Abstract
In Drosophila, the maternal terminal system specifies cell fates at the embryonic poles via the localised stimulation of the Torso receptor tyrosine kinase (RTK). Signalling by the Torso pathway relieves repression mediated by the Capicua and Groucho repressors, allowing the restricted expression of the zygotic terminal gap genes tailless and huckebein. Here we report a novel positive input into tailless and huckebein transcription by maternal posterior group genes, previously implicated in abdomen and pole cell formation. We show that absence of a subset of posterior group genes, or their overactivation, leads to the spatial reduction or expansion of the tailless and huckebein posterior expression domains, respectively. We demonstrate that the terminal and posterior systems converge, and that exclusion of Capicua from the termini of posterior group mutants is ineffective, accounting for reduced terminal gap gene expression in these embryos. We propose that the terminal and posterior systems function coordinately to alleviate transcriptional silencing by Capicua, and that the posterior system fine-tunes Torso RTK signalling output, ensuring precise spatial domains of tailless and huckebein expression.
Keywords: capicua, Drosophila, nanos, tailless, terminal patterning
Introduction
In the Drosophila embryo, four maternal coordinate systems act to specify distinct cell fates along the anteroposterior (A/P) and dorsoventral (D/V) axes: the anterior, posterior, D/V and terminal systems. The head and thorax are patterned by the activity of the anterior bicoid (bcd) morphogen, while abdomen formation is directed by a reciprocal posterior gradient of Nanos (Nos). Asymmetrical nuclear distribution of Dorsal governs D/V axis formation, whereas the nonsegmented termini are patterned by the terminal system. The activities of these systems induce regional-specific transcription of zygotic downstream target genes, the products of which eventually assign the different body parts with their prospective identities (St Johnston and Nusslein Volhard, 1992).
Zygotic transcription in the termini of the embryo is controlled by the terminal system. A key component of this system is the gene torso (tor), which encodes a receptor tyrosine kinase (RTK) (Sprenger et al, 1989; St Johnston and Nusslein Volhard, 1992; Lu et al, 1993; Duffy and Perrimon, 1994). The Torso (Tor) receptor is uniformly distributed throughout the plasma membrane of the early embryo, but is activated only at the poles by a locally processed ligand, where it transmits a signal via the canonical Ras/Raf/MAP-kinase (MAPK) effectors (Casanova and Struhl, 1989, 1993; Sprenger and Nusslein Volhard, 1992; Lu et al, 1993; Duffy and Perrimon, 1994; Casali and Casanova, 2001; Furriols and Casanova, 2003). This signal leads to the restricted expression of two zygotic terminal gap genes, tailless (tll) and huckebein (hkb), at the poles of the embryo. tll and hkb encode transcription factors that, in turn, implement head and tail differentiation programmes (Strecker et al, 1986, 1988; Casanova, 1990; Pignoni et al, 1990; Weigel et al, 1990; Brönner and Jackle, 1991; Brönner et al, 1994; Furriols and Casanova, 2003).
Genetic and molecular studies suggest that the activation of tll and hkb expression by the Tor pathway is indirect, and that Tor signalling allows regional expression of the terminal gap genes by counteracting, at the embryonic poles, general transcriptional repressors (Liaw et al, 1995; Paroush et al, 1997; Jiménez et al, 2000). Two major lines of evidence support this idea. First, a detailed analysis of the tll promoter has defined negative cis-acting regulatory sequences (designated Torso-response elements; TREs), whose deletion leads to ectopic expression throughout the embryo, indicating that tll is normally repressed outside the embryonic poles (Liaw et al, 1995; Rudolph et al, 1997). Second, mutations in genes encoding maternally contributed transcriptional repressors, such as the nuclear HMG-box protein Capicua (Cic) (Jiménez et al, 2000) and the global developmental corepressor Groucho (Gro), result in ectopic, more central, tll and hkb expression, even in the absence of functional Tor signalling (Paroush et al, 1994, 1997; Chen and Courey, 2000). How these factors regulate tll and hkb transcription is not well understood, although it appears that Cic is one of the key elements regulated by the Tor pathway (Jiménez et al, 2000).
At the posterior of the embryo, the activity zone of the terminal system overlaps with that of the posterior system. Posterior group members are localised to the posterior pole, where they function in polar granule assembly and in the formation of the abdomen and germ cells (St Johnston and Nusslein Volhard, 1992). Assembly of the germ plasm occurs in a stepwise manner, with oskar (osk) being the key element of this process (Rongo and Lehmann, 1996). Embryos lacking maternal osk activity fail to form pole cells and to develop abdominal structures. Further, osk dosage dictates the amount of pole plasm assembled, and the consequent number of germ cells produced. Following translation of osk mRNA, the Osk protein localises the Vasa (Vas) DEAD-box RNA-binding helicase and the Tudor (Tud) protein to the posterior pole. These three proteins are then required for the execution of all posterior system functions. Downstream of Tud, the posterior group bifurcates: the product of the germ-cell-less (gcl) gene is required only for pole cell formation, whereas those of nanos (nos) and pumilio (pum) regulate abdomen formation by blocking translation of the anterior determinant hunchback (hb) (Rongo and Lehmann, 1996).
In general, the distinct maternal genetic systems act independently of each other to define discrete portions of the embryonic pattern. Mutations in one maternal coordinate system seem to eliminate a specific body part without grossly affecting the rest of the embryonic pattern. Previous studies, however, have revealed interactions between the anterior, terminal and D/V systems at the most anterior region of the embryo (Pignoni et al, 1992; Liaw and Lengyel, 1993). At the posterior pole, on the other hand, the terminal system has long been thought to act alone in regulating zygotic terminal gap gene transcription, given that expression of tll and hkb is completely absent in embryos devoid of maternal tor activity, or lacking any other constituent of this signalling cascade (Weigel et al, 1990; Brönner and Jackle, 1991).
In this work, we identify a surprising, novel role for maternal posterior group genes in embryonic terminal patterning. We show that tll and hkb expression patterns are spatially reduced in several posterior group mutants, suggesting that posterior group members positively contribute to the transcription of zygotic terminal gap genes. Our data indicate that the positive input from posterior group genes, in particular Nos, into tll and hkb expression converges on the Tor pathway. Thus, minimal TRE sequences, through which terminal system effectors regulate tll expression, also respond to the lack, or to the overactivation, of the posterior system. We further demonstrate that posterior genes act upstream or at the level of Cic, as ectopic accumulation of the Cic repressor is seen in the termini of posterior group mutants, accounting for the reduced tll and hkb expression domains in these embryos. Hence, multiple maternal inputs are required to effectively antagonise ubiquitous negative regulators at the posterior pole, in order to generate the precise patterns of tll and hkb expression. We propose that the posterior system acts to refine Tor RTK signalling output, ensuring that terminal gap gene expression patterns are properly established and consequently cell fates are correctly specified.
Results
Terminal gap gene expression is reduced in oskar, vasa and tudor mutant embryos
By the beginning of cellularisation, tll and hkb are expressed in a nested-set pattern at the pole regions of wild-type Drosophila embryos (Weigel et al, 1990; Brönner et al, 1994). At this developmental stage, the dynamic tll RNA expression pattern has resolved into smaller domains at both the anterior and posterior ends. tll is expressed at the anterior cap in a horseshoe-shaped stripe that extends about two-thirds down along the D/V axis, whereas, posteriorly, tll covers a region comprising approximately 0–16% egg length (EL; 0% being the coordinate of the posterior tip) (Figure 1A) (Pignoni et al, 1990). hkb, on the other hand, is transcribed in the presumptive head region as a cap and, at the posterior, its expression domain overlaps with that of tll, covering about 0–12% EL (Figure 1E) (Brönner and Jackle, 1991).
Figure 1.

Terminal gap gene expression domains are reduced in oskar and tudor mutant embryos. Spatial distribution of tll (A–D) and hkb (E–H) transcripts in wild-type (A, E) and in posterior group mutant embryos (B–D, F–H), detected by in situ hybridisation as described in Materials and methods. Embryos shown are at the syncytial blastoderm stage (stage 4), and are derived from the following females: (A, E) wild type; (B) osk166/osk166; (F) osk346/osk346; (C, G) osk54/Df(3R)p-XT103; (D, H) tudWC/tudB36. Note the reduced posterior tll and hkb expression domains in osk and tud mutants, compared to wild-type embryos. The anterior expression domains of tll and hkb are also affected by the absence of osk (B, C, F, G), tud (D, H) and vasa (not shown) (see Discussion). For quantification of the spatial extent of the posterior terminal gap gene expression domains in these and other mutants, see Table I. Control and mutant embryos were mixed, then fixed and stained simultaneously. In all figures, the views are lateral, anterior is to the left and dorsal side up.
We find that the posterior expression domains of both terminal gap genes are reduced in embryos laid by osk mutant females (Figure 1). Thus, tll and hkb expression is spatially reduced in stage 4 embryos derived from osk166 and osk346 homozygous females, and from null osk54/Df(3R)p-XT103 mutant females (Figure 1B, C, F and G; Table I). Retracted terminal gap gene expression domains are also seen in older, stage 5 embryos (not shown). In these and subsequent experiments, we minimised the effects caused by differential probe concentrations and/or duration of staining reactions by first mixing and then simultaneously fixing and processing wild-type embryos together with mutants, the latter distinguishable by their lack of pole cells. In all cases, the spatial reduction is reproducible and statistically significant (Table I). Thus, we conclude that the posterior group gene osk is required for the full extent of terminal gap gene expression at the posterior pole.
Table 1.
Extent of posterior tailless and huckebein expression domains in posterior group mutants
| Maternal genotype |
tailless |
huckebein |
||
|---|---|---|---|---|
| Stage 4 | Stage 5 | Stage 4 | Stage 5 | |
| osk166/osk166 | 0.82*** | ND | ND | ND |
| osk54/Df(3R)p-XT103 | 0.74*** | 0.61*** | 0.78** | 0.85*** |
| vasPD/vas011 | 0.88** | ND | ND | ND |
| tudWC/tudB36 | 0.61*** | 0.66*** | 1.05 | 1.07 |
| nosL7/nosL7 | 0.89 | 0.96 | ND | ND |
| nosBN/nosBN | 0.72*** | 0.83* | 0.93 | 0.84* |
| nosRC/nosBN | 0.58*** | 0.71*** | 0.52*** | 0.80*** |
| pum680/pum680 | 1.26*** | 1.04 | ND | ND |
| pumET1/In(3R)Msc | 0.99 | 0.98 | 1.24** | 1.02 |
| gclΔ49/gclΔ49 | 0.94 | 1.04 | 1.19* | 1.18*** |
| oskAk/oskAk (4xosk) | 1.75*** | 1.07 | 1.34** | 1.45*** |
| bicS/bicS | 1.39*** | 1.63*** | 1.39*** | ND |
| Values below or above 1 represent a reduction or expansion of gap gene expression domains, respectively, while ratios of around 1 denote normal extent (see Materials and methods). ND: not determined. Statistically significant results are marked by asterisks: ***P<0.01, **P<0.05 and *P<0.1. | ||||
We also examined if mutations in other posterior group genes elicit similar alterations in terminal gap gene patterns. The vasa (vas) gene product is required for the activation of Osk translation (Rongo et al, 1995). We find that vasPD/vas011 mutant embryos show a reduction in tll and hkb expression, comparable to that seen in osk mutants (data not shown; Table I), implying that, like osk, vas also normally contributes to terminal gap gene regulation.
Next we examined the effect of Tud, a posterior group member that mediates the transport of RNA from the mitochondria to polar granules (Amikura et al, 2001). In embryos derived from tudWC/tudB36 mothers, the posterior tll expression domain is reduced, although hkb expression is not significantly altered, perhaps due to the use of hypomorphic alleles (Figure 1D and H; Table I).
In all of the above mutant backgrounds, a lower staining intensity relative to that seen in wild-type embryos (Figure 1) suggests a decline in transcript abundance. This weakened staining is specific to tll and hkb transcripts, as other genes are expressed at normal levels in the posterior and elsewhere of osk mutant embryos (D Gur-Wahnon, unpublished results). Notably, the anterior tll expression domain is also reduced in all of the above posterior group mutants (see Discussion). Thus, the posterior group genes osk, vasa and tud are positively required for the expression of the terminal gap genes tll and hkb.
Nanos positively regulates tailless and huckebein expression
We next established the expression patterns of tll and hkb in embryos mutant for posterior genes acting downstream of osk, vas and tud (Figure 5I) (Rongo and Lehmann, 1996). In embryos produced by gcl homozygous mothers, the tll and hkb expression domains are both normal (data not shown; Table I), suggesting that gcl is not required for terminal gap gene regulation. Given that pole cells do not form in gcl mutants (Jongens et al, 1992), we conclude that the lack of germ cells in osk, vas and tud embryos is not the basis for the reduced tll and hkb expression in these genotypes.
Figure 5.
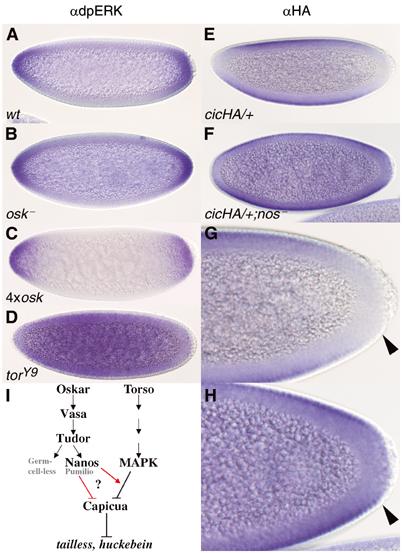
Posterior group genes feed into the Torso pathway upstream of Capicua. (A–D) Staining for dpERK (activated MAPK) at stage 4. (E–H) Distribution of the HA-tagged Cic protein (CicHA) at stage 4, revealed by anti-HA antibody staining. (A) Wild-type embryos; (B) embryos produced by osk54/Df(3R)p-XT103 mutant mothers; (C) embryos produced by 4xosk mothers; (D) embryos produced by torY9 mothers; (E) embryos derived from females heterozygous for the cicHA transgene (cicHA/+); (F) embryos produced by cicHA/+; nosRC/nosBN mothers; (G, H) larger magnifications of the posterior poles of the embryos shown in (E, F), respectively. The arrowheads in (G, H) indicate the same relative point in the two embryos. dpERK levels in osk and 4xosk mutants are identical to those seen in wild-type embryos; however note the ectopic accumulation of CicHA in the terminal regions of nos mutants compared to its exclusion from these regions in wild-type embryos. Note that while HA staining is observed to the left of the arrowhead in nos mutants (H), it is missing at the corresponding point in the wild type (G). (I) Schematic representation of the intersection between the posterior system and the Torso pathway. Nos acts with Pum or some unknown partner, at the level of MAPK or downstream to it, to downregulate the Cic repressor complex at the termini, facilitating tll and hkb transcription (see text for details). Posterior group members that are not involved in terminal gene regulation are presented in light grey.
Embryos derived from nos and pum mutant mothers possess pole cells, but exhibit abdominal defects resulting from the failure of these RNA-binding proteins to repress the translation of maternal hb. We find that Nos participates in tll and hkb regulation. In stage 4 embryos derived from nosRC/nosBN or homozygous nosBN mutant females, the tll and hkb expression domains are significantly reduced (Figure 2B and E; data not shown; Table I), implying that the Nos protein is a positive regulator of terminal gap gene expression. Double in situ hybridisation staining shows that the posterior expression of tll, but not that of the central Krüppel (Kr) stripe, is reduced in nosRC/nosBN embryos compared to wild type (Figure 2M and N), confirming the specificity of this effect. In contrast, we find that pumET1/In(3R)Msc females lay embryos with normal tll and hkb expression (Figure 2C and F; Table I), indicating that pum may not be involved in this regulation.
Figure 2.
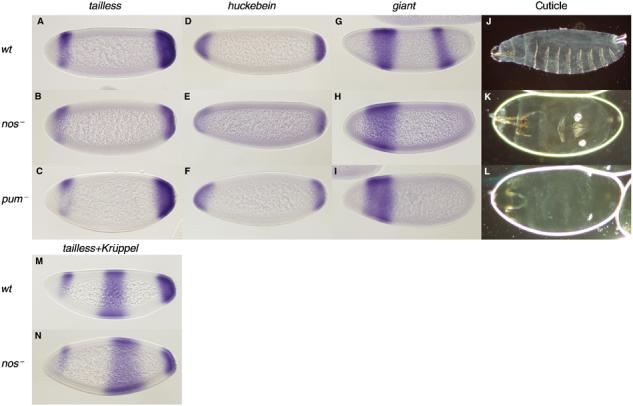
Terminal gap gene expression domains are reduced in nanos but not in pumilio mutants. (A–I, M, N) In situ hybridisation. (J–L) Larval cuticular preparations. (A, D, G, J, M) Wild type; (B, E, H, K, N) nosRC/nosBN; (C, F, I, L) pumET1/In(3R)Msc. Expression domains of tll (A–C), hkb (D–F), gt (G–I) and tll+Kr (M, N) in stage 4 embryos. Note that the posterior tll and hkb expression domains are reduced in nos, but not in pum, mutant embryos. In both nos and pum mutants, abdominal segmentation is defective, as evident by the absence of the posterior giant stripe (H, I; cf. with G) and by the lack of abdominal denticle belts (K, L; cf. with J). The Kr stripe expands posteriorly in nos (cf. N with M) and osk mutant embryos (not shown) (Hulskamp et al, 1990).
The differential effects of Nos and Pum on tll and hkb expression are surprising, given that these two proteins act as partners in abdominal patterning and in other settings. However, two lines of evidence support this conclusion. First, the mutant nos and pum allelic combinations tested show a similar loss of posterior denticle belts (Figure 2K and L) (Irish et al, 1989; Murata and Wharton, 1995) and the absence of abdominal gap gene expression (e.g., the posterior giant stripe; Figure 2H and I), as reported for posterior group mutants (Kraut and Levine, 1991). Second, we find that tll and hkb expression is unaffected in homozygous nosL7 mutant embryos (Table I). This particular allele encodes a Nos protein lacking its carboxy-terminal tail, the domain required for the recruitment of Nos into the Pum/hb mRNA ternary complex (Sonoda and Wharton, 1999). Thus, nosL7 renders Nos inactive as a repressor of hb mRNA translation (a Pum-dependent activity), yet it does not impair Nos' other functions, for example in oogenesis (Arrizabalaga and Lehmann, 1999). Nos could therefore be acting in terminal patterning in conjunction with some other maternally provided factor, perhaps an RNA-binding protein. Given that the pumET1/In(3R)Msc trans-allelic combination might still possess some Pum activity, however, it is premature to completely rule out Pum's contribution to terminal gap gene regulation.
Taken together, our results suggest that Nos is a key posterior group member required for accurate terminal gap gene expression.
Overexpression of oskar or nanos leads to expanded terminal gap gene expression
If posterior group genes have a positive input into terminal gap gene regulation, then the overexpression of these genes should cause broadening of the tll and hkb posterior expression domains. To test this idea, terminal gap gene expression patterns were assessed in offsprings of females with increased osk dosage, or that uniformly express nos. Embryos overexpressing osk were derived from two maternal genetic backgrounds, each containing two extra copies of osk: homozygous females, which harbour a small tandem duplication of the osk locus (referred to herein as bicaudalS; bicS), and homozygous females carrying an osk transgene (4xosk females) (Ephrussi and Lehmann, 1992). To express nos uniformly, we used a nos transgene, in which the nos 3′UTR was replaced with that of tubulin (Gavis and Lehmann, 1994). In all cases, embryos show cuticular phenotypes attributable to osk or nos overexpression: either a pronounced reduction of head structures, or a bicaudal appearance, with posterior terminal telson structures replacing the anterior acron (data not shown). Significantly, all combinations result in expanded posterior domains of tll and hkb (Figure 3; Table I), supporting the notion that osk, nos and other posterior group genes fulfil a positive role in terminal gap gene regulation.
Figure 3.
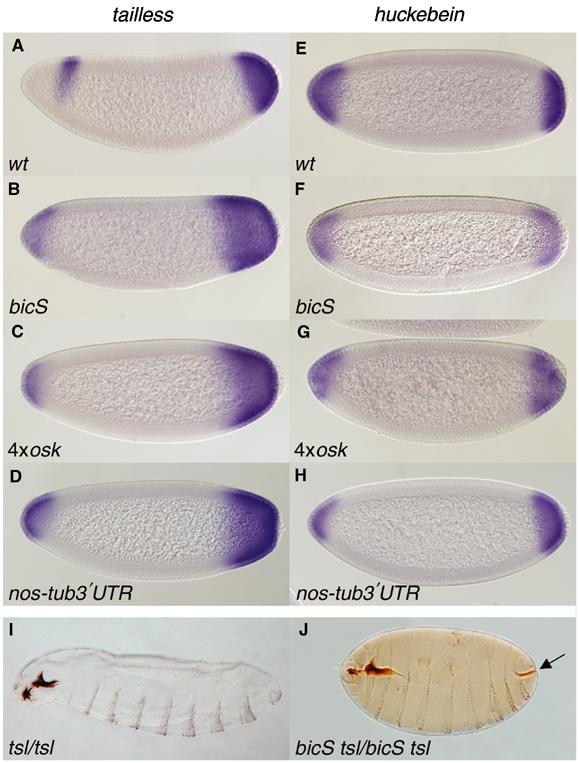
Overexpression of oskar or nanos leads to expanded terminal gap gene expression domains. Expression of tll (A–D) and hkb (E–H) in stage 4 embryos. Shown are embryos produced by the following females: (A, E) wild type; (B, F) homozygous Dp(3;3)bicS (bicS); (C, G) homozygous 4xosk; (D, H) heterozygous nos-tub3′UTR. Note the significant expansion of the tll and hkb posterior expression domains in lines overexpressing osk or nos. In these lines, the anterior tll stripe is also shifted. (I, J) Cuticular phenotypes of embryos produced by homozygous tsl (I) or bicS tsl double-mutant females (J). The arrow in (J) points to a partially restored filzkorper in a bicS tsl embryo, missing in a tsl mutant (I).
Remarkably, overexpression of osk can rescue some of the morphological defects seen in terminal group mutants. Specifically, the terminal filzkorper (FK) structure is never observed in hypomorphic torso-like (tsl691) mutant embryos, but is partially rescued in embryos harbouring this allele that also concomitantly overexpress osk (bicS tsl; Figure 3I and J). Thus, overactivation of the posterior system appears to compensate in part for compromised terminal system activity.
We note that the anterior tll stripe retracts anteriorly when osk or nos are overexpressed (Figure 3B–D), similar to what is seen in bcd mutants (Pignoni et al, 1992) (in both cases, anterior hkb expression does not change). This aspect of tll misexpression likely stems from translational inhibition of bcd mRNA by Nos, which is mislocalised to the anterior in these embryos (Ephrussi and Lehmann, 1992; Gavis and Lehmann, 1994).
Posterior group genes regulate tailless at the transcriptional level
None of the posterior group members encode for transcription factors, so we next asked whether the products of these genes regulate tll and hkb expression at the transcriptional level, or whether they do so post-transcriptionally, for example by stabilising terminal gap gene mRNA. To address this point, we made use of a reporter construct (P1), the expression of which is controlled by extensive tll promoter sequences and resembles that of the endogenous gene (Figure 4A) (Liaw and Lengyel, 1993). We find that P1 lacZ expression is reduced in osk mutants (Figure 4B), while it expands into central parts in embryos laid by females overexpressing osk (Figure 4C), in accordance with endogenous tll expression (Figures 1B and 3C). Moreover, the anterior P1 lacZ stripe is also affected in 4xosk embryos, similar to endogenous tll. Thus, the positive regulatory input from posterior group genes must be acting, presumably indirectly, on tll transcription rather than at a post-transcriptional level.
Figure 4.

Input by the posterior group converges on minimal TREs in the tailless promoter. (A–I) In situ hybridisation for lacZ reporter expression, driven by different tll promoter constructs, in stage 4 embryos: (A–C) P1, a 5.9 kb full-length tll promoter construct; (D–F) K11, tll promoter subregion −2291 to −2770, oligomerised four-fold; (G–I) G22, tll promoter subregion −324 to −200, oligomerised four-fold. (A, D, G) In a wild-type background, all constructs drive reporter expression in a posterior cap. The P1 and G22 constructs also drive expression in the anterior. Expansion of the lacZ expression pattern is apparent for all three constructs when osk is overexpressed (4xosk; C, F, I), while lacZ expression is significantly reduced for the P1 and K11 constructs, although not for G22, in osk166 homozygous flies (B, E, H).
Torso-response elements in the tailless promoter respond to alterations in maternal oskar dosage
We next investigated if posterior group genes act in parallel to, and independently of, the tor signalling cascade, or whether they feed into the terminal pathway. Molecular dissection of the tll promoter has defined two cis-acting TRE sequences that respond to both the lack and the constitutive activation of the tor pathway (Liaw et al, 1995). These DNA regulatory elements mediate transcriptional repression, specifically by the Cic repressor complex, as mutations in either the TREs or in cic lead to derepression (data not shown) (Liaw et al, 1995; Jiménez et al, 2000). We reasoned that if the posterior system regulates terminal gap gene expression independently of the Tor pathway, then its effects are likely to be exerted via distinct cis-acting elements in the tll promoter. If, on the other hand, this maternal system acts jointly with the terminal pathway, then input by the posterior system is expected to converge on the previously defined TREs.
Transgenes containing the minimal distal (K11) and proximal (G22) TRE sequences drive lacZ expression in a tll-like posterior cap, although, as previously published, the intensity of expression is reduced when compared to that of the full-length tll promoter construct (P1), probably as a result of lost activator binding sites (Figure 4D and G; cf. with Figure 4A) (Liaw et al, 1995; Rudolph et al, 1997). We find that lacZ expression, driven by the distal TRE (K11), is reduced in osk mutants (Figure 4E) and expanded in 4xosk embryos (Figure 4F), similar to the P1 reporter. When driven by the proximal TRE (G22), lacZ expression also expands in response to osk overexpression, although it is not significantly reduced in an osk mutant background (Figure 4H and I; cf. with Figure 4G). Given that besides the minimal TREs there are no apparent sequences common to both constructs, it appears that posterior group genes act on the same elements in the tll promoter that mediate terminal system output.
Input from posterior group genes intersects with the torso pathway upstream of Capicua
To test at what level the crosstalk between the posterior and terminal systems occurs, we first monitored the phosphorylation state of MAPK when osk dosage is altered. The doubly phosphorylated form of MAPK (dpERK), which can be reliably detected using a specific antibody (Gabay et al, 1997a), makes a perfect read-out for signals transmitted by RTK pathways via the generic Ras/Raf/MAPK module, including that by the Tor receptor. Thus, MAPK activation is diminished in embryos derived from homozygous tsl mutant females, and is uniformly detected when tor is constitutively activated (Figure 5D) (Gabay et al, 1997b). In contrast, we find that the pattern of anti-dpERK staining is unaltered in osk mutant (Figure 5B) or in 4xosk (Figure 5C) embryos, suggesting that posterior group genes do not converge with the Tor signal transduction cascade upstream of MAPK.
In terminal patterning, the Tor pathway is required to inhibit the Cic repressor, bringing about its post-transcriptional exclusion from the poles. Cic is normally detected throughout the embryo, except at the termini, where the Tor signal is active. Correspondingly, in tor mutants, Cic is found not only in medial regions of the embryo but also at the poles (Jiménez et al, 2000). Using anti-Cic antibodies, we followed the accumulation of Cic at the posterior pole of mutant and normal embryos, finding that it is less effectively removed from the terminal regions of stage 5 osk, nos and tud mutants compared to wild-type embryos (data not shown). Equivalent results were obtained using an HA-tagged cic transgene that rescues cic loss-of-function (see Materials and methods). In normal embryos, tagged Cic is excluded from both the anterior and posterior termini (Figure 5E and G). When introduced into a nosRC/nosBN mutant background, however, ectopic staining is found in the terminal nuclei of stage 4 embryos (Figure 5F and H), although not in those nuclei just adjacent to the pole cells. Note that exclusion of both endogenous and tagged Cic from the anterior pole also appears attenuated in osk and nos mutants (Figure 5F; see Discussion).
These results lead us to conclude that posterior group members act at the level of MAPK or downstream to it, to downregulate the Cic repressor complex (Figure 5I). Importantly, the ectopic accumulation of Cic could account for the reduced tll and hkb expression patterns observed in posterior group mutants.
Repression of tailless is alleviated in CtBP, groucho double-mutant germline clones
We next addressed yet another aspect of tll regulation. In gro or cic maternal mutants, repression of tll is hindered and its expression expands towards the middle of the embryo (Paroush et al, 1997; Jiménez et al, 2000). Notably, tll transcripts are never detected in the centre of these embryos (Figure 6B). What could be the reason for the lack of tll expression at the centre of the embryo, even when these essential repressors are removed? One possibility is that activators of tll are simply absent from this region. However, several other DNA-binding repressors have been implicated in tll silencing (Liaw et al, 1995; Chen et al, 2002), raising the possibility that repressor activities are redundant. Consistent with the latter possibility, we find that the simultaneous removal of both Gro and the C-terminal Binding Protein (CtBP), a second global corepressor that functions at early stages of embryogenesis (Nibu et al, 1998; Poortinga et al, 1998), brings about the uniform (albeit weak) expression of tll throughout the embryo (Figure 6C). This implies that the tll promoter is subjected to multiple repressor mechanisms, at least some of which are CtBP-dependent (and perhaps novel), that inhibit tll expression from spreading to the middle regions of the embryo, thus allowing correct abdominal development.
Figure 6.
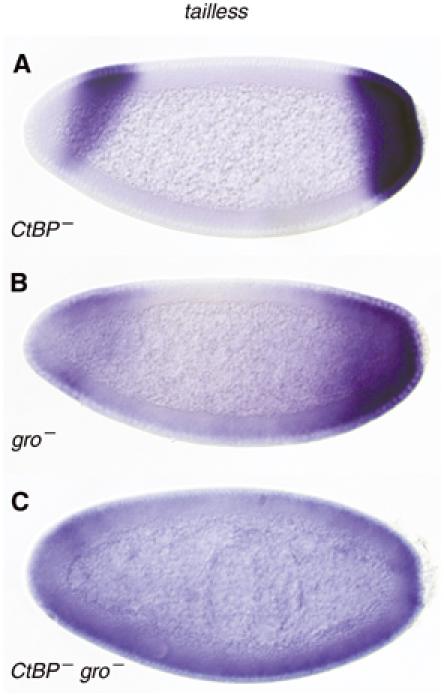
tailless is expressed throughout CtBP, groucho double-mutant germline clones. In either CtBPP1590 (A) or groBX22 (B) single-mutant germline clones, the posterior tll expression domain expands anteriorly (only slightly so in embryos devoid of maternal CtBP), but never reaches the centre. (C) Only when both CtBP and gro are concomitantly removed (CtBPP1590 groBX22) is tll uniformly expressed throughout the embryo (although not in pole cells).
Discussion
Terminal gap gene expression must be tightly regulated for the correct specification of terminal cell fates at the nonsegmented poles. Clearly, the Tor pathway plays a key role in driving tll and hkb transcription, given that terminal gap genes are not expressed at the posterior end of terminal group mutants, and as a result terminal structures such as the FK do not form. In this paper, we reveal a novel biological role for the maternal posterior system, showing that members of this group, in particular Nos, positively regulate transcription of the zygotic subordinate genes of the terminal system. We find that TREs in the tll upstream regulatory region, which are derepressed in cic mutants, also respond to alterations in maternal osk dosage, and that the Cic repressor is not excluded from the termini of posterior group mutants. Our results are consistent with the posterior system feeding into the Tor signalling pathway, upstream of or at the level of the Cic repressor (Figure 5I). We suggest that the concerted activities of both the terminal and posterior systems, in their spatially overlapping zones of action, generate accurate domains of terminal gap gene expression at the posterior.
Crosstalk between maternal coordinate systems
It was originally proposed that the four maternal systems that pattern the early Drosophila embryo act largely independently of each other (St Johnston and Nusslein Volhard, 1992). Recent work, however, demonstrated interactions between the Tor pathway and the anterior and D/V systems. For example, tll has been shown to respond to the anterior determinant Bicoid (Bcd) even when Tor signalling is genetically blocked. Indeed, cis-acting DNA elements responsive to these three maternal systems have been found in the tll upstream regulatory region (Liaw and Lengyel, 1993). Our results now link the terminal and posterior systems, previously thought to be independent of each other, in terminal gap gene regulation, reinforcing the idea that maternal systems that pattern the early embryo act in a coordinated manner.
Why has the positive input, by posterior group genes into terminal patterning, been largely overlooked to date? Classical segmentation studies mostly involved phenotypic analyses at the cuticular level. For this reason, and when taking into account the primary contribution of the terminal system, the delicate input by the posterior group has gone unnoticed. Thus, the unextended FK that develops in posterior group mutant background, which may arise from decreased terminal gap gene expression, had largely been attributed to pleiotropic effects arising from abdominal defects. We have been able to detect the relatively subtle changes in tll and hkb gene expression patterns only by investigating terminal gap gene regulation at the molecular level. In fact, at least one other molecular study had previously reported reduced terminal gap gene expression in osk mutant embryos (Brönner and Jackle, 1991).
Posterior group input impinges on RTK signalling
One emerging concept is that, for the refinement of the expression levels and spatial extents of RTK signalling targets, it is also imperative to integrate accurately information originating from other, non-RTK sources (Simon, 2000). In many cases this integration occurs at the level of target gene enhancers, with various effectors of distinct signalling pathways binding to specific DNA elements to regulate transcription (Flores et al, 2000; Halfon et al, 2000; Xu et al, 2000). For example, D-Pax2 expression in the cone and pigment cells of the developing eye is regulated by effectors of the EGFR RTK pathway, such as Pointed P2 and Yan, and also by the Notch signalling component Suppressor of Hairless, as well as by the transcription factor Lozenge (Flores et al, 2000). Here we have shown that terminal gap gene expression requires not only Tor RTK pathway activity but also a contribution from the posterior system. In this instance, inputs from these two maternal coordinate systems are interpreted and linked not at the level of terminal gap gene promoters but at the level of the Cic repressor. Thus, Cic functions as an integrator of multiple regulatory inputs, with both the posterior and terminal systems acting to relieve transcriptional silencing mediated by this repressor.
Regulation of terminal gap gene expression by Nanos
Surprisingly, we find that anterior tll and hkb expression is also reduced in posterior group mutants (Figures 1 and 2). Similarly, others have reported prolonged bcd expression and head defects in pum mutants (Gamberi et al, 2002). We can only speculate that low levels of Osk and Nos, which escape translational repression, similarly regulate terminal gap gene expression via Cic removal at the anterior. In accordance with this, the dismissal of Cic from the anterior pole of posterior group mutants is also ineffective (Figure 5F).
How does Nos, which has been assigned the role of a translational repressor, positively regulate tll and hkb transcription? Our results suggest that Nos does so indirectly, by downregulating the accumulation of the Cic repressor at the termini. The exact mechanism by which the Tor pathway mediates the exclusion of Cic from terminal regions has not been established, but one model argues that phosphorylation of Cic by MAPK causes degradation of the protein, as in the case of Yan (Rebay and Rubin, 1995; Jiménez et al, 2000). Thus, Nos could be affecting this process in one of several possible ways, at the level or downstream of MAPK. For example, Nos could be facilitating the translocation of phosphorylated MAPK into the nucleus. In posterior group mutants, then, activated MAPK would remain in the cytoplasm rather than enter the nucleus, impeding Cic phosphorylation and degradation. Alternatively, Nos may be modulating MAPK activity, or regulating adaptor proteins that promote Cic phosphorylation by nuclear MAPK. Nos may also be controlling the translation of factors that are involved in the nuclear trafficking (import/export) or degradation of Cic, or perhaps may even be acting on the cic message itself. Future studies will distinguish between these possibilities, and may shed new light on the molecular mechanisms underlying Nos' role in other developmental processes, for example, the establishment/maintenance of transcriptional quiescence in pole cells (Deshpande et al, 1999).
Multiple layers of terminal gap gene regulation
We view the positive input by the posterior group genes as evolving to modulate terminal pathway activity, merging with other varied modes of Tor regulation to ultimately ensure accurate tll and hkb expression and, consequently, precise cell fate determination.
The Tor signal transduction pathway is under multiple tiers of regulation, outside and inside the nucleus. For instance, internalisation and trafficking of the activated Tor receptor to the lysosome for degradation attenuates the signal, as evident by the spatial broadening and temporal prolonging of Tor activation in mutants for hrs, a component of the endosomal recycling machinery (Lloyd et al, 2002). Yet another level of control is provided by the tyrosine phosphatase corckscrew, which sharpens the gradient of Tor activity (Cleghon et al, 1998). Additionally, multiple cytoplasmic adaptor proteins take part in transducing the Tor signal (Luschnig et al, 2000), conceivably buffering against surplus or deficiency in signalling.
In the nucleus, tll and hkb are subjected to silencing by several repressors. Derepression of tll is observed in grainy-head and tramtrack69 (ttk69) mutants, and the proteins encoded by these genes bind tll promoter sequences (Liaw et al, 1995; Chen et al, 2002). Cic and Gro appear to play a leading role in terminal gap gene silencing, given that mutations in cic and gro bring about a significant expansion of the tll and hkb expression domains (Figure 6B) (Paroush et al, 1997; Jiménez et al, 2000). Intriguingly, however, tll expression never reaches the middle of the embryo in these mutants. We find that tll is uniformly expressed, albeit weakly, throughout the embryo only when both the developmental corepressors Gro and CtBP are removed concomitantly (Figure 6C). This broadened tll expression likely stems from the fact that there is a redundancy in the activities that normally restrict terminal gap gene transcription from inappropriately spreading into the central portion of the embryo; by jointly removing the Gro and CtBP coregulators, activity of the above repressors is compromised. Alternatively, CtBP might be acting in conjunction with a novel, unidentified repressor that prevents tll transcription in the middlemost region of the embryo.
So what is the purpose of the input by the posterior group genes into tll and hkb transcription? Quantitative differences in Tor receptor activity have to be eventually interpreted and translated into distinct cell fates at the termini. Strong Tor activation induces both hkb and tll expression, whereas weaker Tor activation only brings about tll expression. We surmise that the precision endowed by the Tor RTK cascade may not suffice for the complex patterning of the termini, given that mere two-fold fluctuations in Tor signalling result in defective embryonic development (Strecker et al, 1989; Furriols et al, 1996; Greenwood and Struhl, 1997). For example, mutants with reduced Tor RTK activity show partial tll expression and the complete loss of hkb. These mutants consequently develop incomplete terminal structures and die at the larval stage. Conversely, overactivation of the Tor pathway leads to anterior expansion of the posterior tll expression domain, perturbing segmentation in central body parts, likely as a result of downregulation of abdominal gap genes by the Tll protein (Steingrímsson et al, 1991; Paroush et al, 1997). Thus, the precise spatial confinement of terminal gap gene expression domains requires the coordinated integration of regulatory inputs, coming from two maternal systems and converging on the same effector protein, Cic.
Materials and methods
Fly culture
Flies were cultured and crossed on standard yeast–cornmeal–molasses–malt extract–agar medium at 25°C.
Fly stocks and germline clones
The following mutant alleles were used: osk166, osk54, osk346, vasPD, vas011, tudWC, tudB36, gclΔ49, nosRC, nosBN, nosL7, nos-tub3′UTR, pumET1, pum680, torY9 and tsl691 (FlyBase). Df(3R)p-XT103 and In(3R)Msc are deficiencies that uncover the osk and pum loci, respectively (FlyBase). OregonR and yw stocks served as wild-type controls.
Flies carrying the P1, G22 and K11 tor-RE-lacZ transgenes (Liaw et al, 1995; Rudolph et al, 1997) were a kind gift from Judith Lengyel. For increasing maternal osk dosage, two lines were used: oskAk (4xosk), kindly provided by Anne Ephrussi (Ephrussi and Lehmann, 1992), and Dp(3;3)bicS, a small tandem duplication of the osk locus, which will be described in detail elsewhere.
Embryos expressing HA-tagged Cic were obtained from females carrying a modified version of the rescuing construct containing the complete cic coding sequence (Jiménez et al, 2000). The tagged transgene is identical to the parental construct, except that it includes three tandem copies of the HA (also known as Flu) epitope inserted at the C-terminal region of the protein. Details on the construction of the plasmid are available on request.
Embryos lacking maternal gro and/or CtBP activities were derived from mosaic gromat− (groE48 or groBX22) and CtBPmat− (CtBPP1590) single- and double-mutant germ lines, obtained using the FLP-DFS technique (Chou et al, 1993). Standard recombination techniques were used to generate the double-mutant chromosomes FRT[82B] CtBPP1590 groBX22, FRT[82B] CtBPP1590 groE48 and bicS 3014tsl691.
In situ hybridisation and antibody staining of Drosophila embryos
Wild-type or mutant embryos (1–3.5 h collections) were dechorionated in bleach and fixed in 4% formaldehyde/PBS/heptane for 15–20 min. Expression patterns were visualised by whole-mount in situ hybridisation using digoxygenin-UTP-labelled antisense RNA probes and anti-digoxygenin antibodies conjugated to alkaline phosphatase (Boehringer Mannheim).
Immunohistochemical detection of activated MAPK, in freshly fixed embryos (10% formaldehyde/PBS/heptane buffer), was achieved with preabsorbed monoclonal antibodies against the diphosphorylated form of Erk (dpERK) (1:100; Sigma). Secondary antibodies were conjugated to biotin (1:2000; Chemicon), and visualised by addition of streptavidin alkaline phosphatase (1:500; Chemicon). For viewing the endogenous Cic protein, a preabsorbed polyclonal antibody (1:1000) was used as previously described (Jiménez et al, 2000). In this case, a preabsorbed alkaline phosphatase-coupled secondary antibody was utilised (1:1500; Jackson). HA-tagged Cic was followed by using an anti-HA monoclonal antibody (1:200; Convance). Incubations of primary and secondary antibodies were performed in 0.2% NaN3. Blocking was performed in a 0.1% PBS, 0.1% Tween, 10% BSA, 5% normal goat serum and 0.2% NaN3 buffer.
In Table I, the spatial extent of the posterior terminal gap gene expression domains, in embryos at stages 4 (syncytial blastoderm) and 5 (cellular blastoderm), was calculated as follows: for a given embryo, tll and hkb expression domains were measured, then divided by the embryo's length to calculate the domain's size as percent of EL. Values were averaged for the respective mutant and corresponding wild-type embryos, and are represented as a mutant to wild-type ratio. Thus for each genetic background, values below or above 1 represent a reduction or expansion of gap gene expression domains, respectively, while ratios of around 1 denote normal extent. Please note that differences between the strengths of alleles used for each mutation likely account for the varying degrees of reduction/expansion of tll and hkb expression domains.
Cuticle preparation
Unhatched larvae (24–48 h) were dechorionated in bleach, transferred into 50% lactic acid and 50% hoyers medium and baked at 70°C for 2 h.
Acknowledgments
We thank members of our laboratory for continued help and encouragement during this project, in particular Yuval Cinnamon and Rona Grossman for technical assistance. We also thank Peleg Hasson, Benny Shilo, Talila Volk and Joel Yisraeli for their insightful comments on the manuscript, and Jordi Casanova, Claude Desplan, Anne Ephrussi, Marc Furiolls, Liz Gavis, Tom Jongens, Iris Koch, Ruth Lehmann, Judith Lengyel, Willis Li, Gwo-Jen Liaw, Norbert Perrimon, Kajan Ratnakumar, Benny Shilo, Steve Small and Uwe Walldorf for DNA constructs, antibodies, reagents and fly stocks. The work was supported by grants from the Israel Cancer Research Fund, Israel Science Foundation (116/00-1), United States–Israel Binational Science Foundation (96-108) and the Jan M and Eugenia Król Charitable Foundation. ZP is a Joseph H and Belle R Braun Lecturer in Medicine.
We wish to dedicate this paper to the dear memory of Judith Lengyel, a true friend and colleague, who supported us throughout this study.
References
- Amikura R, Hanyu K, Kashikawa M, Kobayashi S (2001) Tudor protein is essential for the localization of mitochondrial RNAs in polar granules of Drosophila embryos. Mech Dev 107: 97–104 [DOI] [PubMed] [Google Scholar]
- Arrizabalaga G, Lehmann R (1999) A selective screen reveals discrete functional domains in Drosophila Nanos. Genetics 153: 1825–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brönner G, Chu-LaGraff Q, Doe CQ, Cohen B, Weigel D, Taubert H, Jäckle H (1994) Sp1/egr-like zinc-finger protein required for endoderm specification and germ-layer formation in Drosophila. Nature 369: 664–668 [DOI] [PubMed] [Google Scholar]
- Brönner G, Jackle H (1991) Control and function of terminal gap gene activity in the posterior pole region of the Drosophila embryo. Mech Dev 35: 205–211 [DOI] [PubMed] [Google Scholar]
- Casali A, Casanova J (2001) The spatial control of Torso RTK activation: a C-terminal fragment of the Trunk protein acts as a signal for Torso receptor in the Drosophila embryo. Development 128: 1709–1715 [DOI] [PubMed] [Google Scholar]
- Casanova J (1990) Pattern formation under the control of the terminal system in the Drosophila embryo. Development 110: 621–628 [DOI] [PubMed] [Google Scholar]
- Casanova J, Struhl G (1989) Localized surface activity of torso, a receptor tyrosine kinase, specifies terminal body pattern in Drosophila. Genes Dev 3: 2025–2038 [DOI] [PubMed] [Google Scholar]
- Casanova J, Struhl G (1993) The torso receptor localizes as well as transduces the spatial signal specifying terminal body pattern in Drosophila. Nature 362: 152–155 [DOI] [PubMed] [Google Scholar]
- Chen G, Courey AJ (2000) Groucho/TLE family proteins and transcriptional repression. Gene 249: 1–16 [DOI] [PubMed] [Google Scholar]
- Chen YJ, Chiang CS, Weng LC, Lengyel JA, Liaw GJ (2002) Tramtrack69 is required for the early repression of tailless expression. Mech Dev 116: 75–83 [DOI] [PubMed] [Google Scholar]
- Chou TB, Noll E, Perrimon N (1993) Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development 119: 1359–1369 [DOI] [PubMed] [Google Scholar]
- Cleghon V, Feldmann P, Ghiglione C, Copeland TD, Perrimon N, Hughes DA, Morrison DK (1998) Opposing actions of CSW and RasGAP modulate the strength of Torso RTK signaling in the Drosophila terminal pathway. Mol Cell 2: 719–727 [DOI] [PubMed] [Google Scholar]
- Deshpande G, Calhoun G, Yanowitz JL, Schedl PD (1999) Novel functions of nanos in downregulating mitosis and transcription during the development of the Drosophila germline. Cell 99: 271–281 [DOI] [PubMed] [Google Scholar]
- Duffy JB, Perrimon N (1994) The torso pathway in Drosophila: lessons on receptor tyrosine kinase signaling and pattern formation. Dev Biol 166: 380–395 [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Lehmann R (1992) Induction of germ cell formation by oskar. Nature 358: 387–392 [DOI] [PubMed] [Google Scholar]
- Flores GV, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U (2000) Combinatorial signaling in the specification of unique cell fates. Cell 103: 75–85 [DOI] [PubMed] [Google Scholar]
- Furriols M, Casanova J (2003) In and out of Torso RTK signalling. EMBO J 22: 1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furriols M, Sprenger F, Casanova J (1996) Variation in the number of activated torso receptors correlates with differential gene expression. Development 122: 2313–2317 [DOI] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo BZ (1997a) In situ activation pattern of Drosophila EGF receptor pathway during development. Science 277: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo BZ (1997b) MAP kinase in situ activation atlas during Drosophila embryogenesis. Development 124: 3535–3541 [DOI] [PubMed] [Google Scholar]
- Gamberi C, Peterson DS, He L, Gottlieb E (2002) An anterior function for the Drosophila posterior determinant Pumilio. Development 129: 2699–2710 [DOI] [PubMed] [Google Scholar]
- Gavis ER, Lehmann R (1994) Translational regulation of nanos by RNA localization. Nature 369: 315–318 [DOI] [PubMed] [Google Scholar]
- Greenwood S, Struhl G (1997) Different levels of Ras activity can specify distinct transcriptional and morphological consequences in early Drosophila embryos. Development 124: 4879–4886 [DOI] [PubMed] [Google Scholar]
- Halfon MS, Carmena A, Gisselbrecht S, Sackerson CM, Jimenez F, Baylies MK, Michelson AM (2000) Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell 103: 63–74 [DOI] [PubMed] [Google Scholar]
- Hulskamp M, Pfeifle C, Tautz D (1990) A morphogenetic gradient of hunchback protein organizes the expression of the gap genes Kruppel and knirps in the early Drosophila embryo. Nature 346: 577–580 [DOI] [PubMed] [Google Scholar]
- Irish V, Lehmann R, Akam M (1989) The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature 338: 646–648 [DOI] [PubMed] [Google Scholar]
- Jiménez G, Guichet A, Ephrussi A, Casanova J (2000) Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev 14: 224–231 [PMC free article] [PubMed] [Google Scholar]
- Jongens TA, Hay B, Jan LY, Jan YN (1992) The germ cell-less gene product: a posteriorly localized component necessary for germ cell development in Drosophila. Cell 70: 569–584 [DOI] [PubMed] [Google Scholar]
- Kraut R, Levine M (1991) Spatial regulation of the gap gene giant during Drosophila development. Development 111: 601–609 [DOI] [PubMed] [Google Scholar]
- Liaw GJ, Lengyel JA (1993) Control of tailless expression by bicoid, dorsal and synergistically interacting terminal system regulatory elements. Mech Dev 40: 47–61 [DOI] [PubMed] [Google Scholar]
- Liaw GJ, Rudolph KM, Huang JD, Dubnicoff T, Courey AJ, Lengyel JA (1995) The torso response element binds GAGA and NTF-1/Elf-1, and regulates tailless by relief of repression. Genes Dev 9: 3163–3176 [DOI] [PubMed] [Google Scholar]
- Lloyd TE, Atkinson R, Wu MN, Zhou Y, Pennetta G, Bellen HJ (2002) Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell 108: 261–269 [DOI] [PubMed] [Google Scholar]
- Lu X, Perkins LA, Perrimon N (1993) The torso pathway in Drosophila: a model system to study receptor tyrosine kinase signal transduction. Dev Suppl 47–56 [PubMed] [Google Scholar]
- Luschnig S, Krauss J, Bohmann K, Desjeux I, Nusslein Volhard C (2000) The Drosophila SHC adaptor protein is required for signaling by a subset of receptor tyrosine kinases. Mol Cell 5: 231–241 [DOI] [PubMed] [Google Scholar]
- Murata Y, Wharton RP (1995) Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80: 747–756 [DOI] [PubMed] [Google Scholar]
- Nibu Y, Zhang H, Levine M (1998) Interaction of short-range repressors with Drosophila CtBP in the embryo. Science 280: 101–104 [DOI] [PubMed] [Google Scholar]
- Paroush Z, Finley RL Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D (1994) Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79: 805–815 [DOI] [PubMed] [Google Scholar]
- Paroush Z, Wainwright SM, Ish-Horowicz D (1997) Torso signalling regulates terminal patterning in Drosophila by antagonising Groucho-mediated repression. Development 124: 3827–3834 [DOI] [PubMed] [Google Scholar]
- Pignoni F, Baldarelli RM, Steingrímsson E, Diaz RJ, Patapoutian A, Merriam JR, Lengyel JA (1990) The Drosophila gene tailless is expressed at the embryonic termini and is a member of the steroid receptor superfamily. Cell 62: 151–163 [DOI] [PubMed] [Google Scholar]
- Pignoni F, Steingrimsson E, Lengyel JA (1992) bicoid and the terminal system activate tailless expression in the early Drosophila embryo. Development 115: 239–251 [DOI] [PubMed] [Google Scholar]
- Poortinga G, Watanabe M, Parkhurst S (1998) Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J 17: 2067–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay I, Rubin GM (1995) Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell 81: 857–866 [DOI] [PubMed] [Google Scholar]
- Rongo C, Gavis ER, Lehmann R (1995) Localization of oskar RNA regulates oskar translation and requires Oskar protein. Development 121: 2737–2746 [DOI] [PubMed] [Google Scholar]
- Rongo C, Lehmann R (1996) Regulated synthesis, transport and assembly of the Drosophila germ plasm. Trends Genet 12: 102–109 [DOI] [PubMed] [Google Scholar]
- Rudolph KM, Liaw GJ, Daniel A, Green P, Courey AJ, Hartenstein V, Lengyel JA (1997) Complex regulatory region mediating tailless expression in early embryonic patterning and brain development. Development 124: 4297–4308 [DOI] [PubMed] [Google Scholar]
- Simon MA (2000) Receptor tyrosine kinases: specific outcomes from general signals. Cell 103: 13–15 [DOI] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP (1999) Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev 13: 2704–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger F, Nusslein Volhard C (1992) Torso receptor activity is regulated by a diffusible ligand produced at the extracellular terminal regions of the Drosophila egg. Cell 71: 987–1001 [DOI] [PubMed] [Google Scholar]
- Sprenger F, Stevens LM, Nüsslein-Volhard C (1989) The Drosophila gene torso encodes a putative receptor tyrosine kinase. Nature 338: 478–483 [DOI] [PubMed] [Google Scholar]
- St Johnston D, Nusslein Volhard C (1992) The origin of pattern and polarity in the Drosophila embryo. Cell 68: 201–219 [DOI] [PubMed] [Google Scholar]
- Steingrímsson E, Pignoni F, Liaw GJ, Lengyel JA (1991) Dual role of the Drosophila pattern gene tailless in embryonic termini. Science 254: 418–421 [DOI] [PubMed] [Google Scholar]
- Strecker TR, Halsell SR, Fisher WW, Lipshitz HD (1989) Reciprocal effects of hyper- and hypoactivity mutations in the Drosophila pattern gene torso. Science 243: 1062–1066 [DOI] [PubMed] [Google Scholar]
- Strecker TR, Kongsuwan K, Lengyel JA, Merriam JR (1986) The zygotic mutant tailless affects the anterior and posterior ectodermal regions of the Drosophila embryo. Dev Biol 113: 64–76 [DOI] [PubMed] [Google Scholar]
- Strecker TR, Merriam JR, Lengyel JA (1988) Graded requirement for the zygotic terminal gene, tailless, in the brain and tail region of the Drosophila embryo. Development 102: 721–734 [DOI] [PubMed] [Google Scholar]
- Weigel D, Jurgens G, Klingler M, Jäckle H (1990) Two gap genes mediate maternal terminal pattern information in Drosophila. Science 248: 495–498 [DOI] [PubMed] [Google Scholar]
- Xu C, Kauffmann RC, Zhang J, Kladny S, Carthew RW (2000) Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell 103: 87–97 [DOI] [PubMed] [Google Scholar]


