form hierarchical vasculature networks. Compared to the traditional PDMS molding technique, 3D stamping greatly improved the capability of engineering complex vasculature tissues. However, multiple molding and transferring steps as well as accurate alignment of the parts were required to produce these complex scaffolds, which is labor-intensive and not suitable for live cell encapsulation during the fabrication process. Moreover, both PDMS molding and 3D stamping require the manufacturing of new physical molds per each design change, which is costly and laborious. Thus, more sophisticated fabrication approaches featuring excellent flexibility, speed and versatility are still of great interest to the field of vasculature tissue engineering.
With proven flexibility and versatility, nozzle-based 3D printers have also been adopted to build perfusable 3D tissues with sacrificial inks, involving the extra process of dissolving the sacrificial network and perfusing the endothelial cells [1,3,13,14]. Challenges such as clogging and bursting might occur during perfusion when this approach is used to engineer large-scale tissues with intricate vascular network featuring small vessel branches. Recently, nozzle-based bioprinters have been adapted to print endothelial networks directly with cell-encapsulated bioinks, which is potentially useful for engineering large-scale vascularized tissues [15,16]. Laser based SLA systems have also been modified to write 3D structures with cell-laden hydrogels [17]. However, with such serial line by line writing approaches, printing large-scale tissues is presumably time-consuming. Moreover, the mechanical integrity of the tissues printed in a serial fashion by nozzle or laser -based bioprinters is a concern especially at the interfaces of the lines. Digital light processing (DLP) based 3D printing has emerged as a next generation microfabrication technique, offering superior speed, resolution, scalability and flexibility for printing various complex 3D architectures with micrometer resolution [18,19]. No physical masks or molds are needed in DLP based 3D printing process. Instead, a digital micromirror array device (DMD) is able to take digital 3D designs from a computer to control the 3D fabrication of complex structures. Compared to the raster scanning of nozzle based 3D printers, DLP based 3D printers are capable to continuously project and alter entire planes of photo-masks to fabricate 3D objects without artificial interfaces, which provides better mechanical integrity [18,20]. More importantly, since the printing is based on the photopolymerization of a solution, a wide range of biomaterials as well as cells, nanoparticles, and biomolecules can be incorporated into the printed tissue constructs [21–24].
In this work, we present a rapid DLP based bioprinting method - microscale continuous optical bioprinting (μCOB) - to create prevascularized tissue constructs directly with unprecedented speed and resolution. The 3D-printed prevascularized tissues have complex microarchitecture and precisely controlled distribution of multiple cell types and biomaterial compositions. To simplify and speed up the tissue engineering process, endothelial cells and mesenchymal cells were printed directly into the designed vascular channels without the use of sacrificial material or perfusion. These cells formed lumen-like structures and functional endothelial networks spontaneously both in vitro and in vivo, which provides a much simpler and more efficient platform for engineering tissues with complex structures and functions.
2. Material and methods
2.1. Polymer and photoinitiator synthesis
Glycidal methacrylate-hyaluronic acid (GM-HA) was synthesized according to a protocol modified from previous work [25]. 1g of hyaluronic acid was first dissolved in 100ml of acetone/water (50/50) solution at room temperature overnight. 7.2ml tri-ethylamine (Sigma–Aldrich) and 7.2ml glycidyl methacrylate (Sigma–Aldrich) were added dropwise both at 20-fold excess in succession until thoroughly mixed. The solution was covered with aluminum foil and stirred overnight at room temperature. The resulting solution was then dialyzed against DI water with 3.5 kDa tubing (Spectrum Labs) at room temperature. The DI water was changed after 2 hours, 4 hours and 24 hours. The dialyzed solution was frozen overnight at −80 °C and then lyophilized for 48 hours at 0.040 mbar and −50 °C. The lyophilized GM-HA was stored at −80 °C for future use.
Gelatin methacrylate (GelMa) was synthesized according to a protocol adapted from previous work [26]. Briefly, 10% (w/v) porcine skin gelatin (Sigma–Aldrich) was dissolved into Dulbecco’s phosphate-buffered saline (DPBS) by stirring at 60 °C. Methacrylate anhydride (Sigma–Aldrich) was added to the solution at a rate of 0.5 ml/min until the final concentration of 8% (v/v) MA was reached. The reaction continued for 3 hours at 60 °C with constant stirring. After 3 hours, the resulted solution was diluted 1:1 with warm DPBS and dialyzed against DI water with 13.5 kDa tubing for 1 week at 40 °C. The dialyzed solution was then frozen at − 80 °C and lyophilized for 1 week. The lyophilized GelMa was store at − 80 °C for future use.
Photoinitiator lithium phenyl-2,4,6 trimethylbenzoylphosphinate (LAP) was synthesized according to previously published work [27]. Briefly, 3.2g (0.018mol) of 2,4,6-trimethylbenzoyl chloride (Sigma–Aldrich) was added dropwise to an equal molar amount of dimethyl phenylphosphonite (3g, Acros Organics) with continuous stirring at room temperature under argon. After 18 hours, 6.1g lithium bromide (Sigma–Aldrich) dissolved in 100ml of 2-butanone (Sigma–Aldrich) was added into the previous mixture at 4-fold excess. The reaction was then heated to 50 °C and a solid precipitate was formed after 10 min. The mixture was allowed to cool down to room temperature and rest for overnight before filtration. 2-butanone was used to wash the filtrate and remove the unreacted lithium bromide. After 3 times wash and filtration, the excess solvent was removed by vacuum, leaving LAP in a white solid chunk state which was pestled into powder. LAP was stored at − 80 °C under argon for future use.
2.2. Cell and tissue cultures
Human umbilical vein endothelial cells (HUVECs, Lonza) were maintained in endothelial cell growth medium (EGM-2, Lonza) and C3H/10T1/2 cells (10T1/2s, ATCC) were maintained in Dulbecco’s modified eagle medium (DMEM, Gibco) supplemented with 10% fetal bovine serum. HepG2 cells were maintained in DMEM (Gibco) supplemented with 10% fetal bovine serum. All cell cultures were passaged per the protocol from the respective vendors. HUVECs from passage 3–6 were used for the bioprinting experiments. The bioprinted tissue constructs were cultured in EGM-2 medium and the medium was changed every other day.
2.3. 3D bioprinting of tissues
Before bioprinting, HUVECs and 10T1/2 cells were digested by 0.05% trypsin-EDTA and 0.25% trypsin-EDTA, respectively, and they were mixed at a ratio of 50:1 (40 million/ml HUVECs and 800,000/ml 10T1/2s) suggested by previous work [28]. Prepolymer A was prepared with 5% (w/v) GelMa and 0.15% (w/v) LAP. Prepolymer B was prepared with 5% (w/v) GelMa, 2% (w/v) HA and 0.3%(w/v) LAP. Prepolymer A was first used to print the isolated hexagonal regions (base layer) with the first mask on the left in Fig. 1A. After 15 seconds of UV exposure (88mW/cm2), the unpolymerized part of prepolymer A was removed and washed off with DPBS. For the prevascularized tissue constructs, prepolymer B was mixed at a ratio of 1:1 with the prepared cell suspension of HUVECs and 10T1/2, giving a final composition of 2.5% (w/v) GelMa, 1%(w/v) HA, 0.15%(w/v) LAP, 20 million/ml HUVECs and 40,000/ml 10T1/2s. The cell-laden prepolymer solution was loaded to the fabrication stage and exposed to the UV pattern (15 seconds) with the vascular channel mask (vascular layer) in the middle in Fig. 1A. The unpolymerized part of the cell-laden prepolymer solution was then washed off. Prepolymer A was loaded to the fabrication stage and exposed to the UV pattern with the slab mask (top layer) on the right in Fig. 1A to enclose the vascular network (15 seconds). The entire tissue construct measured around 4mm × 5mm in XY plane and 600 μm thick in Z direction. The bioprinted tissue construct was then transferred to a well plate for culturing. For the non-prevascularized tissue construct, the same fabrication process was performed except that prepolymer B was mixed with an equal volume of cell culture medium instead of cells suspensions for the fabrication of the vascular network. To print the tissues in Fig. 1D–1I, HepG2 cells were digested by 0.25% trypsin-EDTA and mixed with the prepolymer to a final composition of 5%(w/v) GelMa+0.15%(w/v) LAP+20 million/ml HepG2.
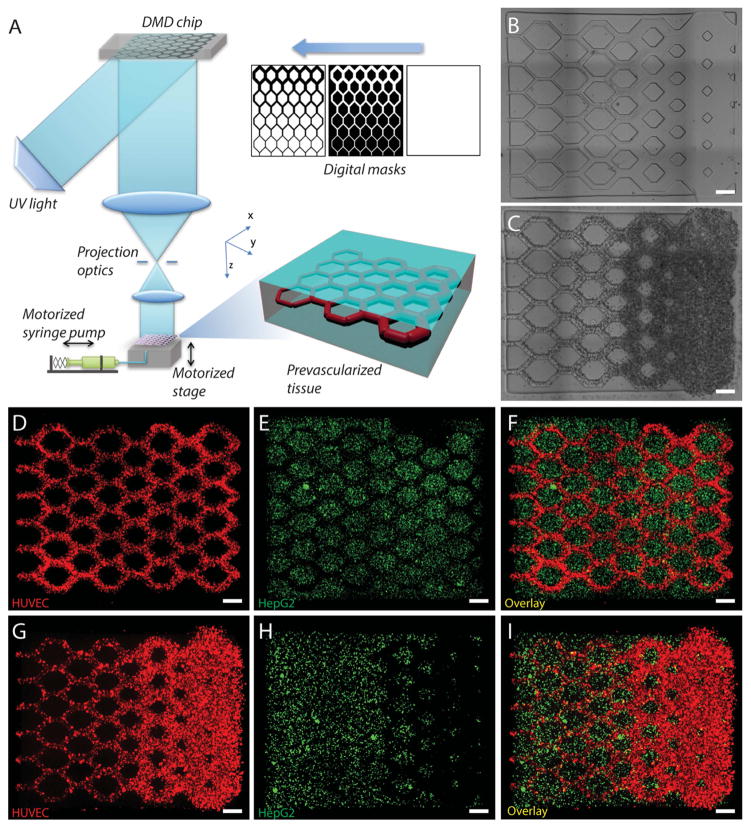
Fig. 1. 3D bioprinting of the prevasularized tissue constructs.
(A) Schematic of the bioprinting platform. (B) Bioprinted acellular construct featuring intended channels with gradient widths. (C) Bioprinted cellular construct with HUVECs and 10T1/2 (50:1) encapsulated in the intended channels. (D–F) Fluorescent images demonstrating the bioprinting of heterogeneous cell-laden tissue constructs with uniform channel width. HUVECs (red) are encapsulated in the intended channels and HepG2 (green) are encapsulated in the surrounding area. (G–I) Fluorescent images demonstrating the bioprinting of heterogeneous cell-laden tissue constructs with gradient channel widths. Scale bars, 250 μm.
An image of the native rat capillary network was adapted to a 3D model (measuring 4mm long, 2mm wide, and 500 μm thick) and it was printed with 5%GelMa + 0.15%LAP. The printed hydrogel construct was imaged by a Leica DMI 6000B microscope (bright field, Leica Microsystems).
2.4. Mechanical measurement
Mechanical properties of the two compositions of hydrogels were investigated by dynamic mechanical analysis (DMA 8000, Perkin Elmer, USA). The samples (n=3 for each composition) were made into a 1 mm × 10 mm × 11 mm cuboid using the same UV exposure parameters. Compression tests with frequency scan measurement mode was used to determine the compression modulus of the hydrogels. The test was conducted at 23 °C with loading rates from 0.1 Hz to 3Hz. The maximum strain was kept constant at 10%. Data were reported as mean ± standard deviation.
2.5. Cell viability assay
Slab tissue constructs (4mmx5mmx600μm) were printed with 1%HA+2.5%GelMa+0.15%LAP encapsulating 20 million/ml HUVECs and 40,000/ml 10T1/2 cells. Cell viability assay (LIVE/DEAD® Viability/Cytotoxicity Kit, Invitrogen) was performed on day 1, day 3 and day 7 after the tissue constructs were printed. The tissue constructs (n=3 for each time points) were washed with DPBS three times after removing the culture medium. The tissue constructs were then stained with 2μM calcein AM (live cell stain) and 4μM ethidium homodimer-1 (dead cell stain) solution at room temperature for 30 minutes. After the incubation, live/dead assay was removed and the samples were washed with DPBS. Z-stack fluorescence images of the samples were taken by a Leica DMI 6000B microscope (10X objective, Leica Microsystems) immediately after the wash for quantification. Live and dead cells were counted manually in a blinded experiment for each sample. Data were reported as mean ± standard deviation.
2.6. Degradation test in vitro
We printed the same hydrogel constructs with fluorescent beads labeling different regions. The base (island) and top layers were printed with 5% GelMa+0.15%LAP mixed with green fluorescent beads. The channel layer was printed with 1%HA+2.5%GelMa+0.15%LAP mixed with red fluorescent beads. The hydrogel constructs were incubated with 100 units/ml hyaluronidase at 37 ºC. Fluorescent images of the hydrogel constructs were taken by Leica DMI 6000B microscope (Leica Microsystems) after 0, 1, 2, 4, 24, and 48 hours. Before the imaging of each time point, the hyaluronidase solution was removed and the samples were washed with DPBS solution for three times. After imaging, the DPBS solution was removed and replaced with fresh 100 units/ml hyaluronidase solution. Then the samples were put back to the incubator for subsequent digestion.
2.7. Immunofluorescence staining and image acquisition of the in vitro cultured tissues
To investigate the endothelial network formation in vitro, the tissue constructs were cultured for a week and then fixed with 4% paraformaldehyde phosphate buffer solution (PFA, Wako) for 30 minutes at room temperature. The fixed samples were blocked and permeabilized by 2% bovine serum albumin (BSA, Gemini Bio-Products) solution with 0.1% Triton X-100 (Promega) for 1 hour at room temperature and then immunostained using primary antibodies against human CD31 (1:100, Thermo Scientific) and alpha-smooth muscle actin (α-SMA, 1:100, Abcam). The samples were then imaged by a Leica SP5 confocal microscope (Leica microsystems) with the z-stack function. The 3D view was reconstructed by Imaris software (Bitplane).
2.8. In vivo implantation of tissue constructs
Severe combined immunodeficiency (SCID) mice were used for the in vivo experiments. The bioprinted tissue constructs were cultured in vitro for 1 day to stabilize before implantation. For the subcutaneous implantation, a dorsal skin incision (1 cm) was created on each side of the back of the SCID mice and a subcutaneous pocket was built by blunt preparation. The prevascularized and non-prevascularized tissue constructs were inserted into the left pocket and the right pocket respectively. Afterwards, the wounds were thoroughly closed with 3-0 polypropylene sutures. The tissue constructs were harvested after 2 weeks post-implantation.
2.9. Immunofluorescence, histology staining and image acquisition of the in vivo grafted tissues
Two weeks after implantation the tissue constructs were harvested from in vivo. The tissue constructs were fixed with 4% PFA immediately for 30 minutes at room temperature and then dehydrated in 30% sucrose solution at 4 °C overnight. The dehydrated samples were embedded in the optimal cutting temperature (OCT) compound and frozen at −80 °C overnight. The frozen samples were then cryosectioned at a thickness of 20 μm. To identify the implanted endothelial network, the sections were stained with nuclei stain (DAPI) and primary antibodies against human-specific Von Willebrand Factor (hVWF, 1:100, Santa Cruz Biotechnology) and human CD31 (1:100, Thermo Scientific). The fluorescent images were taken by a Leica DMI 6000-B microscope.
To investigate the anastomosis between the grafted tissue construct and host circulation, sections were stained with hematoxylin and eosin (H&E) for the identification of red blood cells and blood vessels. Histology images were taken by a Keyence BZ-9000 microscope with multicolor CCD camera.
2.10. Perfusion of mouse and human specific lectins
To distinguish the mouse and human endothelial networks at the interface of the host and grafted tissues, mouse and human specific lectins were injected in vivo two weeks after the implantation according to the previous work [28]. In brief, 200 μl of 500 μg/ml lectin from helix pomatia agglutinin (HPA) FITC conjugate (Sigma–Aldrich), and 100 μg/ml lectin from Ulex europaeus agglutinin-Atto (UEA) 594 conjugate (Sigma-Aldrich) in DPBS was injected via the tail vein of the SCID mice. The grafted tissue constructs were subsequently harvested from the mice and fixed with 4% PFA immediately for 30 minutes at room temperature. The fixed tissue constructs were then dehydrated in 30% sucrose solution at 4 °C overnight. The dehydrated samples were embedded in the OCT compound and frozen at −80 °C overnight. The frozen samples were cryosectioned at a thickness of 20 μm. The sections were imaged by a Leica DMI 6000-B microscope.
2.11. Statistical Analysis and Quantification of Vascularization Parameters
The vascularization parameters were quantified on the imaged H&E sections using FIJI open source software [2,10,29]. Vessel area density was quantified by adding up the area of the individual vessels in a section and normalized to the area of the grafted tissue in the section. Vessel number was quantified by counting individual disconnected vessels within a section and then normalized to the area of grafted tissue in the section. Data was reported as mean values with error bars representing the standard deviations. Comparisons of sample means were performed by t test using Origin software (OriginLab). P < 0.05 was considered statistically significant.
3. Results and Discussion
3.1. 3D bioprinting of prevascularized tissues
The schematic of the μCOB is shown in Fig. 1A. The digital micromirror array device (DMD) features an array of approximately two million micromirrors which can be controlled individually to dictate the optical pattern that is projected to the monomer solution on the fabrication stage. A UV LED (365 nm) was used to induce the photopolymerization of the photosensitive biomaterials. A motorized syringe pump system was used to add and remove the prepolymer solution. The continuous 3D printing process is realized by continuously feeding a series of digital masks to the DMD chip and simultaneously moving the stage. The digital masks can be sliced from 3D models built in computer-aided-design (CAD) software or computed tomography (CT) and magnetic resonance imaging (MRI) scans of the native organs. The entire 3D printing process is digitalized and controlled by a computer, which offers the great reproducibility and flexibility to modify the design and optimize the design parameters.
To 3D print the tissue constructs, we have chosen two biocompatible and photopolymerizable hydrogels as the matrix material: GM-HA and GelMa. Hyaluronic acid (HA) is an immunoneutral biocompatible material that can be found ubiquitously in native tissues and it has important roles in many cellular responses, such as cell signaling, wound healing, and angiogenesis [30]. The addition of methacrylate groups to HA makes it photopolymerizable, while retaining the biological activity of HA to promote endothelial cell proliferation [24,31,32]. GelMa is also a photopolymerizable hydrogel modified from denatured collagen that retains natural cell binding motifs [26]. Besides supporting good cell viability following encapsulation and mediated cell-biomaterial interaction, GelMa has also been used to create perfusable microchannels seeded with endothelial cells for the engineering of microvascular network [26]. Furthermore, both GM-HA and GelMa offer tunable mechanical properties by varying the methacrylation ratio and material concentration [26,31].
For this particular study, we have designed three digital masks (Fig. 1A) to fabricate tissues with gradient channel widths (ranging from 50 μm to 250 μm), mimicking the branching structure of a vasculature network. A non-prevascularized tissue (Fig. 1B) and a prevascularized tissue (Fig. 1C) were fabricated with the same design. With the highly efficient μCOB platform, the printing process for each tissue construct was completed within 1 minute. For both types of tissues, we used a mixture of 1% GM-HA and 2.5% GelMa for the fabrication of the channel region. 5% GelMa was used to fabricate the surrounding region. For the prevascularized tissue, HUVECs and 10T1/2 cells were encapsulated in the same mixture of GM-HA and GelMa at a cell density ratio of 50:1 suggested by previous work [28]. To demonstrate the versatility of 3D printing tissues with complex microarchitecture and controlled cell distribution, we fabricated tissue constructs with uniform channels (Fig. 1, D to F) as well as gradient channels (Fig. 1, G to I). For both tissue constructs, we encapsulated HUVECs (CellTracker™ red labeled) in the channels and liver hepatocellular HepG2 cells (CellTracker™ green labeled) in the surrounding region. Both types of cells were precisely localized to the designated region, which is a key feature for engineering tissue with complex microarchitecture and multiple material compositions. To demonstrate the capability of printing highly complex 3D structures with microscale resolution, we also printed a tissue construct replicating the rat capillary network (Fig. S1). This biomimetic tissue construct features intricate microchannels with diameters ranging from 50μm down to 5μm at different z heights.
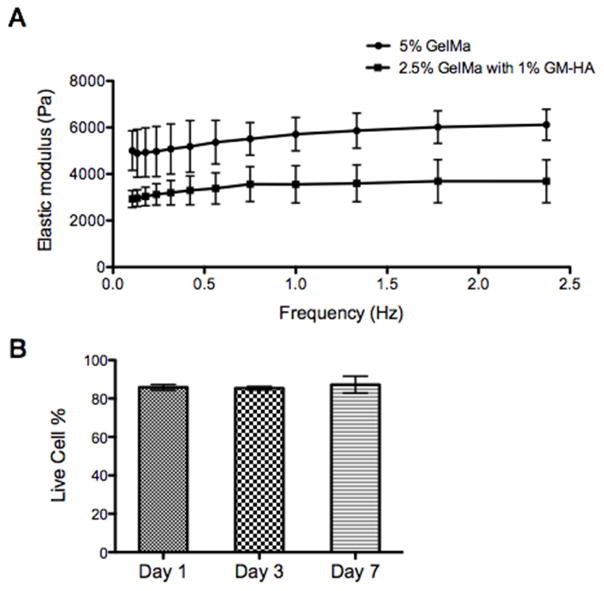
The mechanical property of the two compositions of hydrogels was measured by dynamic mechanical analysis (DMA, Fig. 2A). As shown in Fig. 2A, the hydrogel used for the channel region (2.5% GelMa + 1% GM-HA) has a much lower elastic modulus than the hydrogel used for the surrounding region (5% GelMa). We hypothesize that the difference in mechanical stiffness would induce the lining of the cells to the walls of the channels, which is the interface between the two hydrogels based on previous studies [33–36]. Notably, the mechanical property of the two hydrogels remained stable over different frequencies (from 0.1 to 2.5 Hz), which has been the suggested requirement for mechanical stability needed for the pulsatile flow environment of blood vessels [1].
Fig. 2. In vitro characterization of the bioprinted tissue.
(A) Elastic modulus of the biomaterials used to encapsulate cells measured by DMA: 2.5% GelMa with 1% HA for the channel region and 5% GelMa for the surrounding region. (B) Results of cell-viability assay for the bioprinted tissue constructs encapsulated with HUVECs demonstrating over 85% cell viability. Error bars represent SEM, n=3 for all data points.
3.2. In vitro culture of the prevascularized tissues
We first evaluated the biocompatibility of the μCOB printing process. We performed the cell viability assay on day 1, day 3, and day 7 after printing the tissue constructs based on z-stack imaging of the live/dead staining samples (Fig. 2B). It was found that over 85% viability was achieved, which is similar if not higher compared to the tissues printed by nozzle-based 3D bioprinters [3,15,37]. This can be partly explained by the high efficiency of the μCOB method that cells are exposed to the printing process for very short time (less than 1 min). Also, in μCOB printing, there is no shear stress or dispensing pressure caused by the cell delivery through the nozzle in the conventional 3D bioprinters which is reported to have negative influence on cell viability [38–40]. This highlights the biocompatibility of the μCOB printing process. And optimization of cell handling before and during the printing process can be performed to further increase the cell viability.
To test our hypothesis that the material in the channel region can be biodegraded by enzymes such as hyaluronidase and form hollow channels, we begin with a hydrogel degradation assay in vitro. We labeled the different regions of the tissue constructs with different fluorescent beads. The channel area was labeled with red fluorescence and the other regions were labeled with green fluorescence. The printed tissue constructs were incubated with 100 u/ml hyaluronidase at 37 ºC [31]. Within 48 hours, most hydrogel in the channel area vanished, leaving hollow channels (Fig. S2). We are aware that it is very hard to replicate the in vivo biodegradation environment, given the complex ECM dynamics in vivo and the dramatic differences from tissue to tissue. However, this in vitro degradation assay could support our hypothesis to some extent and encourage us to move on to the in vitro culture with cells and the in vivo implantation.
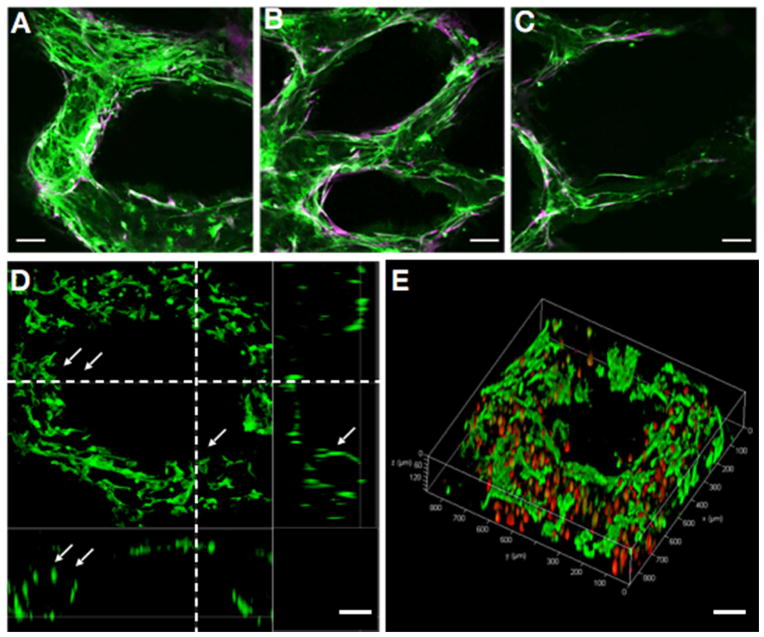
The 3D bioprinted prevascularized tissues with endothelial cells and supportive 10T1/2 cells were then cultured in vitro to investigate the formation of the endothelial network. We performed immunofluorescence staining on the prevascularized tissues after 1-week culture in vitro. Human-specific CD31 staining (green) shows the conjunctive network formation of HUVECs at different patterned channel widths (ranging from ~50 μm to ~250 μm, Fig. 3, A to C). Alpha-smooth muscle actin (α-SMA) positive cells (purple) were observed lining along the channel wall in tight conjunction with the HUVECs, which suggests that 10T1/2 cells were induced into a pericyte phenotype supporting the vessel formation (Fig. 3, A to C) [2]. Cross-sectional view and 3D reconstruction from confocal microscopy revealed the formation of lumen-like structures by CD31-positive HUVECs lining along the channels after 1-week culture in vitro (Fig. 3, D and E), further confirming our degradation hypothesis.
Fig. 3. Endothelial network formation after 1-week culture of the prevascularized tissue construct in vitro.
(A–C) Confocal microscopy images show HUVECs (Green, CD31-positive) and supportive mesenchymal cells (10T1/2, Purple, alpha-smooth muscle actin (α-SMA)-positive) aligned within the patterned gradient channel regions with different vessel sizes. (D) Cross-section view shows the endothelial cells (CD31-positive) form lumen-like structures (highlighted by arrows) along the bioprinted channels. (E) 3D view of the endothelial cells lining along the printed microchannel walls by confocal microscopy. Endothelial cells were labeled by fluorescent cell tracker (red) and stained by CD31 (green). Scale bars: 100 μm.
3.3. Endothelial network formation in vivo
To investigate the formation of the endothelial network in vivo, we implanted both the prevascularized tissues and the non-prevascularized tissues (control group) under the dorsal skin of SCID mice. In the prevascularized tissues, a mixture of HUVECs and 10T1/2 cells (50:1) were printed into the gradient channels (Fig. 1C presents the prevascularized tissue before implantation). In the non-prevascularized tissues, no cells were printed into the samples, only hydrogels with same compositions were used (Fig. 1B presents the non-prevascularized tissue before implantation).
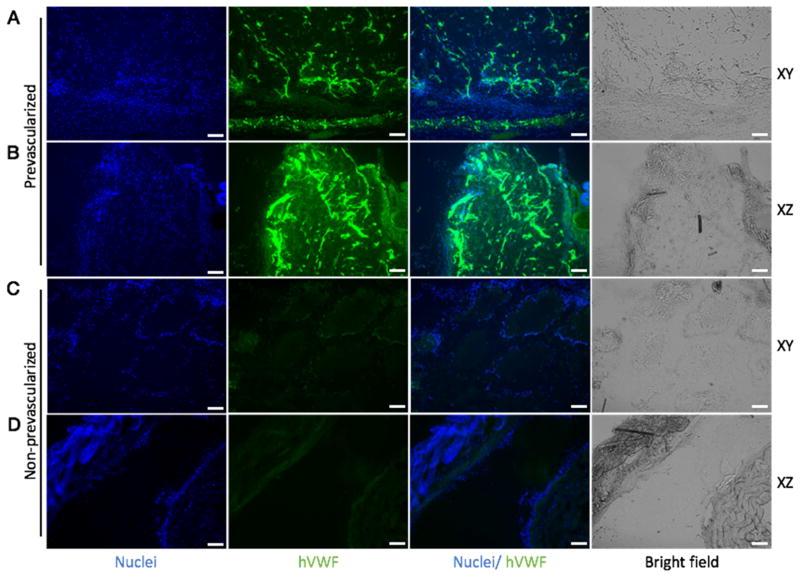
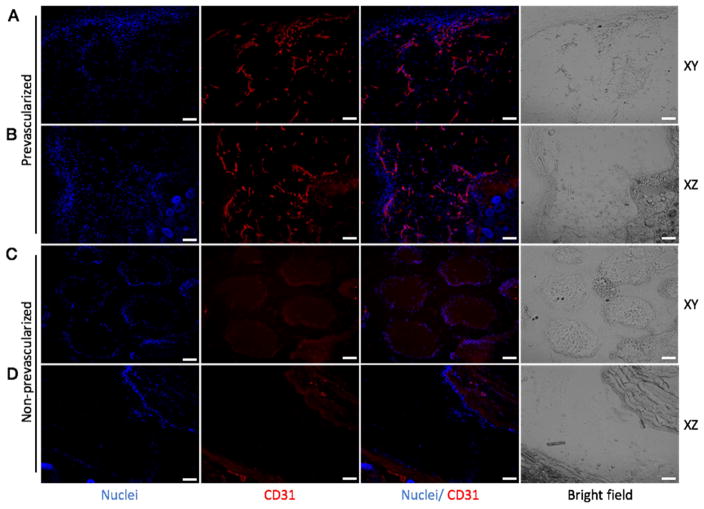
After 2 weeks of subcutaneous implantation, the grafted tissues were harvested and cryosectioned for immunofluorescence staining. To inspect the interconnection of the endothelial network, each group of tissues was sectioned in two directions: longitudinally (xy-plane, denoted in Fig. 1A) and transversely (xz-plane, denoted in Fig. 1A). hVWF staining revealed the survival and progressive formation of the endothelial network in the group of prevascularized tissues, while no positive staining of hVWF was found in the control group (Fig. 4). Similarly, staining of human-specific CD31 confirmed the survival and dense formation of the endothelial network in the prevascularized tissues (Fig. 5). Notably, from Fig. 4C and Fig. 5C, we observed that the printed patterns were well preserved in the non-prevascularized tissues after 2 weeks of in vivo implantation. In the prevascularized tissues, however, the hydrogels were observed to have lost the printed patterns. These results suggested that the prevascularized tissues could survive and form progressive endothelial network in vivo over the 2-week period.
Fig. 4. Endothelial network formation in the prevascularized tissues (A, B) and non-prevascularized tissues (C, D) after 2-week subcutaneous implantation shown by hVWF staining.
In the prevascularized tissues, a mixture of HUVECs and supportive 10T1/2 cells were printed into the gradient channels (Fig. 1C presents the prevascularized tissue before implantation). In the non-prevascularized tissues, no cells were printed into the samples, only biomaterials were used (Fig. 1B presents the non-prevascularized tissue before implantation). The bioprinted tissues were implanted under the dorsal skin of SCID mice for two weeks. The samples harvested from the implantation were cryosectioned in both xy and xz planes (xyz axises were designated in Fig. 1A). DAPI was used to stain the nuclei (blue) and hVWF was used to stain the endothelial network (green). Scale bars, 100 μm.
Fig. 5. Endothelial network formation in the prevascularized tissues (A, B) and non-prevascularized tissues (C, D) after 2-week subcutaneous implantation shown by CD31 staining.
In the prevascularized tissues, a mixture of HUVECs and supportive 10T1/2 cells were printed into the gradient channels (Fig. 1C presents the prevascularized tissue before implantation). In the non-prevascularized tissues, no cells were printed into the samples, only biomaterials were used (Fig. 1B presents the non-prevascularized tissue before implantation). The bioprinted tissues were implanted under the dorsal skin of SCID mice for two weeks. The samples harvested from the implantation were cryosectioned in both xy and xz planes (xyz axises were designated in Fig. 1A). DAPI was used to stain the nuclei (blue) and CD31 was used to stain the endothelial network (red). Scale bars, 100 μm.
3.4. Anastomosis of the prevascularized tissue with the host circulation
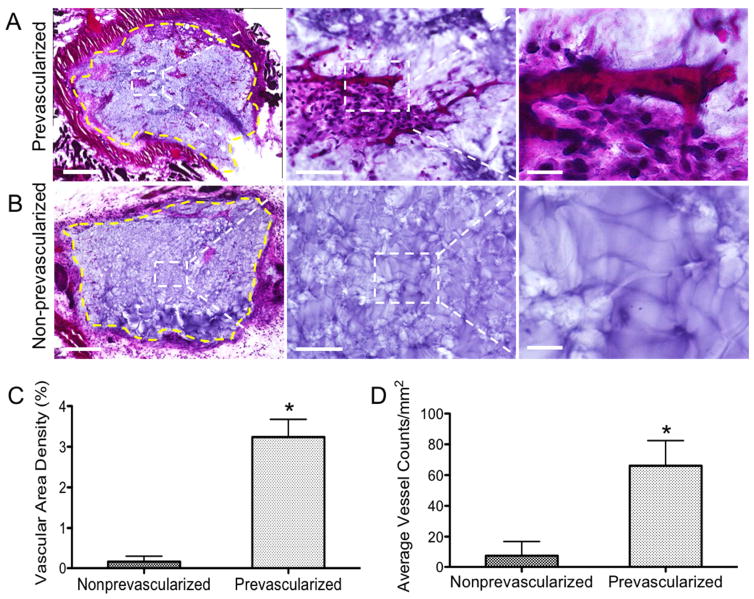
To investigate the anastomosis of the implanted tissue construct with the host vasculature, we performed H&E staining to the tissue grafts harvested from the 2-week subcutaneous implantation (Fig. 6). In the prevascularized tissues, significant amount of endothelial vessels with red blood cells were found, indicating successful anastomosis of the preformed vasculature with the host blood vessels (Fig. 6A). In the non-prevascularized tissues, very limited endothelial networks were found only near the periphery of the implanted tissue construct, and a majority of the imaged graft area remained as the biomaterial matrix without cells (Fig. 6B). To better assess the benefits of prevascularization, we quantified the vascular area density and the average vessel counts per area on the H&E sections (Fig. 6C&D) [2,10]. The quantifications suggested that compared to the non-prevascularized tissues, the prevascularized tissues were characterized by significantly increased vascular area density (3.24% ± 0.44% compared to 0.17% ± 0.14%, in the non-prevascularized group) and a significantly higher number of vessels per area (66.10 ± 16.32/mm2, compared to 7.66 ± 9.25/mm2 in the non-prevascularized group).
Fig. 6. H&E staining of the grafted tissues after 2-week subcutaneous implantation and quantification of vasculature parameters.
(A) Representative H&E stained images of the prevascularized tissues showing significant amount of endothelial vessels with red blood cells were found. Yellow dash line marks the interface between the graft and host tissue. (B) Representative H&E stained images of the non-prevascularized tissues, showing limited endothelial vessels. Yellow dash line marks the interface between the graft and host tissue. (C) Quantification of vascular area density in the grafted tissues. (D) Quantification of average vessel counts per area in the grafted tissue. Error bars represent SEM, n=6 for all data points. * indicates significant difference between the prevascularized group and the nonprevascularized group, p < 0.05. Scale bars: (A) 500μm (left), 100μm (middle), 25μm (right); (B) 500μm (left), 100μm (middle), 25μm (right).
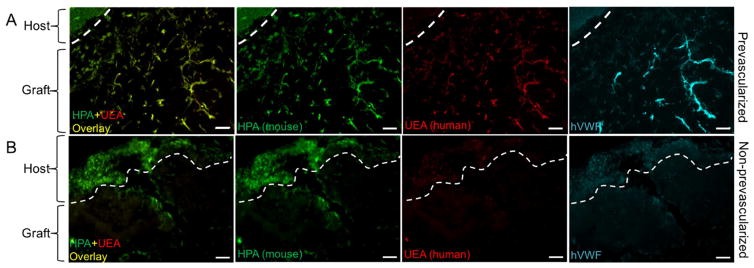
To further investigate the origins of the vasculature at the interface between the host and the grafted tissue, we injected the mice with mouse-specific lectin (HPA) and human-specific lectin (UEA) via tail vein after two weeks post-implantation based on previous work [2]. Previous studies have demonstrated that these lectins bind specifically to mouse or human endothelial cells respectively [2,41,42]. Fluorescent images exhibited that in the prevascularized tissues, large numbers of the endothelial networks in the graft area were co-labeled with both HPA and UEA, suggesting the chimeric compositions of host and grafted cells in these vessels (Fig. 7A). In the non-prevascularized tissues, only the mouse-specific HPA stained the host tissue and minor region of the graft area (Fig. 7B). Notably, the host tissues in both prevascularized and non-prevascularized groups were only stained with HPA and no UEA stain was observed in the graft area of the non-prevascularized group, which confirms the binding specificity of the two lectins. Further staining of hVWF confirmed the contribution of the human-origin HUVECs to the endothelial networks in the graft of prevascularized tissues (Fig. 7A). Together, these results indicated that the human endothelial networks in the prevascularized tissues were perfused after implantation and anastomosed with the host circulation.
Fig. 7. Perfusion of mouse and human specific lectins after two-week subcutaneous implantation.
(A) In the prevascularized tissue, mouse-specific lectin (HPA) and human-specific lectin (UEA) are chimeric in the graft area, and the host tissue is only stained with mouse-specific lectin. Further staining of hVWF confirms the endothelial network formed by human-origin HUVECs. (B) In the nonprevascularized tissue, HPA stains the host tissue and minor regions of the graft area, no UEA or hVWF staining is observed. Scale bars: 100μm.
4. Conclusion
Vascularization has been the bottleneck for engineering large-scale or highly metabolic tissues for decades [43–45]. While considerable amount of work has been carried out in the tissue engineering field, there remains an urgent need for a versatile and efficient approach that simultaneously offers speed, resolution, flexibility and scalability to build complex tissues integrated with functional vascular network [1–3,44]. We have presented a new platform for engineering vascularized tissues with naturally derived biomaterials based on our rapid 3D printing platform – μCOB. This computer-aided photopolymerization-based 3D bioprinting system offers superior speed, resolution, flexibility and scalability over the conventional bioprinters. Its digital nature also provides the flexibility to easily investigate different designs, which is a key to studying the architectural features of the vasculature network. The prevascularized tissues printed by μCOB demonstrated high cell viability and successful endothelial network formation both in vitro and in vivo. Anastomosis between the grafted prevascularized tissues and the host vasculature was observed indicating the formation of functional vasculature in engineered tissues. This platform can be further extended to engineer other tissues that feature complex microarchitectures, such as liver, heart and nerve tissues. By incorporating the prevascularization technique with other primary or stem cells we can potentially engineer functional large-scale tissues for drug testing or even organs for transplantations. With the high resolution and rapid printing speed, it’s relatively easy to scale up to print large hollow vessels for blood flow. Future work can be done to print large vessels and micro-vasculature network together for large tissue constructs. Also, the large scale tissue can be integrated into fluidic devices/bioreactors to simulate blood flow and promote diffusion. Although we used two types of hydrogels (GM-HA and GelMa), a variety of other biomaterials can also be modified and incorporated into the bioprinted tissue constructs to promote tissue maturation and functions, including growth factors, nanoparticles and other biomolecules. With its versatility and biocompatibility, the presented engineering strategy of building vascularized 3D tissues can be broadly applied to promote the development and translation of tissue engineering and regenerative medicine.
Supplementary Material
Scale bars: 250μm(upper), 50μm(lower).
The base (island) and top layers were printed with 5% GelMa+0.15%LAP mixed with green fluorescent beads. The channel layer was printed with 1%HA+2.5% GelMa+0.15%LAP mixed with red fluorescent beads. The hydrogel constructs were incubated with 100 u/ml hyaluronidase at 37 ºC. After 48-hour incubation, most of the hydrogel in the channel was degraded, leaving minimal red fluorescent beads attached to the edges of the channels. Scale bars: 250μm.
Acknowledgments
The work was supported in part by grants from the California Institute for Regenerative Medicine (RT3-07899), National Institutes of Health (R01EB021857) and National Science Foundation (CMMI-1332681 and CMMI-1644967). The UCSD Neuroscience Microscopy Shared Facility was supported by Grant P30 (NS047101). We thank Dr. Shu Chien of UC San Diego for providing HUVECs as a gift and we thank Shangting You for his assistance.
Footnotes
Author contributions
W.Z., X.Q., M. G., Y.X., K.Z. and S.C. conceived and initiated the project. W.Z., X.Q., J. Z. and X.M. designed and performed the experiments. S.P. and C.L. contributed to the sample processing and characterization. J.L. contributed to the material synthesis. P.W. contributed to the mechanical measurement. W.Z., X.Q., J.Z., X.M. and S.C. wrote the paper. Y.X., K.Z. and S.C. supervised the project.
Competing interests. The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen D-HT, Cohen DM, Toro E, Chen Aa, Galie Pa, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat Mater. 2012;11:768–74. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baranski JD, Chaturvedi RR, Stevens KR, Eyckmans J, Carvalho B, Solorzano RD, Yang MT, Miller JS, Bhatia SN, Chen CS. Geometric control of vascular networks to enhance engineered tissue integration and function. Proc Natl Acad Sci. 2013;110:7586–7591. doi: 10.1073/pnas.1217796110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolesky DB, Truby RL, Gladman aS, Busbee Ta, Homan Ka, Lewis Ja. 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs. Adv Mater. 2014;26:3124–3130. doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 4.Lee KY, Peters MC, Anderson KW, Mooney DJ. Controlled growth factor release from synthetic extracellular matrices. Nature. 2000;408:998–1000. doi: 10.1038/35050141. [DOI] [PubMed] [Google Scholar]
- 5.Fischbach C, Mooney DJ. Polymers for pro- and anti-angiogenic therapy. Biomaterials. 2007;28:2069–2076. doi: 10.1016/j.biomaterials.2006.12.029. [DOI] [PubMed] [Google Scholar]
- 6.Richardson TP, Peters MC, Ennett aB, Mooney DJ. Polymeric system for dual growth factor delivery. Nat Biotechnol. 2001;19:1029–1034. doi: 10.1038/nbt1101-1029. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Aledia AS, Ghajar CM, Griffith CK, Putnam AJ, Hughes CCW, George SC. Prevascularization of a fibrin-based tissue construct accelerates the formation of functional anastomosis with host vasculature. Tissue Eng Part A. 2009;15:1363–1371. doi: 10.1089/ten.tea.2008.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahota PS, Burn JL, Brown NJ, MacNeil S. Approaches to improve angiogenesis in tissue-engineered skin. Wound Repair Regen. 2004;12:635–642. doi: 10.1111/j.1067-1927.2004.12608.x. [DOI] [PubMed] [Google Scholar]
- 9.Levenberg S, Rouwkema J, Macdonald M, Garfein ES, Kohane DS, Darland DC, Marini R, van Blitterswijk Ca, Mulligan RC, D’Amore Pa, Langer R. Engineering vascularized skeletal muscle tissue. Nat Biotechnol. 2005;23:879–884. doi: 10.1038/nbt1109. [DOI] [PubMed] [Google Scholar]
- 10.Caspi O, Lesman A, Basevitch Y, Gepstein A, Arbel G, Huber I, Habib M, Gepstein L, Levenberg S. Tissue engineering of vascularized cardiac muscle from human embryonic stem cells. Circ Res. 2007;100:263–272. doi: 10.1161/01.RES.0000257776.05673.ff. [DOI] [PubMed] [Google Scholar]
- 11.Koike N, Fukumura D, Gralla O, Au P, Schechner JS, Jain RK. Tissue engineering: creation of long-lasting blood vessels. Nature. 2004;428:138–139. doi: 10.1038/428138a. [DOI] [PubMed] [Google Scholar]
- 12.Zhang B, Montgomery M, Chamberlain MD, Ogawa S, Korolj A, Pahnke A, Wells LA, Massé S, Kim J, Reis L, Momen A, Nunes SS, Wheeler AR, Nanthakumar K, Keller G, Sefton MV, Radisic M. Biodegradable scaffold with built-in vasculature for organ-on-a-chip engineering and direct surgical anastomosis. Nat Mater. 2016;1:1–42. doi: 10.1038/nmat4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertassoni LE, Cecconi M, Manoharan V, Nikkhah M, Hjortnaes J, Cristino AL, Barabaschi G, Demarchi D, Dokmeci MR, Yang Y, Khademhosseini A. Hydrogel bioprinted microchannel networks for vascularization of tissue engineering constructs. Lab Chip. 2014;14:2202–11. doi: 10.1039/c4lc00030g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA. Three-dimensional bioprinting of thick vascularized tissues. Proc Natl Acad Sci U S A. 2016;113:3179–84. doi: 10.1073/pnas.1521342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia W, Gungor-Ozkerim PS, Zhang YS, Yue K, Zhu K, Liu W, Pi Q, Byambaa B, Dokmeci MR, Shin SR, Khademhosseini A. Direct 3D bioprinting of perfusable vascular constructs using a blend bioink. Biomaterials. 2016;106:58–68. doi: 10.1016/j.biomaterials.2016.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang YS, Arneri A, Bersini S, Shin SR, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell’Erba V, Bishop C, Shupe T, Demarchi D, Moretti M, Rasponi M, Dokmeci MR, Atala A, Khademhosseini A. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials. 2016;110:45–59. doi: 10.1016/j.biomaterials.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan V, Zorlutuna P, Jeong JH, Kong H, Bashir R. Three-dimensional photopatterning of hydrogels using stereolithography for long-term cell encapsulation. Lab Chip. 2010;10:2062. doi: 10.1039/c004285d. [DOI] [PubMed] [Google Scholar]
- 18.Zhang aP, Qu X, Soman P, Hribar KC, Lee JW, Chen S, He S. Rapid Fabrication of Complex 3D Extracellular Microenvironments by Dynamic Optical Projection Stereolithography. Adv Mater. 2012;24:4266–4270. doi: 10.1002/adma.201202024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tumbleston JR, Shirvanyants D, Ermoshkin N, Janusziewicz R, Johnson AR, Kelly D, Chen K, Pinschmidt R, Rolland JP, Ermoshkin A, Samulski ET, DeSimone JM. Continuous liquid interface production of 3D objects. Science (80- ) 2015;347:1349–1352. doi: 10.1126/science.aaa2397. [DOI] [PubMed] [Google Scholar]
- 20.Tumbleston JR, Shirvanyants D, Ermoshkin N, Janusziewicz R, Johnson AR, Kelly D, Chen K, Pinschmidt R, Rolland JP, Ermoshkin A, Samulski ET, DeSimone JM. Continuous liquid interface production of 3D objects. Science (80- ) 2015;347:1349–1352. doi: 10.1126/science.aaa2397. [DOI] [PubMed] [Google Scholar]
- 21.Suri S, Han LH, Zhang W, Singh A, Chen S, Schmidt CE. Solid freeform fabrication of designer scaffolds of hyaluronic acid for nerve tissue engineering. Biomed Microdevices. 2011;13:983–93. doi: 10.1007/s10544-011-9568-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gou M, Qu X, Zhu W, Xiang M, Yang J, Zhang K, Wei Y, Chen S. Bio-inspired detoxification using 3D-printed hydrogel nanocomposites. Nat Commun. 2014;5:3774. doi: 10.1038/ncomms4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu W, Li J, Leong YJ, Rozen I, Qu X, Dong R, Wu Z, Gao W, Chung PH, Wang J, Chen S. 3D-Printed Artificial Microfish. Adv Mater. 2015;27:4411–4417. doi: 10.1002/adma.201501372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma X, Qu X, Zhu W, Li YS, Yuan S, Zhang H, Liu J, Wang P, Lai CSE, Zanella F, Feng GS, Sheikh F, Chien S, Chen S. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc Natl Acad Sci. 2016;113:2206–2211. doi: 10.1073/pnas.1524510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leach JB, Bivens KA, Patrick CW, Schmidt CE. Photocrosslinked hyaluronic acid hydrogels: Natural, biodegradable tissue engineering scaffolds. Biotechnol Bioeng. 2003;82:578–589. doi: 10.1002/bit.10605. [DOI] [PubMed] [Google Scholar]
- 26.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Cell-laden microengineered gelatin methacrylate hydrogels. Biomaterials. 2010;31:5536–5544. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Photoinitiated polymerization of PEG-diacrylate with lithium phenyl-2,4,6-trimethylbenzoylphosphinate: polymerization rate and cytocompatibility. Biomaterials. 2009;30:6702–7. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baranski JD, Chaturvedi RR, Stevens KR, Eyckmans J, Carvalho B, Solorzano RD, Yang MT, Miller JS, Bhatia SN, Chen CS. Geometric control of vascular networks to enhance engineered tissue integration and function. Proc Natl Acad Sci. 2013;110:7586–7591. doi: 10.1073/pnas.1217796110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burdick JA, Prestwich GD. Hyaluronic acid hydrogels for biomedical applications. Adv Mater. 2011;23:H41–56. doi: 10.1002/adma.201003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burdick Ja, Chung C, Jia X, Randolph Ma, Langer R. Controlled degradation and mechanical behavior of photopolymerized hyaluronic acid networks. Biomacromolecules. 2005;6:386–91. doi: 10.1021/bm049508a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baier Leach J, Bivens Ka, Patrick CW, Schmidt CE. Photocrosslinked hyaluronic acid hydrogels: natural, biodegradable tissue engineering scaffolds. Biotechnol Bioeng. 2003;82:578–89. doi: 10.1002/bit.10605. [DOI] [PubMed] [Google Scholar]
- 33.Hribar KC, Choi YS, Ondeck M, Engler AJ, Chen S. Digital plasmonic patterning for localized tuning of hydrogel stiffness. Adv Funct Mater. 2014;24:4922–4926. doi: 10.1002/adfm.201400274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi YS, Vincent LG, Lee AR, Kretchmer KC, Chirasatitsin S, Dobke MK, Engler AJ. The alignment and fusion assembly of adipose-derived stem cells on mechanically patterned matrices. Biomaterials. 2012;33:6943–6951. doi: 10.1016/j.biomaterials.2012.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vincent LG, Choi YS, Alonso-Latorre B, del Álamo JC, Engler AJ. Mesenchymal stem cell durotaxis depends on substrate stiffness gradient strength. Biotechnol J. 2013;8:472–484. doi: 10.1002/biot.201200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isenberg BC, DiMilla PA, Walker M, Kim S, Wong JY. Vascular smooth muscle cell durotaxis depends on substrate stiffness gradient strength. Biophys J. 2009;97:1313–1322. doi: 10.1016/j.bpj.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy SV, Atala A. 3D bioprinting of tissues and organs. Nat Biotechnol. 2014;32:773–785. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 38.Chang R, Nam J, Sun W. Effects of dispensing pressure and nozzle diameter on cell survival from solid freeform fabrication-based direct cell writing. Tissue Eng Part A. 2008;14:41–48. doi: 10.1089/ten.a.2007.0004. [DOI] [PubMed] [Google Scholar]
- 39.Smith CM, Christian JJ, Warren WL, Williams SK. Characterizing environmental factors that impact the viability of tissue-engineered constructs fabricated by a direct-write bioassembly tool. Tissue Eng. 2007;13:373–383. doi: 10.1089/ten.2007.13.ft-338. [DOI] [PubMed] [Google Scholar]
- 40.Aguado BA, Mulyasasmita W, Su J, Lampe KJ, Heilshorn SC. Improving Viability of Stem Cells During Syringe Needle Flow Through the Design of Hydrogel Cell Carriers. Tissue Eng Part A. 2012;18:806–815. doi: 10.1089/ten.tea.2011.0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Debbage PL, Sölder E, Seidl S, Hutzler P, Hugl B, Öfner D, Kreczy A. Intravital lectin perfusion analysis of vascular permeability in human micro- and macro- blood vessels. Histochem Cell Biol. 2001;116:349–359. doi: 10.1007/s004180100328. [DOI] [PubMed] [Google Scholar]
- 42.Debbage PL, Griebel J, Ried M, Gneiting T, DeVries A, Hutzler P. Lectin Intravital Perfusion Studies in Tumor-bearing Mice: Micrometer-resolution, Wide-area Mapping of Microvascular Labeling, Distinguishing Efficiently and Inefficiently Perfused Microregions in the Tumor. J Histochem Cytochem. 1998;46:627–639. doi: 10.1177/002215549804600508. [DOI] [PubMed] [Google Scholar]
- 43.Rouwkema J, Rivron NC, van Blitterswijk CA. Vascularization in tissue engineering. Trends Biotechnol. 2008;26:434–441. doi: 10.1016/j.tibtech.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 44.Novosel EC, Kleinhans C, Kluger PJ. Vascularization is the key challenge in tissue engineering. Adv Drug Deliv Rev. 2011;63:300–311. doi: 10.1016/j.addr.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 45.Caspi O, Lesman A, Basevitch Y, Gepstein A, Arbel G, Huber I, Habib M, Gepstein L, Levenberg S. Tissue engineering of vascularized cardiac muscle from human embryonic stem cells. Circ Res. 2007;100:263–272. doi: 10.1161/01.RES.0000257776.05673.ff. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scale bars: 250μm(upper), 50μm(lower).
The base (island) and top layers were printed with 5% GelMa+0.15%LAP mixed with green fluorescent beads. The channel layer was printed with 1%HA+2.5% GelMa+0.15%LAP mixed with red fluorescent beads. The hydrogel constructs were incubated with 100 u/ml hyaluronidase at 37 ºC. After 48-hour incubation, most of the hydrogel in the channel was degraded, leaving minimal red fluorescent beads attached to the edges of the channels. Scale bars: 250μm.