Abstract
Integration of viral DNA into the host genome is an essential step in retroviral replication. The viral DNA made by reverse transcription is a component of the preintegration complex (PIC) that also contains the viral integrase protein, the enzyme that integrates the viral DNA. Several other viral and cellular proteins are present in the PIC, but their functional roles are less well established. Barrier-to-autointegration factor (BAF) is a cellular protein component of the PIC that blocks autointegration of the viral DNA and stimulates intermolecular integration. In uninfected cells, BAF interacts with members of the LEM family of inner nuclear membrane and nucleoplasmic proteins. Here, we demonstrate that one of the LEM proteins, lamina-associated polypeptide 2α (LAP2α), is a component of the PIC. LAP2α stabilizes the association of BAF with the PIC to stimulate intermolecular integration and suppress autointegration. To further understand the role of LAP2α, we established LAP2α-knockdown cell lines. Depletion of LAP2α significantly inhibited viral replication. Our results demonstrate a critical contribution of LAP2α to the nucleoprotein organization of the PIC and to viral replication.
Keywords: barrier-to-autointegration factor, integration, lamina-associated polypeptide 2α, moloney murine leukemia virus, preintegration complex
Introduction
DNA integration, an essential step for retroviruses to establish infection, is mediated by a large nucleoprotein complex derived from the core of the infecting virion (Goff, 1992; Brown, 1997; Asante-Appiah and Skalka, 1999; Craigie, 2001). This preintegration complex (PIC) is comprised of a copy of viral DNA, which is synthesized by reverse transcription of the viral RNA genome, and a number of viral and cellular proteins. For Moloney murine leukemia virus (MoMLV), capsid protein, reverse transcriptase and integrase have been reported as viral protein components of the PIC (Bowerman et al, 1989; Fassati and Goff, 1999).
PICs isolated from virus-infected cells efficiently integrate their viral DNA into an exogenously added target DNA in vitro (intermolecular integration); on the other hand, intramolecular integration into the viral DNA itself (autointegration) is avoided (Brown et al, 1987, 1989; Fujiwara and Mizuuchi, 1988; Bowerman et al, 1989; Lee and Craigie, 1994). This strong preference for intermolecular integration is a hallmark of the PIC. Autointegration would lead to destruction of viral DNA before it is able to migrate to the nucleus and integrate into chromosomal DNA. Barrier-to-autointegration factor (BAF) is a cellular component of MoMLV and human immunodeficiency virus type 1 (HIV-1) PICs, which blocks autointegration and stimulates intermolecular integration activity in vitro (Chen and Engelman, 1998; Lee and Craigie, 1998; Suzuki and Craigie, 2002; Lin and Engelman, 2003). This 89 amino-acid protein is highly conserved among multicellular eukaryotes with 60% sequence identity between human and Caenorhabditis elegans homologs (Cai et al, 1998). BAF exists as a dimer in solution (Cai et al, 1998) and binds double-stranded DNA, but not single-stranded DNA or RNA, with no detectable sequence specificity (Zheng et al, 2000). BAF bridges DNA molecules in a large nucleoprotein complex (Zheng et al, 2000), and we have proposed that the resulting compaction of the viral DNA within the PIC makes it inaccessible as a target for autointegration (Lee and Craigie, 1998; Suzuki and Craigie, 2002).
The function of BAF for the host cell is not well understood, but several lines of evidence point to a role in chromatin organization. BAF has profound effects on chromatin decondensation and nuclear growth in Xenopus extracts in vitro (Segura-Totten et al, 2002). It is associated with chromatin in a cell-cycle-dependent manner (Furukawa, 1999; Haraguchi et al, 2001; Furukawa et al, 2003) and the RNA interference (RNAi) phenotype in C. elegans revealed that knockdown of BAF caused a defect in chromatin segregation during mitosis (Zheng et al, 2000). In multicellular eukaryotes, BAF interacts with members of the LEM protein family (Lin et al, 2000), which includes Lamina-associated polypeptide 2 (LAP2), emerin and MAN1 proteins (Furukawa, 1999; Lin et al, 2000; Foisner, 2001; Lee et al, 2001; Shumaker et al, 2001). LEM proteins are components of the nuclear lamina structure at the nuclear periphery and of the lamin complexes in the nuclear interior (Foisner, 2001). This protein family is defined by a highly conserved motif of approximately 40 amino-acid residues near the N-terminus, the LEM domain (Lin et al, 2000), that directly interacts with BAF (Furukawa, 1999; Cai et al, 2001; Lee et al, 2001; Shumaker et al, 2001). As BAF binds to both DNA and LEM domain proteins, it has been proposed that BAF plays a key role in chromatin organization and may be involved in anchoring chromosomal DNA to the inner nuclear membrane (Furukawa, 1999; Dechat et al, 2000b; Shumaker et al, 2001; Furukawa et al, 2003; Segura-Totten and Wilson, 2004).
In murine cells, LAP2 and emerin have been identified as LEM proteins (Berger et al, 1996; Small et al, 1997). LAP2 is a well-characterized LEM protein. To date, six isoforms of the LAP2 have been identified in mammals; they are generated from one gene by alternative splicing (Harris et al, 1994; Furukawa et al, 1995; Berger et al, 1996). All of the isoforms share a conserved 186 amino-acid N-terminal domain (LAP2 common domain) and this common domain contains the LEM domain (Berger et al, 1996). Lamina-associated polypeptide 2α (LAP2α) is the largest isoform and, unlike most isoforms, it lacks a transmembrane region (Berger et al, 1996). Consequently, it is not membrane-bound and is localized predominantly throughout the nuclear interior, while a lesser amount has been shown to be present in the cytoplasm at stages of mitosis (Dechat et al, 1998; Vlcek et al, 1999).
We have previously demonstrated that BAF is a component of the MoMLV PICs that enhances intermolecular integration and blocks autointegration (Lee and Craigie, 1998; Suzuki and Craigie, 2002). The interaction of BAF with the LEM protein family gives rise to the new questions: (i) are any LEM proteins also associated with PICs? and (ii) if so, what is the role of the LEM proteins in the retroviral PIC and for infection? Here, we show that one of the LEM proteins, LAP2α, is a component of MoMLV PIC and that LAP2α stimulates the intermolecular integration activity of the PICs in collaboration with BAF. By analyzing the salt stability of a DNA complex with BAF and LAP2α, and PICs from LAP2α-knockdown cells, we find that the association of BAF with DNA is stabilized by LAP2α. Furthermore, MoMLV replication is significantly inhibited in the LAP2α-knockdown cells. Thus, our data suggest that LAP2α plays an important role in the nucleoprotein organization of the PIC and in virus replication.
Results
LAP2α associates with MoMLV PICs
To determine if any LEM proteins are associated with MoMLV PICs, murine LAP2α(FLAG-LAP2α), LAP2β(FLAG-LAP2β), LAP2 common domain (FLAG-LAP2c) and emerin (FLAG-emerin) proteins fused to the FLAG epitope (Figure 1A) were transiently expressed in NIH3T3 cells. Expression of the LEM proteins was confirmed by western blotting (Figure 1B). The transfected cells were cocultivated with the MoMLV-producing cell line clone 4 and cytoplasmic extract containing PICs was isolated. Anti-FLAG antibody immunoprecipitated the PICs derived from the FLAG-LAP2α-expressing cells, but not PICs from the FLAG-LAP2β- or FLAG-emerin-expressing cells (Figure 1B, lanes 1, 2 and 4 of the lower panel). Also, PICs derived from the FLAG-LAP2c-expressing cells were not recovered by immunoprecipitation with the anti-FLAG antibody (lane 3). These data suggest that only one of the LEM proteins, LAP2α, is associated with PICs and the N-terminal LAP2 common domain alone is insufficient for this association.
Figure 1.
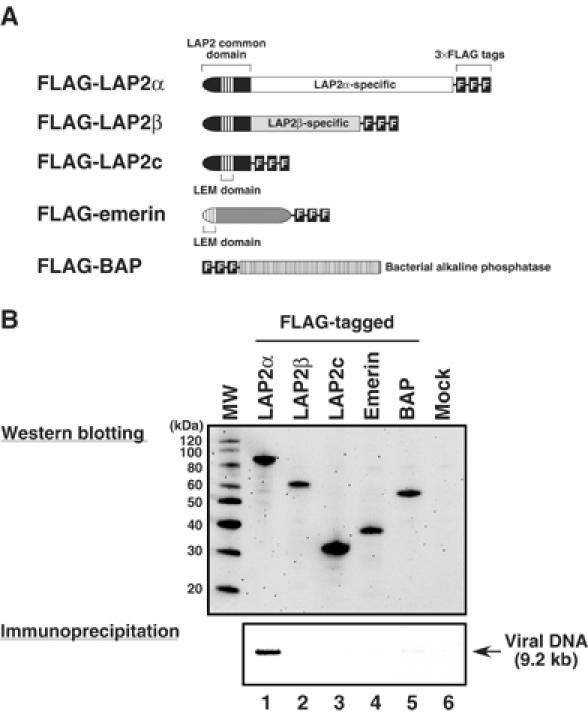
Immunoprecipitation of MoMLV PICs from FLAG-tagged LEM protein-expressing cells. (A) FLAG-tagged LEM protein constructs. (B) Expression of the FLAG-tagged LEM proteins in NIH3T3 cells and immunoprecipitation of PICs from these cells. NIH3T3 cells were transfected with the expression vectors encoding FLAG-tagged proteins and, at 24 h after transfection, the cells were cocultured with MoMLV-producing cells. Cytoplasmic extracts from these cells were immunoprecipitated with an anti-FLAG monoclonal antibody. The weak band in lane 5 of panel B is background that is not reproducibly observed. Viral DNA was extracted from the captured immunocomplex and detected by Southern blotting (lower panel). Protein expression in the cocultured cells was analyzed by western blotting using anti-FLAG monoclonal antibody (upper panel). MW: molecular weight marker. Control experiments demonstrated that the anti-FLAG antibody immunoprecipitates each of the LAP2 isoforms with similar efficiency (Supplementary Figure S1).
To confirm the association of LAP2α with the PICs, anti-LAP2α polyclonal IgG was purified from a rabbit serum raised against hexahistidine (His)-tagged LAP2α-specific domain (His-LAP2α/ΔN186) and immunoprecipitation analysis with this antibody was carried out on PICs from virus-infected NIH3T3 cells. PICs were efficiently recovered by immunoprecipitation with the anti-LAP2α antibody, whereas immunoprecipitation with control rabbit IgG yielded very little recovery, demonstrating that endogenous LAP2α is associated with PICs (Figure 2, upper panels); Western blotting controls with whole-cell lysates from NIH3T3 cells revealed that the anti-LAP2α rabbit IgG specifically reacted with endogenous LAP2α, but not with the other isoforms of LAP2 protein (Supplementary Figure S2). Interestingly, whereas stripping BAF from the PICs by high-salt treatment impaired the recovery of PICs by immunoprecipitation with anti-BAF serum as expected (Suzuki and Craigie, 2002), salt-stripped PICs were efficiently immunoprecipitated with anti-LAP2α antibody (Figure 2, lower panels), indicating that LAP2α remained associated with PICs after salt-stripping. We conclude that LAP2α is a stable component of the MoMLV PIC and BAF is not essential for this association.
Figure 2.
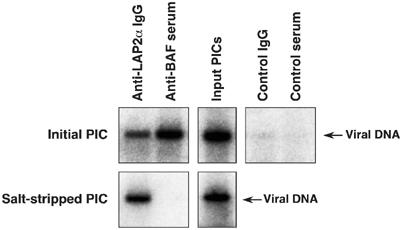
Association of endogenous LAP2α with MoMLV PICs. Initial PICs (upper panels) and salt-stripped PICs (lower panels), made by coculture of NIH3T3 cells with MoMLV producer cells, were immunoprecipitated with the indicated antibodies and viral DNA was detected by Southern blotting.
As a final test of the association of LAP2α with the PIC, we incubated glutathione S transferase (GST) fusion recombinant LAP2α (GST-LAP2α), LAP2 common domain (GST-LAP2c) and LAP2α-specific domain (GST-LAP2α/ΔN186) proteins (Figure 3A) with PICs and assayed for capture by glutathione beads. Incubation of PICs with GST-LAP2α, but not with the truncated proteins or beads alone, resulted in recovery of PICs in this assay (Figure 3B). These data demonstrate that LAP2α is able to associate with PICs in vitro, and suggests that both the LAP2 common and LAP2α-specific domains are necessary for this association.
Figure 3.
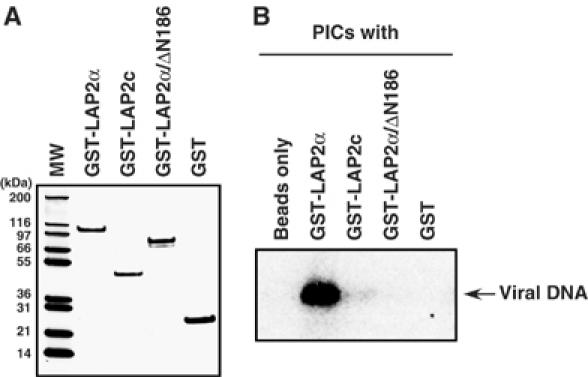
In vitro association of LAP2α with MoMLV PICs. (A) PAGE of the purified GST-LAP2α fusion proteins. (B) GST pulldown assay of PICs. Cytoplasmic extract containg PICs was incubated with the indicated GST-LAP2α fusion proteins and precipitated by glutathione beads. The viral DNA in the bound fraction was detected by Southern blotting.
LAP2α stimulates integration activity of PICs in vitro
To test if LAP2α stimulates intermolecular integration, or blocks autointegration, we performed in vitro PIC integration assays in the presence of recombinant LAP2α (Figure 4). Like BAF, His-LAP2α stimulated the intermolecular integration activity of initial PICs (lanes 1–5); similar stimulation was also observed with GST-LAP2α (data not shown). However, unlike the effect of BAF, stimulation of intermolecular integration by LAP2α was not accompanied by a decrease in autointegration (compare lanes 4 and 5).
Figure 4.
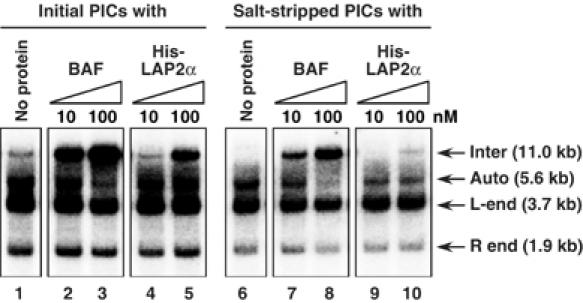
Stimulation of intermolecular integration of MoMLV PICs by LAP2α. Initial PICs (lanes 1–5) and salt-stripped PICs (lanes 6–10) were incubated with BAF or His-LAP2α and then added to the integration reaction mixture containing Φ × 174 RFI target DNA. After incubation, DNA products from the reaction were digested with BamHI and detected by Southern blotting. The 11.0-kb band results from intermolecular integration of the viral DNA into Φ × 174 RFI DNA (inter). The 5.6-kb band and the smear below it result from autointegration of the viral DNA into itself (auto). The 3.7- and 1.9-kb bands are the unreacted viral DNA containing 5′ LTR (L-end) or 3′ LTR (R-end), respectively (Lee and Craigie, 1994).
We next examined whether LAP2α can restore the intermolecular integration activity of salt-stripped PICs. As expected, BAF efficiently restored the intermolecular integration preference and reduced the autointegration activity of salt-stripped PICs (Figure 4, lanes 6–8). In contrast, addition of His-LAP2α (or GST-LAP2α, data not shown) resulted in only minimal stimulation of intermolecular integration of salt-stripped PICs and did not suppress autointegration (Figure 4, lanes 9 and 10). Since immunoprecipitation of salt-stripped PICs shows that most of the BAF is dissociated by salt-stripping (Figure 2) (Suzuki and Craigie, 2002), this result suggests that the stimulation of intermolecular integration by LAP2α requires BAF.
LAP2α stabilizes association of BAF with DNA
A high salt concentration (greater than 500 mM KCl) is required to disrupt the protection of the PIC against autointegration (Lee and Craigie, 1994). At 400 mM KCl, PICs retain the ability to integrate intermolecularly, indicating that BAF remains associated with the PIC at a surprising high salt concentration (Figure 5A). We therefore tested whether the DNA-binding properties of BAF alone are sufficient to explain this behavior. We formed complexes of BAF with linearized Φ × 174 DNA in presence of 115 mM salt and then challenged these complexes with 150 or 400 mM salt concentration. The complexes were then sedimented in sucrose gradients containing 150 or 400 mM KCl, respectively, and the position of DNA in the gradient was monitored. Figure 5B shows that DNA alone remained in the top fractions of the gradient (panel 1), while the BAF/DNA complex sedimented faster in presence of 150 mM KCl (panel 2). However, in the presence of 400 mM KCl, the sedimentation rate was indistinguishable from that of free DNA, indicating that BAF was dissociated (Figure 5B, panel 3). The simple BAF/DNA complex is therefore unstable to high-salt challenge (Figure 5B, panel 3). We also tested if circular DNA could stabilize the BAF/DNA complex against the high-salt challenge, since both ends of viral DNA must be topologically closed by integrase within the PIC. However, the BAF/DNA complex with circular Φ × 174 DNA was also unstable to 400 mM KCl (Figure 5B, panels 4–6). These data indicate that, in contrast to the stable association of BAF with PIC, the complex of BAF with DNA alone is not stable to high salt and that the DNA-binding activity of BAF alone is insufficient to account for its incorporation into the PIC.
Figure 5.
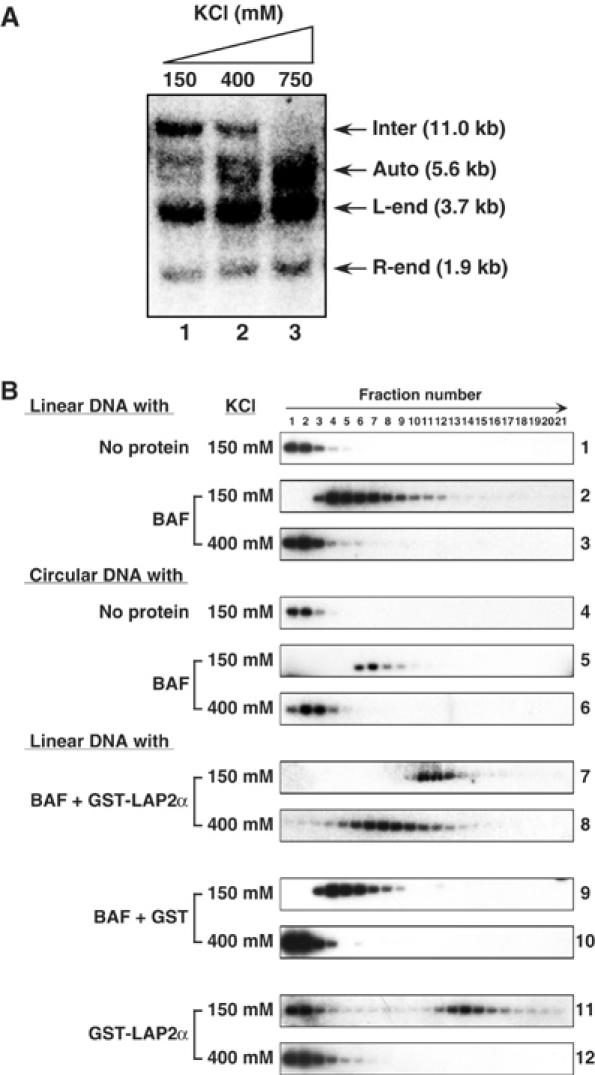
Stabilization of BAF/DNA complex by LAP2α in vitro. (A) Retention of intermolecular integration activity of MoMLV PICs after treatment with 400 mM KCl. The PIC fractions were incubated in the presence of 150 mM (lane 1), 400 mM (lane 2), or 750 mM (lane 3) KCl and, after gel filtration, samples were added to integration reaction mixtures containing Φ × 174 RFI DNA. Products were digested with BamHI and detected by Southern blotting. (B) Velocity sedimentation of BAF/DNA complexes with or without LAP2α. BAF and LAP2α were incubated with Φ × 174 DNA in buffer containing 115 mM KCl as indicated and then challenged with 150 or 400 mM KCl. After centrifugation in a sucrose gradient containing the same concentration of KCl, gradients were fractionated and the DNA was detected by Southern blotting.
To determine if LAP2α enhances the salt resistance of the association of BAF with DNA, we performed the sedimentation assay in the presence of GST-LAP2α. The BAF/LAP2α/DNA complex sedimented to near the middle of the gradient in the presence of 150 mM KCl (Figure 5B, panel 7). Surprisingly, even after treatment with 400 mM KCl, the complex did not dissociate and still sedimented much faster than free DNA (Figure 5B, panel 8). In control experiments, GST did not stabilize the BAF/DNA complex (Figure 5B, panels 9 and 10). Interestingly, GST-LAP2α alone was able to form a complex with DNA (Figure 5B, panel 11), although the affinity was lower than for BAF, as evidenced by free DNA at the top of the gradient. However, this GST-LAP2α/DNA complex was also unstable at 400 mM KCl (Figure 5B, panel 12). These results demonstrate that BAF and LAP2α together form a complex with DNA that is resistant to high salt.
Both the common and α-specific domains of LAP2α are necessary for stabilization of the BAF/DNA complex
To determine the functional domains of LAP2α that confer the stable association of BAF with DNA, we generated a series of deletion mutants lacking a part of the N- or C-terminal domain of LAP2α as fusion proteins with GST and tested their ability to form a salt-resistant DNA complex with BAF and DNA (Figure 6). Both the LAP2 common domain, which contains LEM domain that interacts with BAF, and the LAP2α-specific domain alone failed to stabilize the BAF/DNA complex in the presence of 400 mM KCl (Figure 6, panels 4 and 6). These data demonstrate that the common domain or α-specific domain alone is insufficient to form a stable complex with DNA and BAF. In the presence of 150 mM KCl, the LAP2α deletion mutant missing the C-terminal half of the LAP2α-specific domain (GST-LAP2α/ΔC253) formed a complex with BAF/DNA that sedimented slightly faster than the complex of BAF alone with DNA (compare panel 2 of Figure 5 and panel 7 of Figure 6), but failed to form a salt-stable complex. In contrast, another deletion mutant (GST-LAP2α/ΔN112), that contains the LEM domain and α-specific domain, but lacks the N-terminal 112 amino acids of common domain, did not alter the sedimentation behavior of the BAF/DNA complex (Figure 6, panel 9). These results suggest that, not only the LEM domain but also the N-terminal part of the common domain contributes to functional interactions with BAF and/or DNA. However, none of these deletion mutants were able to stabilize the BAF/DNA complex in the presence of 400 mM KCl (Figure 6, panels 8 and 10), demonstrating that the N-terminal part of the common domain and the C-terminal part of α-specific domain are necessary for the stabilization. We finally tested whether the independent proteins of the LAP2 common domain and LAP2α-specific domain could together reconstitute the function of LAP2α in stabilization of the BAF/DNA complex. Surprisingly, when the GST-LAP2c and GST-LAP2α/ΔN186 were mixed together and incubated with BAF and DNA, the majority of the resulting complex was stable in the presence of 400 mM KCl (Figure 6, panels 11 and 12). We conclude that both the common domain and α-specific domains of LAP2α are required to form a stable complex with BAF and DNA.
Figure 6.
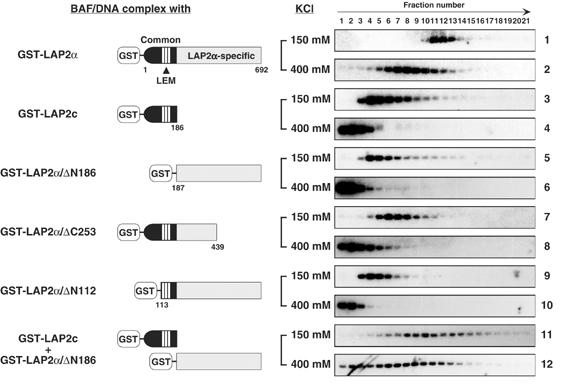
Stabilization of the BAF/DNA complex by LAP2α requires both the LAP2 common and LAP2α-specific domains. Complexes of DNA, BAF and deletion derivatives of LAP2α were analyzed as in Figure 5B.
Salt stability of the PIC is diminished by in vivo depletion of LAP2α
Velocity sedimentation of the complex of DNA with BAF and GST-LAP2α revealed that association of BAF with DNA is reinforced by LAP2α in vitro (Figure 5). Since the association of BAF with the PIC regulates the preference for intermolecular integration of the viral DNA (Suzuki and Craigie, 2002), depletion of LAP2α would be expected to render the intermolecular integration preference more sensitive to salt challenge. Immunoprecipitation analysis of salt-stripped PICs using anti-LAP2α serum reveals that LAP2α is not efficiently removed from the PIC by high-salt treatment (Figure 2). We therefore used the small interference RNA (siRNA) gene-silencing technique (Brummelkamp et al, 2002) to make a stable NIH3T3 cell line with substantially reduced levels of LAP2α from which PICs were isolated (Figure 7). Growth of this cell line was indistinguishable from that of NIH3T3, although other cell lines with greater reductions in LAP2α exhibited impaired growth (data not shown). We initially asked whether LAP2α was depleted from PICs made in the LAP2α-knockdown cells. Immunoprecipitation with anti-LAP2α antibody shows that recovery of PICs derived from LAP2α-knockdown cells was significantly decreased compared with PICs from NIH3T3 and siRNA control cells (Figure 7B). These data demonstrate that reduced levels of LAP2α are associated with PICs from LAP2α-knockdown cells.
Figure 7.

Depletion of LAP2α in vitro renders MoMLV PICs more sensitive to high salt. (A) Establishment of LAP2α-knockdown cells. NIH3T3 cells were transfected with a U6 promoter-driven siRNA expression vector encoding hairpin siRNA against LAP2α or noninteracting siRNA (siRNA control), and stable cell lines were selected. Cell lysate from each cell line was subjected to western blotting analysis using anti-LAP2 common domain (upper panel) or anti-lamins A/C (lower panel) monoclonal antibodies. MW: molecular weight markers. (B) Reduced levels of LAP2α are associated with MoMLV PICs from the LAP2α-knockdown cell line. The PIC fractions from each cell line were immunoprecipitated (IP, upper panel) with anti-LAP2α antibody and the recovered PICs were detected by Southern blotting. The lower panel shows the input PIC fractions without immunoprecipitation. (C) Diminished salt stability of PICs from the LAP2α-knockdown cells. The PIC fractions from each cell line were treated with the indicated concentration of KCl and, after gel filtration, assayed for integration activity. Although there was some quantitative variation between experiments, with residual intermolecular integration activity sometimes being observed after treatment of PICs from NIH3T3 cells with 750 mM KCl, PICs that derived the LAP2α-knockdown cells were consistently diminished in stability to 400 mM KCl.
We then examined whether the intermolecular integration activity of PICs from the LAP2α-knockdown is more sensitive to salt challenge. PICs isolated from NIH3T3 cells, the knockdown cell line and control cell were treated with 400 or 750 mM KCl and assayed for integration activity. As predicted, although initial PICs from LAP2α-knockdown cells were able to carry out intermolecular integration, this activity was mostly abolished after treatment with 400 mM KCl (Figure 7, lanes 4–6). In contrast, PICs from NIH3T3 and siRNA control cells still retained intermolecular integration activity after treatment with 400 mM KCl (Figure 7C, lanes 1–3 and 7–9).
Replication of MoMLV is inhibited in LAP2α-knockdown cells
To evaluate the biological importance of LAP2α in MoMLV replication, we tested whether the depletion of LAP2α had an effect on virus replication. NIH3T3, LAP2α-knockdown and siRNA control cells were infected with MoMLV for 2 h and, after washing, aliquots of culture supernatants were collected at 3, 6 and 9 days post infection and assayed for exogenous RT to monitor viral spread in the infected cultures. As shown in Figure 8, virus replication was drastically inhibited in LAP2α-knockdown cells, but not in cultures of NIH3T3 and siRNA control cells. The observed restriction of virus replication was the greatest when cells were infected with virus at low m.o.i. This demonstrates that LAP2α also contributes to efficient replication of MoMLV.
Figure 8.
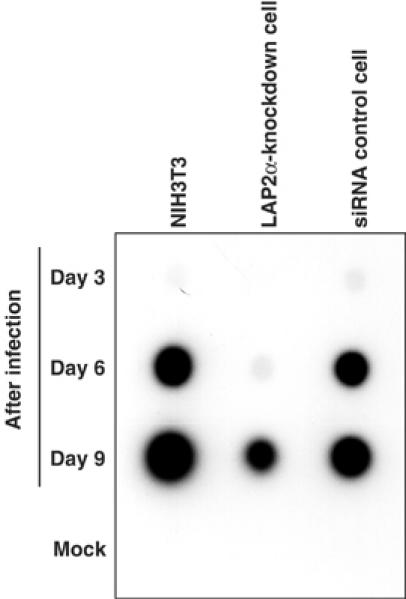
Inhibition of MoMLV replication in LAP2α-knockdown cells. Each cell line was infected with MoMLV at low multiplicity for 2 h in the presence of polybrene and, after twice washing, cells were cultured in fresh culture medium. Culture supernatants were collected 3, 6, and 9 days after infection and virus spreading was monitored by RT activity assay.
Discussion
Chromosomal DNA, the normal target DNA for retroviral integration in infected cells, is enclosed by the nuclear membrane that forms the boundary between the nucleus and cytoplasm. Chromatin is attached to the inner nuclear membrane through a lamina structure, composed of lamin proteins and lamina-associated proteins, that is thought to be important for nuclear organization and function (Holaska et al, 2002). LEM proteins are the largest family of lamina-associated proteins. In the present study, we have reported that one of the LEM proteins, LAP2α, is a stable component of MoMLV PIC. Furthermore, we have demonstrated a functional role for LAP2α in the nucleoprotein organization of the PIC and for retroviral replication.
Why is only LAP2α, and not other LEM proteins, associated with the PIC? LAP2α is unique among LEM proteins in that it lacks a transmembrane domain and consequently is distributed throughout the nuclear interior rather than at the nuclear membrane (Dechat et al, 1998). Although LAP2α is predominantly found within the nucleus, some LAP2α also localizes in the cytoplasm, at least at the early stages of mitosis (Dechat et al, 1998; Vlcek et al, 1999). Indeed, when we checked the subcellular distribution of LAP2 isoforms by Western blotting of the pellet and supernatant fractions of NIH3T3 cells extracted with digitonin, a significant fraction of LAP2α was observed in the cytoplasmic extract (data not shown). Therefore, it is likely that the presence of LAP2α in cytoplasm enables its association to PIC, whereas the other isoforms sequestered by the nuclear envelope. However, the unique C-terminal domain of LAP2α plays a crucial role in its recruitment since the FLAG-tagged common domain of LAP2α, which binds BAF, did not associate with PICs.
How does LAP2α promote the intermolecular integration activity of the PIC in vitro? Although His-LAP2α stimulated intermolecular integration activity of initial PICs, when salt-stripped PICs were incubated with the His-LAP2α, only minimal stimulation of intermolecular integration was observed (Figure 4). As immunoprecipitation with anti-BAF serum shows that most of the BAF is removed from the PIC by salt-stripping (Figure 2) (Suzuki and Craigie, 2002), these data indicate that LAP2α requires BAF for the stimulation of integration activity. However, unlike stimulation of intermolecular integration by BAF, stimulation by LAP2α does not appear to be accompanied by a decrease in autointegration.
An important clue as to the role of LAP2α for the PIC is the stabilization of the BAF/DNA complex by LAP2α. The difference in stability between the simple BAF/DNA complex and the association of BAF with the PIC suggests that the DNA-binding property of BAF alone is not sufficient for efficient incorporation into the PIC. Our results show that the salt resistance of the BAF/DNA complex was increased by the presence of LAP2α, implying that LAP2α may contribute to the efficient acquisition of BAF by the PIC. Shumaker et al (2001) have demonstrated that the LAP2 common domain prefers to bind BAF in a nucleoprotein complex with DNA rather than BAF alone. The structures of the BAF dimer, and the LEM and ‘LEM-like' domains of the LAP2 common domain, have been reported (Cai et al, 1998, 2001; Umland et al, 2000). While the LEM domain directly interacts with BAF (Cai et al, 2001) and BAF binds DNA (Zheng et al, 2000), the interactions involving the other domains and DNA are unclear. The detailed structural organization of the complex of BAF, LAP2α and DNA remains to be elucidated.
An important unanswered question remains as to how LAP2α is retained in the PIC after salt-stripping? Most of the BAF is dissociated from the PIC by salt-stripping (Suzuki and Craigie, 2002). However, in the absence of BAF, the association of LAP2α with DNA is unstable at 400 mM KCl. Therefore, unless a residual undetectable level of BAF in the salt-stripped PIC is sufficient to promote the association of LAP2α, we speculate that another cellular and/or viral factor(s) may be involved in the stable association of LAP2α with PIC. Preliminary immunoprecipitation experiments indicate that lamins A/C, which bind LAP2α (Dechat et al, 2000a), appear not to be associated with MoMLV PICs (data not shown). The possible interactions between BAF and LAP2α and other viral proteins present in the PIC need to be investigated. Indeed, there is relatively little biochemical data on the interactions among the protein components of the PIC. Experiments are in progress to address this issue.
Finally, our data show that MoMLV replication is significantly inhibited in LAP2α-knockdown cells, implicating a critical contribution of LAP2α in virus infection. The step at which replication is inhibited in LAP2α-knockdown cells remains to be determined; furthermore, the possibility that knockdown of LAP2α indirectly affects viral replication should not be ignored. Unlike lentiviruses such as HIV-1, MoMLV can infect only dividing cells (Roe et al, 1993; Lewis and Emerman, 1994; Hatziioannou and Goff, 2001), because MoMLV PICs lack the appropriate nuclear localization signals to enter the intact nucleus of nondividing cells (Fouchier and Malim, 1999). Thus, it is commonly believed that MoMLV requires mitosis, during which the nuclear envelope is disassembled, for the PIC to access the chromosomal DNA. However, the molecular mechanisms for import of the PIC into the nucleus in dividing cells, and in nondividing cells, remain to be unambiguously established. Even if disassembly of the nuclear envelope during mitosis is a prerequisite for nuclear import of MoMLV PIC, why are PICs localized to the nuclear compartment when the envelope reforms? In the course of mitosis, the localization of LAP2α in the cytoplasm and nucleus is dynamically changed (Dechat et al, 1998). In interphase and early stages of mitosis, LAP2α is predominantly found in the nucleus. When disassembly of the nuclear envelope begins at metaphase and anaphase, LAP2α is observed throughout the cell, without an apparent association with the chromosomal DNA. However, in the initial stage of nuclear reassembly at early telophase, LAP2α redistributes to the nuclear interior (Dechat et al, 1998). Similarly, BAF has been shown to be diffusely nonlocalized at metaphase and anaphase, but specifically localized at chromatin core regions at telophase (Haraguchi et al, 2001). Thus, this re-localization of BAF and LAP2α during the cell cycle may play a role in directing the PIC to the chromosomal DNA. In addition, these inner nuclear proteins may also contribute to the retention of the PIC within the nucleus in addition to blocking autointegration and stimulating intermolecular integration. Therefore, it will be interesting to determine whether the nuclear entry step of MoMLV replication is blocked in the LAP2α-knockdown cell lines. Determining the step(s) in which LAP2α is involved during the virus replication cycle will be of help in understanding how retroviruses locate and direct integration to chromosomal DNA.
Materials and methods
Preparation of MoMLV PICs
NIH3T3 cells and the MoMLV-producing cell line clone 4 were grown in Dulbecco's modified Eagle medium containing high glucose, sodium pyruvate, 100 U/ml penicillin, 100 μg/ml streptomycin and 10% fetal calf serum (GibcoBRL). Standard preparation of MoMLV PICs was carried out as described previously (Fujiwara and Mizuuchi, 1988; Lee and Craigie, 1994). Cytoplasmic extract containing PICs was isolated with buffer A (20 mM Hepes-NaOH, pH 7.5, 5 mM MgCl2, 150 mM KCl, 10 mM DTT) containing 20 μg/ml aprotinin (Sigma) in the presence of 0.025% digitonin (Sigma) and stored at −80°C in buffer B (20 mM Hepes-NaOH, pH 7.5, 5 mM MgCl2, 150 mM KCl, 10 mM DTT, 6 mM EDTA, 6% sucrose) until use (fraction I). In most experiments, fraction I was first subjected to gel filtration through a spin column of Sephacryl S-1000 Superfine (Amersham-pharmacia) equilibrated with buffer B containing 0.1% BSA (fraction II).
High-salt treatment of PICs
For salt-stripping the PICs, fraction I was mixed with KCl to a final concentration of 750 mM and incubated on ice for 1 h. Salt-stripped PICs were separated from free components by gel filtration on a spin column of Sephacryl S-1000 equilibrated with buffer B containing 750 mM KCl and 0.1% BSA, and subjected to a second gel filtration on a spin column equilibrated with buffer B containing 0.1% BSA (Lee and Craigie, 1998). In some experiments, to test the salt stability of PICs, fraction I was also mixed with KCl to a final concentration of 400 mM KCl and subjected to gel filtration on a spin column equilibrated with buffer B containing 400 mM KCl and 0.1% BSA, followed by a second spin column equilibrated with buffer B containing 0.1% BSA.
Immunoprecipitation of PICs derived from FLAG-tagged protein-expressing cells
The cDNAs encoding full-length LAP2α (residues 1–692), LAP2β (residues 1–451), the LAP2 common domain (LAP2c, residues 1–186) and emerin (residues 1–258) were amplified from a murine spleen cDNA library (Clontech) by PCR using Pfu polymerase (Stratagene) in the presence of 10% DMSO and cloned into the EcoRV site of p3XFLAG-CMV-14 mammalian expression vector (Sigma). In all, 20 μg of the recombinant plasmid or control plasmid DNA, p3XFLAG-CMV-7-BAP (Sigma), was introduced into 1 × 106 of NIH3T3 cells by the calcium phosphate transfection method in a 10 cm diameter dish. At 24 h after transfection, the cells were washed once and cocultured with 5 × 105 of clone 4 cells in the presence of polybrene for 18 h. Cytoplasmic extract was prepared from infected cells using 500 μl of buffer A with 0.025% digitonin and stored in buffer B as fraction I.
Fraction I was subjected to gel filtration through a spin column of Sephacryl S-1000 Superfine equilibrated with buffer B containing 0.1% BSA (fraction II), and 20 μl of fraction II was incubated with 25 μg of anti-FLAG monoclonal antibody M2 (Sigma) in 500 μl of buffer C (20 mM Hepes-NaOH, pH 7.5, 5 mM MgCl2, 150 mM KCl, 6 mM EDTA, 0.04% BSA) containing 0.1% Nonidet P-40 (NP-40) at 4°C for 1 h. Then, 30 μl of protein A/G agarose beads (Santa Cruz Biotechnology) was added to the mixture and incubation was continued at 4°C for 3 h. Immune complex was pelleted by centrifugation, washed three times with buffer C containing 0.1% NP-40 and deproteinized by treatment with 1 mg/ml proteinase K and 1% SDS at 37°C for 1 h. Viral DNA was recovered by phenol/chloroform extraction and ethanol precipitation, suspended in TE containing 20 μg/ml RNase and detected by Southern blotting analysis using 32P-labeled probe for MoMLV LTR sequence (Lee and Craigie, 1994).
Preparation of bacterially expressed proteins
Human BAF protein was expressed and purified from Escherichia coli as described previously (Zheng et al, 2000). For purification of GST-fusion proteins, the cDNA encoding full-length LAP2α (GST-LAP2α) and its deletion mutants, GST-LAP2c (LAP2 common domain), GST-LAP2αΔN186 (LAP2α-specific domain, residues 187–692), GST-LAP2αΔC253 (residues 1–439) and GST-LAP2αΔN112 (residues 113–692) were amplified by PCR and cloned into the SmaI site of pGEX-2T vector (Amersham-Pharmacia) and the recombinant plasmids were transformed into E. coli, strain BL21 (DE3) (Stratagene). An overnight culture from a single colony was diluted 1:50 in fresh LB medium containing 100 μg/ml ampicillin, incubated at 37°C and, upon reaching an OD600 of 0.5, protein expression was induced under the control of T7 RNA polymerase by addition of isopropyl β-D-thiogalactoside (IPTG) to 1 mM. Bacteria were harvested 5 h after induction and frozen in 1/10 volume of suspension buffer (50 mM Tris–HCl, pH 8.0, 500 mM NaCl, 2 mM EDTA). The cells were then thawed on ice, lysed by addition of 0.4 mg/ml lysozyme, 0.028% β-mercaptoethanol and proteinase inhibitors (1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 5 μg/ml aprotinin (Sigma)) at 4°C for 1 h and centrifuged at 30 000 r.p.m. for 1 h in a Beckman 45 Ti rotor. GST-fusion protein was bound to a glutathione Sepharose 4B column (Amersham-Pharmacia) and washed with phosphate-buffered saline (PBS) containing 500 mM NaCl, eluted with protein buffer (20 mM Tris–HCl, pH 7.5, 500 mM NaCl, 0.1 mM EDTA, 5 mM DTT, 10% (wt/vol) glycerol) containing 15 mM reduced glutathione (Sigma). Fractions containing GST-fusion protein were further purified by gel filtration on a Superdex 200 column (Amersham-Pharmacia) in the same buffer and stored at −80°C.
To prepare His-tagged protein, the cDNAs of full-length LAP2α (His-LAP2α) and LAP2α-specific domain (His-LAP2α/ΔN186) were amplified by PCR and cloned into pET15b vector (Novagen) that had been digested with NdeI, and treated with Mung Bean Nuclease (New England BioLabs) for blunt end ligation. Expression of protein and lysis of bacteria were carried out by the same procedure as for GST-fusion proteins. The soluble fraction containing His-LAP2α was loaded onto a Ni2+-affinity chromatography column (Amersham-pharmacia) and the column was washed with PBS containing 500 mM NaCl and 20 mM imidazole. His-tagged LAP2α was eluted by a 60–1000 mM imidazole gradient in PBS containing 500 mM NaCl and 10% glycerol and further purified by gel filtration on a Superdex 200 column equilibrated with protein buffer.
For the purification of His-LAP2α/ΔN186, which was mostly found in insoluble fraction, the pellet after lysis was solubilized by 1/20 of original culture volume of urea buffer (50 mM Tris–HCl, pH 8.0, 500 mM NaCl, 7 M urea) and incubated at room temperature for 30 min. After homogenization, the lysate was centrifuged at 30,000 r.p.m. for 1 h in a Beckman 45 Ti rotor and the supernatant was then loaded onto a Ni2+ affinity column. The His-tagged LAP2α-specific domain protein was washed with PBS containing 500 mM KCl and 1 M urea, and eluted with a 60–1000 mM imidazole gradient in PBS containing 500 mM NaCl, 10% glycerol and 1 M urea. Fractions containing the His-LAP2α/ΔN186 were pooled and dialyzed against the protein buffer containing 1 M urea.
Immunoprecipitation of PICs with anti-LAP2α antibody
The His-LAP2α/ΔN186 purified under partially denaturing conditions was used as an antigen to generate a rabbit serum against LAP2α. Immunizations and serum production were carried out by Washington Biotechnology, Inc. Anti-LAP2α polyclonal IgG was purified from the antisera using MabTrap Kit (Amersham-Pharmacia).
For immunoprecipitation, PIC fraction I was gel-filtrated through a spin column of Sephacryl S-1000 Superfine equilibrated with buffer B containing 0.1% BSA (fraction II) and 20 μl of the fraction II was incubated with anti-LAP2α polyclonal IgG (5 μl), anti-BAF rabbit serum (20 μl, Suzuki and Craigie, 2002), control rabbit IgG, or control rabbit serum in 500 μl of buffer C containing 0.5% NP-40 at 4°C for 1 h. After adding 30 μl of protein A/G agarose beads, incubation was continued at 4°C for 3 h and the immune complex was washed three times with buffer C containing 0.5% NP-40. Finally, viral DNA was extracted from the immune complex and detected by Southern blotting using 32P-labeled probe for MoMLV LTR sequence.
GST-pulldown assay
A volume of 20 μl of PIC fraction II was incubated with 100 nM GST-LAP2α, GST-LAP2c, GST-LAP2α/ΔN186, or GST in 100 μl of buffer D (20 mM Hepes-NaOH, pH 7.5, 5 mM MgCl2, 400 mM KCl, 6 mM EDTA, 0.04% BSA, 40% Nycodenz, 10 mM (NH4)2SO4) on ice. After 1 h incubation, 30 μl of glutathione Sepharose 4B beads (50% slurry equilibrated with PBS, Amersham-pharmacia) was added to the mixture and the incubation was continued on ice for 30 min. Beads were washed with buffer C containing 0.05% NP-40 three times and deproteinized with proteinase K and SDS. Viral DNA from captured PICs was isolated and detected by Southern blotting using 32P-labeled probe for MoMLV LTR sequence.
PIC integration activity assay
A volume of 20 μl of PIC fraction II or salt-stripped PIC was incubated with 10 and 100 nM BAF or His-LAP2α in 100 μl of buffer D on ice for 1 h (Lee and Craigie, 1998). Integration activity of the PICs was evaluated by the previously described integration activity assay (Lee and Craigie, 1994, 1998).
Velocity sedimentation assay
DNA substrates were Φ × 174 DNA linearized with XhoI or a circular form of Φ × 174 DNA (replicative form II, New England Biolabs). BAF/DNA complex, with or without GST-fusion LAP2α proteins or its deletion mutants, was formed with 10 nM proteins and 0.1 pM substrate DNA in 100 μl of a reaction mixture (20 mM Hepes-NaOH, pH 7.5, 115 mM KCl, 100 ng/ml BSA, 5 mM DTT) at 30°C for 1 h. To check the salt stability of the nucleoprotein complex, KCl was added to a final concentration of 150 or 400 mM and the mixture was incubated on ice for 1 h. Continuous sucrose gradients (2 ml) were made by layering 15, 20, 25 and 30% sucrose solutions in buffer A containing KCl (150 or 400 mM) and 6 mM EDTA at 4°C overnight. The gradient was overlayered with the salt-challenged reaction (100 μl) mixture and centrifuged at 30 000 r.p.m. at 4°C for 1 h in a Beckman TLS55 rotor. The gradient was fractionated from the top into 21 fractions and each fraction was deproteinized by proteinase K and SDS. The DNA in each fraction was isolated by phenol/chloroform extraction and ethanol precipitation, and detected by Southern blotting using 32P-labeled probe for Φ × 174 DNA sequence.
Establishment of LAP2α-knockdown cell lines and isolation of PICs
A 19-bp siRNA sequence was selected from nucleotide positions 1063–1081 (5′-AGA GAA GUA CUG CAG GAG U-3′) in the open reading frame of LAP2α mRNA and synthesized DNA of this sequence was inserted into the pSilencer 2.1-U6 neo siRNA expression vector (Ambion) according to the manufacturer's protocol. As a negative control, we used the pSilencer 2.1-U6 neo Negative Control plasmid (Ambion), whose hairpin siRNA sequence is not found in the mouse genome database. The vectors were digested with XmnI to linearize the plasmid DNA and transfected into NIH3T3 cells by the calcium phosphate transfection method. To establish stable siRNA-expressing cell lines, the transfected cells were selected and maintained in culture medium containing 800 μg/ml neomycin (Invitrogen). Reduction in expression of LAP2α protein expression was analyzed by western blotting using anti-LAP2 monoclonal antibody as described below. To isolate the PICs, 2 × 106 of cells were cultured with 10 ml of supernatant from clone 4 cells in the presence of 8 μg/ml polybrene for 5 h, and cytoplasmic extract was prepared from the infected cells using 500 μl of buffer A containing digitonin and stored in buffer B.
Western blotting analysis
Cells were trypsinized, washed with PBS and lysed in SDS sample buffer (62.5 mM Tris–HCl, pH 6.8, 2% SDS, 5% glycerol, 0.003% bromophenol blue, 0.9% β-mercaptoethanol) by boiling. Samples were loaded on a NuPAGE 4–12% Bis–Tris gel (Invitrogen) and transferred to Invitrolon PVDF membrane (Invitrogen). The blot was then subjected to immunoblotting using WesternBreeze chemiluminescent immunodetection kit (Invitrogen) and proteins were detected by a luminescence image analyzer, LAS 1000 (Fujifilm). Primary antibodies used for the western blotting analysis were anti-FLAG mouse monoclonal IgG (Clone M2, Sigma) for detection of FLAG-tagged proteins, anti-LAP2α rabbit polyclonal IgG for detection of LAP2α, anti-LAP2 mouse monoclonal IgG (Clone 27, BD Transduction Laboratories) for detection of LAP2 isoforms and anti-lamins A/C mouse monoclonal IgM (Clone 346, Santa Cruz Biotechnology).
Analysis of MoMLV replication
To prepare MoMLV virus stock, culture supernatant from clone 4 cells was harvested, filtered through a membrane (0.45 μm pore size) and stored at −80°C until use. The titer of the viral stock was determined by end-point titration of 10-fold dilution on cultured NIH3T3 cells. For an infection experiment, NIH3T3, LAP2α-knockdown and siRNA control cell lines were exposed to 1 × 103 infectious virus particles per 2 × 105 cells in 60 mm diameter plates with culture medium containing 8 μg/ml polybrene at 37°C for 2 h. After washing twice with fresh medium to remove residual free virus, the infected cells were maintained in culture medium and split 1:5 every 3 days. Virus production in culture supernatant from the MoMLV-infected cells at 3, 6 and 9 days after infection was monitored by an exogenous RT activity assay using 32P-labeled dTTP as described previously (Telesnitsky et al, 1995).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Acknowledgments
We thank Christina Bradley, Ying Huang and Jian-Yong Wang for helpful suggestions, and Christina Bradley and Kiyoshi Mizuuchi for their comments on the manuscript. This work was supported in part by the NIH Intramural AIDS Targeted Antiviral Program.
References
- Asante-Appiah E, Skalka AM (1999) HIV-1 integrase: structural organization, conformational changes, and catalysis. Adv Virus Res 52: 351–369 [DOI] [PubMed] [Google Scholar]
- Berger R, Theodor L, Shoham J, Gokkel E, Brok-Simoni F, Avraham KB, Copeland NG, Jenkins NA, Rechavi G, Simon AJ (1996) The characterization and localization of the mouse thymopoietin/lamina-associated polypeptide 2 gene and its alternatively spliced products. Genome Res 6: 361–370 [DOI] [PubMed] [Google Scholar]
- Bowerman B, Brown PO, Bishop JM, Varmus HE (1989) A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev 3: 469–478 [DOI] [PubMed] [Google Scholar]
- Brown PO (1997) Integration. In Retroviruses, Coffin JM, Hughes SH, Varmus HE (eds), pp 161–203. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [PubMed] [Google Scholar]
- Brown PO, Bowerman B, Varmus HE, Bishop JM (1987) Correct integration of retroviral DNA in vitro. Cell 49: 347–356 [DOI] [PubMed] [Google Scholar]
- Brown PO, Bowerman B, Varmus HE, Bishop JM (1989) Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci USA 86: 2525–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Cai M, Huang Y, Ghirlando R, Wilson KL, Craigie R, Clore GM (2001) Solution structure of the constant region of nuclear envelope protein LAP2 reveals two LEM-domain structures: one binds BAF and the other binds DNA. EMBO J 20: 4399–4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Huang Y, Zheng R, Wei SQ, Ghirlando R, Lee MS, Craigie R, Gronenborn AM, Clore GM (1998) Solution structure of the cellular factor BAF responsible for protecting retroviral DNA from autointegration. Nat Struct Biol 5: 903–909 [DOI] [PubMed] [Google Scholar]
- Chen H, Engelman A (1998) The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc Natl Acad Sci USA 95: 15270–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R (2001) HIV integrase, a brief overview from chemistry to therapeutics. J Biol Chem 276: 23213–23216 [DOI] [PubMed] [Google Scholar]
- Dechat T, Gotzmann J, Stockinger A, Harris CA, Talle MA, Siekierka JJ, Foisner R (1998) Detergent-salt resistance of LAP2α in interphase nuclei and phosphorylation-dependent association with chromosomes early in nuclear assembly implies functions in nuclear structure dynamics. EMBO J 17: 4887–4902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Korbei B, Vaughan OA, Vlcek S, Hutchison CJ, Foisner R (2000a) Lamina-associated polypeptide 2α binds intranuclear A-type lamins. J Cell Sci 113: 3473–3484 [DOI] [PubMed] [Google Scholar]
- Dechat T, Vlcek S, Foisner R (2000b) Review: lamina-associated polypeptide 2 isoforms and related proteins in cell cycle-dependent nuclear structure dynamics. J Struct Biol 129: 335–345 [DOI] [PubMed] [Google Scholar]
- Fassati A, Goff SP (1999) Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J Virol 73: 8919–8925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foisner R (2001) Inner nuclear membrane proteins and the nuclear lamina. J Cell Sci 114: 3791–3792 [DOI] [PubMed] [Google Scholar]
- Fouchier RA, Malim MH (1999) Nuclear import of human immunodeficiency virus type-1 preintegration complexes. Adv Virus Res 52: 275–299 [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Mizuuchi K (1988) Retroviral DNA integration: structure of an integration intermediate. Cell 54: 497–504 [DOI] [PubMed] [Google Scholar]
- Furukawa K (1999) LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2–chromatin interaction. J Cell Sci 112: 2485–2492 [DOI] [PubMed] [Google Scholar]
- Furukawa K, Pante N, Aebi U, Gerace L (1995) Cloning of a cDNA for lamina-associated polypeptide 2 (LAP2) and identification of regions that specify targeting to the nuclear envelope. EMBO J 14: 1626–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Sugiyama S, Osouda S, Goto H, Inagaki M, Horigome T, Omata S, McConnell M, Fisher PA, Nishida Y (2003) Barrier-to-autointegration factor plays crucial roles in cell cycle progression and nuclear organization in Drosophila. J Cell Sci 116: 3811–3823 [DOI] [PubMed] [Google Scholar]
- Goff SP (1992) Genetics of retroviral integration. Annu Rev Genet 26: 527–544 [DOI] [PubMed] [Google Scholar]
- Haraguchi T, Koujin T, Segura-Totten M, Lee KK, Matsuoka Y, Yoneda Y, Wilson KL, Hiraoka Y (2001) BAF is required for emerin assembly into the reforming nuclear envelope. J Cell Sci 114: 4575–4585 [DOI] [PubMed] [Google Scholar]
- Harris CA, Andryuk PJ, Cline S, Chan HK, Natarajan A, Siekierka JJ, Goldstein G (1994) Three distinct human thymopoietins are derived from alternatively spliced mRNAs. Proc Natl Acad Sci USA 91: 6283–6287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Goff SP (2001) Infection of nondividing cells by Rous sarcoma virus. J Virol 75: 9526–9531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holaska JM, Wilson KL, Mansharamani M (2002) The nuclear envelope, lamins and nuclear assembly. Curr Opin Cell Biol 14: 357–364 [DOI] [PubMed] [Google Scholar]
- Lee KK, Haraguchi T, Lee RS, Koujin T, Hiraoka Y, Wilson KL (2001) Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J Cell Sci 114: 4567–4573 [DOI] [PubMed] [Google Scholar]
- Lee MS, Craigie R (1994) Protection of retroviral DNA from autointegration: involvement of a cellular factor. Proc Natl Acad Sci USA 91: 9823–9827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Craigie R (1998) A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci USA 95: 1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PF, Emerman M (1994) Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol 68: 510–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Engelman A (2003) The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J Virol 77: 5030–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Blake DL, Callebaut I, Skerjanc IS, Holmer L, McBurney MW, Paulin-Levasseur M, Worman HJ (2000) MAN1, an inner nuclear membrane protein that shares the LEM domain with lamina-associated polypeptide 2 and emerin. J Biol Chem 275: 4840–4847 [DOI] [PubMed] [Google Scholar]
- Roe T, Reynolds TC, Yu G, Brown PO (1993) Integration of murine leukemia virus DNA depends on mitosis. EMBO J 12: 2099–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura-Totten M, Kowalski AK, Craigie R, Wilson KL (2002) Barrier-to-autointegration factor: major roles in chromatin decondensation and nuclear assembly. J Cell Biol 158: 475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura-Totten M, Wilson KL (2004) BAF: roles in chromatin, nuclear structure and retrovirus integration. Trends Cell Biol 14: 261–266 [DOI] [PubMed] [Google Scholar]
- Shumaker DK, Lee KK, Tanhehco YC, Craigie R, Wilson KL (2001) LAP2 binds to BAF·DNA complexes: requirement for the LEM domain and modulation by variable regions. EMBO J 20: 1754–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small K, Wagener M, Warren ST (1997) Isolation and characterization of the complete mouse emerin gene. Mamm Genome 8: 337–341 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Craigie R (2002) Regulatory mechanisms by which barrier-to-autointegration factor blocks autointegration and stimulates intermolecular integration of Moloney murine leukemia virus preintegration complexes. J Virol 76: 12376–12380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesnitsky A, Blain S, Goff SP (1995) Assays for retroviral reverse transcriptase. Methods Enzymol 262: 347–362 [DOI] [PubMed] [Google Scholar]
- Umland TC, Wei SQ, Craigie R, Davies DR (2000) Structural basis of DNA bridging by barrier-to-autointegration factor. Biochemistry 39: 9130–9138 [DOI] [PubMed] [Google Scholar]
- Vlcek S, Just H, Dechat T, Foisner R (1999) Functional diversity of LAP2α and LAP2β in postmitotic chromosome association is caused by an alpha-specific nuclear targeting domain. EMBO J 18: 6370–6384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R, Ghirlando R, Lee MS, Mizuuchi K, Krause M, Craigie R (2000) Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc Natl Acad Sci USA 97: 8997–9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2


