Abstract
The molecular motor, myosin, undergoes conformational changes in order to convert chemical energy into force production. Based on kinetic and structural considerations, we assert that three crystal forms of the myosin V motor delineate the conformational changes that myosin motors undergo upon detachment from actin. First, a motor domain structure demonstrates that nucleotide-free myosin V adopts a specific state (rigor-like) that is not influenced by crystal packing. A second structure reveals an actomyosin state that favors rapid release of ADP, and differs from the rigor-like state by a P-loop rearrangement. Comparison of these structures with a third structure, a 2.0 Å resolution structure of the motor bound to an ATP analog, illuminates the structural features that provide communication between the actin interface and nucleotide-binding site. Paramount among these is a region we name the transducer, which is composed of the seven-stranded β-sheet and associated loops and linkers. Reminiscent of the β-sheet distortion of the F1-ATPase, sequential distortion of this transducer region likely controls sequential release of products from the nucleotide pocket during force generation.
Keywords: β-sheet distortion, cytoskeleton motility, myosin motor, X-ray crystallography
Introduction
Myosin V functions as a two-headed processive actin-based motor owing to a number of structural and kinetic differences from conventional myosin (myosin II) (De La Cruz et al, 1999; Mehta et al, 1999). Each head has a long ‘lever arm' that allows myosin V to walk hand-over-hand with 36 nm steps along actin filaments (Forkey et al, 2003; Yildiz et al, 2003). The kinetic properties of this motor allow processivity (multiple steps along F-actin filament without detaching), which involves the heads remaining bound strongly to actin in an ADP state for the majority of the overall ATPase cycle (high duty ratio). The structural alterations (chemo-mechanical coupling) in the myosin motor that occur upon actin binding and product release (Pi followed by MgADP) are communicated to the long myosin V lever arm, generating a swing of approximately 25 nm. Although the kinetics and lever arm length of myosin V are unusual, its overall mechanism for force generation is thought to be common to all myosins (summarized in Figure 1), and is known as the lever arm hypothesis (Geeves and Holmes, 1999).
Figure 1.
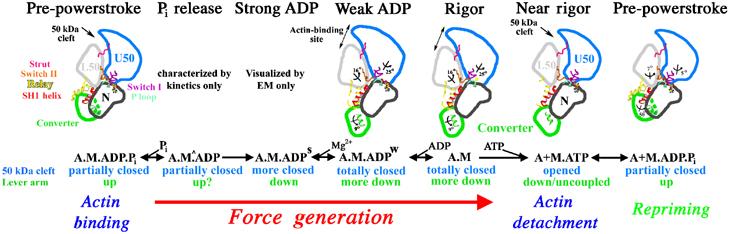
Chemo-mechanical transduction in the actin–myosin ATPase cycle. Structural transitions in the actomyosin ATPase cycle are depicted in terms of characterized subdomain positions. For each state, the biochemical composition, the status of the large 50 kDa cleft and lever arm position are noted. The depiction begins with the pre-powerstroke bound to actin. An uncharacterized structural isomerization (seen kinetically (Rosenfeld and Sweeney, 2004), and denoted as M^) opens an exit route for phosphate. This is followed by an essentially irreversible isomerization (Rosenfeld and Sweeney, 2004) to create a state seen in cryo-EM (Whittaker et al, 1995) and characterized kinetically (strong ADP binding) (Rosenfeld et al, 2000). Most of force generation and lever arm movement on actin are partitioned between these two as yet unresolved structural transitions. Based on the new structure presented in this paper (weak ADP binding; Figure 7), the next isomerization (Rosenfeld et al, 2000) destroys Mg2+ binding, has a lever arm swing associated with it and greatly weakens ADP affinity. ADP then dissociates, forming the rigor state. Binding of ATP to the rigor state opens the 50 kDa cleft, causing myosin to dissociate from actin in the post-rigor state. The post-rigor state isomerizes into the pre-powerstroke conformation, repriming the lever arm and allowing hydrolysis of ATP. The hydrolysis products are trapped until the myosin rebinds to actin, which induces the product release transitions. Note that the attributes of the myosin structural states characterized to date at high resolution are described in Supplementary Table 7.
Prior to our publication of a nucleotide-free myosin V crystal structure (1OE9; Coureux et al, 2003), all other high-resolution myosin structures appeared to represent weak actin-binding states, and therefore yielded limited information about the mechanical coupling. Based on kinetic studies, we argued that this state was representative of the structure of the final force-generating state in the actomyosin cycle (the rigor state). We compared this structure to the high-resolution structures of myosin II that were trapped in states that interact weakly with actin (Fisher et al, 1995; Smith and Rayment, 1996). The first comparison was with the pre-powerstroke or transition state (crystals with ATP or ADP.Pi analogs), a state in which ATP is hydrolyzed to create a ‘primed' position of the myosin lever arm and represents the beginning of the force-generating cycle. The second state to which the myosin V was compared was the near-rigor state, which represents the ATP state of myosin that rapidly forms as ATP binds to myosin in the actomyosin rigor complex and causes the myosin to dissociate from actin. Note that the pre-powerstroke and near-rigor states have also been called closed and open states, respectively (Geeves and Holmes, 1999), in reference to the conformation of switch II. This nomenclature is however too restrictive since several distinct conformations of switch II must be adopted in the motor cycle as demonstrated by the rigor-like state of myosin V (PDB code: 1OE9). Since the near-rigor state is ‘near' rigor only in terms of lever arm position, but corresponds in the cycle to the state that follows rigor and allows dissociation of the rigor complex, it has recently been renamed the post-rigor state. A table comparing the essential characteristics of the myosin structural states characterized to date at high resolution is provided as Supplementary data. In summary: post-rigor=near-rigor=open; pre-powerstroke=transition-state=closed.
In order to demonstrate that this new rigor-like state of myosin V was not influenced by crystal packing forces, we crystallized the myosin V motor domain, lacking all IQ motifs (PDB code: 1W8J) in the absence of nucleotide. Even though the crystal contacts and packing were different from our earlier published crystals of the motor domain plus light chain (MDE), the motor domain structures were identical. We also sought to crystallize the myosin V motor with an ATP analog bound, which is essential for a detailed understanding of the structural changes associated with MgATP-induced dissociation of the actin–myosin complex. Furthermore, this structure was needed to show that myosin V does populate the same structural states as other myosins, but likely with minor isoform-specific modifications that underlie the differences in its kinetic tuning. Herein, we report the crystal structure of myosin V MDE in the so-called post-rigor state (which contains MgADP.BeFx at the active site) solved at 2 Å resolution (PDB code: 1W7J). By comparing this new structure to the post-rigor structure of myosin II and the rigor-like structure of myosin V (at 2 Å resolution), new insights into the myosin motor mechanism have emerged, as well as indications of isoform-specific tuning within the myosin superfamily.
Our initial comparisons of the subdomain positions of the myosin V rigor-like state to those found in the post-rigor and pre-powerstroke states revealed the general features of the subdomain rearrangements that must be driven by myosin binding to actin (Coureux et al, 2003). The motor domain of the myosin molecule can be most simply described as being composed of subdomains and connectors between them. The subdomains move as units, subject to constraints imposed by the rearrangements of the connectors or ‘joints' (Houdusse et al, 1999). As shown in Figure 1, the subdomains are the N-terminal, upper 50 kDa (U50), lower 50 kDa (L50) subdomains and the converter (to which the lever arm is attached). Also depicted in this figure are the connectors that facilitate rearrangement of these subdomains, which are referred to as the ‘strut', switch II, relay and SH1 helix. These motor domain rearrangements explained the fact that strong binding to actin and strong binding to ATP are reciprocal. That is, the structure revealed that actin must induce closure of the large cleft in the myosin molecule between the U50 and L50 subdomains and in doing so moves the elements of the nucleotide-binding pocket into positions that can no longer strongly coordinate nucleotide. When ATP binds, the post-rigor state is formed by the nucleotide-binding elements being repositioned in order to coordinate MgATP, and in doing so the cleft is reopened, which disrupts the actin–myosin interface causing dissociation. This provides a general outline of the mechanism of chemo-mechanical coupling in the actomyosin force-producing cycle.
In this paper, we provide a detailed description of these structural changes, based on comparisons with our new structure of myosin V in the post-rigor state. We call attention to a key region of the molecule, which we name the transducer. It allows chemo-mechanical transduction induced by actin binding. We also introduced MgADP into myosin V rigor-like MDE crystals to observe possible rearrangements in the nucleotide-binding elements. We assert that this has revealed the structure of an actin-bound myosin state to which ADP binds weakly (PDB code: 1W7I). Such a kinetic state has been described for a number of members of the myosin superfamily. Its existence has implications for the manner in which MgADP is released from actomyosin.
Results
Overview of the rigor-like state
Crystallization of a nucleotide-free motor domain of myosin V generated a 2.7 Å structure (PDB code: 1W8J). The four molecules in the asymmetric unit, each with different crystal contacts from each other and from those of the previously published motor domain plus essential light chain (MDE), are in the same rigor-like state that we previously described for MDE (Coureux et al, 2003). Taken together, the four motor domains of the MD crystal and the MDE motor domain superimpose very well with an r.m.s. deviation for 592 Cα atoms of respectively 0.522, 0.458, 0.753 and 0.815 Å for molecules A, B, C and D of the MD model (Figure 2). Thus, this structural state, which we referred to as the rigor-like (or closed cleft) state of myosin V, is not affected by crystal packing. The hallmarks of this unique structural state are: (1) closure of the large cleft between the upper and lower 50 kDa subdomains; (2) specific interactions between elements of the active site that prevent high-affinity nucleotide binding; and (3) a switch II position that allows interaction with the N-terminal subdomain and the SH1 helix to rigidly couple the subdomains. In solution, nucleotide-free myosin V is unique among all myosins that have been examined in that its binding to actin appears to be diffusion limited, suggesting that it is in a structural state essentially equivalent to the rigor state on actin.
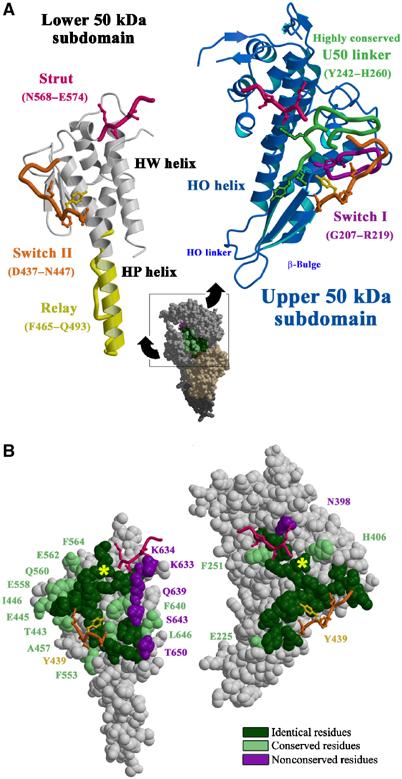
Figure 2.

Overall view of the myosin V structures. The myosin V MDE NF structure (Coureux et al, 2003) (red) and the myosin V MDE ADP-BeFx structure (green) are compared to various myosin V and Dictyostelium myosin II structures by superimposing atoms of the lower 50 kDa subdomains so that the degree of cleft closure between the U50 and L50 subdomains can be visualized. To compare the structural states of these motor domains, we have selected 393 Cα atoms corresponding to 117, 167 and 109 atoms of the N-terminal, U50 and L50 subdomains. Using these atoms, the r.m.s. differences between myosin V MDE NF (in red) and myosin V MD NF (in gray) are only 0.507, 0.407, 0.520 and 0.593 Å for molecules A, B, C and D, respectively, while the r.m.s. differences between myosin V MDE NF and myosin II MD NF (Reubold et al, 2003) (in blue) is 1.298 Å. Using these same atoms, an r.m.s. difference of only 0.840 Å is obtained between the two post-rigor states myosin V MDE ADP-BeFx and myosin II MD ADP-BeFx (in purple; PDB code: 1MMD), while an r.m.s. difference of 3.047 Å is obtained for the two myosin V MDE structures in the rigor-like (in red) and post-rigor state (in green). Note that the selected Cα residues are (70–78, 85–94, 97–102, 107–116, 119–128, 135–161, 167–183, 655–668, 673–686), (199–224, 237–265, 304–340, 354–364, 366–380, 388–422, 575–582, 587–592) and (440–467, 493–501, 504–513, 519–531, 535–540, 546–569, 635–653) for the N-terminal, U50 and L50 subdomains, respectively.
While a recently published structure of Dictyostelium nucleotide-free myosin II (DdNF) (PDB code: 1Q5G; Reubold et al, 2003) has a β-sheet geometry that is similar to that seen in the myosin V rigor-like structure (and quite distorted from that of either post-rigor or pre-powerstroke structures discussed below), none of the hallmark features listed above are seen in the DdNF structure. In particular, switch II is not in position to facilitate cleft closure. Thus, the cleft is only slightly more closed than in other Dictyostelium myosin II structures, and would be predicted to have a weak actin-binding affinity. This is consistent with kinetic studies on nucleotide-free Dictyostelium myosin II that suggest that it populates a weak actin-binding state, and not a rigor-like state in the absence of actin.
Myosin V complexed with MgADP.BeFx reveals the post-rigor structure of myosin V
Crystals of myosin V MDE complexed with MgADP.BeFx diffracted to 2.0 Å resolution (PDB code: 1W7J). As compared to the analogous structure of Dd myosin II MD (PDB code: 1MMD; Fisher et al, 1995), the subdomains are in essentially the same positions (superimpose with an r.m.s. deviation of 0.767 Å for 442 Cα atoms), with the exception of the converter (Figure 2). However, the converter is somewhat flexible in the post-rigor state, and thus its position in a crystal can be altered by packing. There are also nonconserved loops within the myosin superfamily found in different orientations in myosin V as compared to myosin II. The nucleotide coordination in the active site is the same as originally reported for Dd myosin II MD (PDB code: 1MMD; Fisher et al, 1995), the cleft is open and there is no rigid coupling between the subdomains.
Closure of the major cleft in myosin
The most striking difference in the post-rigor and rigor-like structures is the state of the major cleft between the 50 kDa subdomains. In the myosin II and V post-rigor structures, the cleft is found in a totally open conformation. In comparison, it is closed both near the actin interface and near the nucleotide-binding pocket in the rigor-like structures (see Supplementary Movie 1). The closed conformation in the rigor-like state is stabilized by a number of specific interactions not present in the post-rigor state involving a very conserved linker region (residues Y242–E256) of the U50 subdomain with the beginning of the HP and the HW helices of the L50 subdomain (see Figure 3). Partial cleft closure, near the nucleotide-binding pocket but not near actin, is seen in the pre-powerstroke states (Smith and Rayment, 1996) and the DdNF (Reubold et al, 2003) structures. Comparison of the closure of the cleft near switch II shows that different sets of interactions can be observed in this region depending on the switch II conformation, which differs in all these states. The unique closure near the actin-binding site seen in the rigor-like structures requires a conformational change in a connector between the U50 and L50 subdomains (the strut) that allows it to interact specifically with both subdomains (Coureux et al, 2003). The main change in conformation of the strut occurs for residue D570 (chicken myosin V sequence), which belongs to the helix HT in the pre-powerstroke and post-rigor structures but is elongated both in the rigor-like and the DdNF structures (Reubold et al, 2003). Thus, the strut conformational change alone is not sufficient to trigger cleft closure in the nucleotide-free Dictyostelium myosin II.
Figure 3.
Cleft closure. (A) The lower and upper 50 kDa subdomains of myosin V MDE have been pulled apart exposing the surface interactions allowing cleft closure. Note (in green on the right) the U50 highly conserved linker that interacts with the HW and HP helices (white on the left) of the L50 subdomain. Switch II (orange) and the strut (pink) are two connectors between the subdomains that help mediate the interactions between these two surfaces and they are shown on both sides. In particular, a hydrophobic residue of switch II (Y439, yellow ball and stick) is a serine or alanine in all myosin II isoforms. This difference may account in part for the difference in the kinetics of cleft closure for the two molecules. (B) A surface CPK representation of the residues involved in this interface is presented in the same orientation as in A). Depending on the conservation in the sequence of these residues in the myosin superfamily, different colors are used (absolutely conserved (green), conservative changes (pale green) and nonconserved (purple)). A yellow star indicates how to reposition the two surfaces to reconstruct the interface.
The specific conformations and interactions of the strut and switch II connectors are critical for the myosin V cleft closure, but a large number of direct interactions between the two 50 kDa subdomains also contribute to this closure (see Figure 3). Many of these interactions seen in myosin V correspond to conserved residues in the myosin superfamily. This led us to predict that in the true actomyosin rigor state of all myosins, the same interactions as seen in myosin V without actin will probably allow cleft closure near actin and the strut (Coureux et al, 2003). The importance of the strut in cleft closure has been verified by a mutagenesis study that demonstrated that alterations in the strut, including removal or repositioning of the D570 equivalent in Dictyostelium myosin II (D590), destroyed strong binding to actin (Sasaki et al, 2000).
However, there are a number of nonconserved interactions that may account for the ability of myosin V to maintain a closed cleft in the absence of actin and nucleotide (Figure 3). The lack of these interactions in Dictyostelium myosin II could in part account for the fact that it does not completely close the cleft in the absence of actin and nucleotide, while myosin V does. Many of these interactions may exist in myosin V to allow rapid cleft closure near the actin interface, which might promote phosphate release and a rapid transition from weak to strong actin-binding states. These kinetic changes from myosin II are necessary for the processivity of myosin V.
Control of subdomain positioning
Comparing the known myosin structures, it is becoming clear that rigid-body subdomain movements are allowed by sliding on hydrophobic patches. This sliding on ‘greasy patches' is a general feature of subdomain rearrangements and allows them to maintain interactions and yet move relative to each other. Two examples are the sliding of the L50 subdomain at the bottom of the 50 kDa cleft near the position of Tyr553 over the bend at the junction of the SH1 and SH2 helices (Figure 4a and Supplementary Movie 2), and the sliding at the relay/SH1 helix interface described in detail below (Figure 4b and Supplementary Movie 3). Another example is the differential interactions between the U50 and L50 subdomains near the nucleotide pocket in the pre-powerstroke versus the rigor-like structures, which implies that these subdomains will remain coupled as myosin progresses through the force-generating states on actin. However, this interface is broken in the state that dissociates myosin from actin (post-rigor). The hydrophobic surface of the seven-stranded β-sheet (shared by the U50 and N-terminal subdomains) provides another example of sliding on hydrophobic residues (Figure 5 and Supplementary Movie 4). This surface allows different interactions to be made with the HW helix of the L50 subdomain, which are critical for the motor domain cohesion. (All of these points are most easily visualized in the animations supplied as Supplementary data.)
Figure 4.
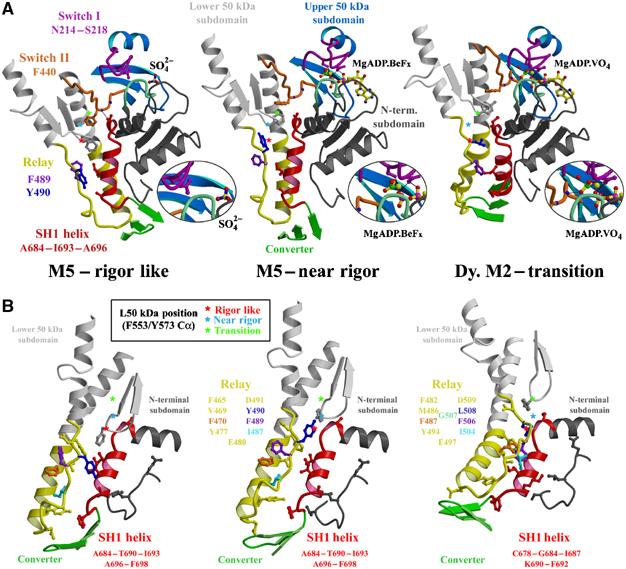
Switch II controls converter position by inducing conformational changes at the interface between the relay and the SH1 helix. (A) The region around switch II for myosin V MDE rigor-like and post-rigor states and Dictyostelium myosin II pre-powerstroke state are shown, positioned by overlaying the N-terminal subdomains. Changes in the switch II conformation that depend on the nucleotide state of the active site (see the magnification of this region in the inset) control the movements of the L50 subdomain (gray). In order to demonstrate this movement, stars denote the Cα position of one residue of the L50 subdomain (F553 in myosin V; Y573 in Dictyostelium myosin II) in each of the three states. The star is red, cyan and green for the rigor-like, post-rigor and pre-powerstroke states, respectively. Note that from the rigor-like position, the star moves ‘up' in the post-rigor state, changing the interface between the relay and the SH1 helix, but without drastically changing the overall conformation of the relay. This results in minimal change in position of the converter (green) and thus the lever arm. The additional upward movement seen in the pre-powerstroke state structure results in steric hindrance between the relay and the SH1 helix. To accommodate the clash, the relay helix bends and positions the converter and lever arm in the pre-powerstroke conformation. In the rigor-like structure, switch II directly interacts with the beginning of the SH1 helix, contributing, along with the greater relay–SH1 helix interactions, to stabilization of this region. This would likely create a more rigid coupling of the converter to the motor domain, which would be necessary to bear force in an actomyosin rigor state. (B) The interface between the SH1 helix and the relay is shown for myosin V rigor-like and post-rigor states and Dictyostelium myosin II pre-powerstroke state, positioned by overlaying the SH1 helix. Colored residues of the relay show how they change the orientation of their side chain to accommodate the sliding at the interface between the two connectors. Note, in particular, the differences between how the beginning of the SH1 helix interacts with hydrophobic side chains of the relay and the L50 subdomain while in the rigor-like state and how the beginning of the SH1 helix interacts with the L50 subdomain near residue F553. In the post-rigor state, the upward movement of this subdomain weakens these interactions leading to a state from which the SH1 helix may unwind more easily. In the pre-powerstroke state, a bending in the relay allows additional contacts between the SH1 helix and the relay to be created via several conserved hydrophobic side chains of the relay.
Figure 5.
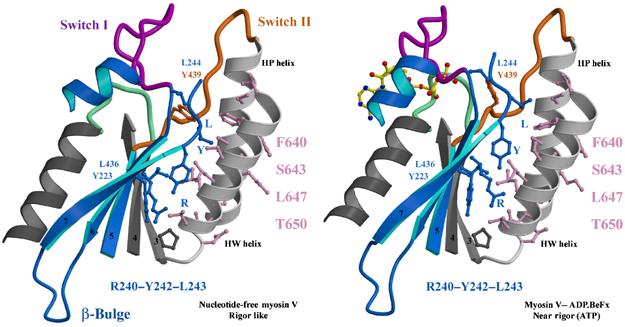
Sliding on hydrophobic patches. The surfaces of the HP and HW helices of the L50 subdomain make variable contacts with the seven-stranded β-sheet, as shown for myosin V MDE rigor-like and post-rigor states (positioned by overlaying the L50 subdomains). Note in particular the sliding of two residues of the β-sheet (Y223 and L436) on the surface of the HW helix near T650, as well as three residues (R240, Y242 and L243; labelled RYL) found near the beginning of the highly conserved U50 linker involved in closing the cleft.
While this sliding accommodates reorganization of the subdomains without loss of their contacts, rearrangements of the connectors help specify the subdomain positions. Furthermore, a major and unexpected finding from the rigor-like structure is that the β-sheet itself can distort to allow large movement of subdomains (Coureux et al, 2003). This finding highlights the limitations of a simple description of rigid-body movements of subdomains. It clearly shows that the region between the N-terminal and U50 subdomains (which we now refer to as the transducer—see below) undergoes coupled distortion to accommodate the relative repositioning of the two subdomains. This underscores that there may be as yet unseen deformable linkages within the subdomains necessary to accomplish product release steps on actin. Ultimately, the coupling of the subdomain rearrangements allows communication between the actin interface, nucleotide pocket and the lever arm, and is thus the basis for chemo-mechanical transduction.
Transducer region
The distortion of the β-sheet is essential to allow the rearrangements within the nucleotide pocket, which distinguish the post-rigor and rigor-like myosin V structures. This unexpected rearrangement provides the most profound new mechanistic insights derived from the rigor-like structure. It forms the structural basis for communication between the actin interface and nucleotide-binding pocket (Coureux et al, 2003). Now that we have the rigor-like and post-rigor states for myosin V, we can precisely describe how this distortion occurs. It is clear that the large rotation of the N-terminal and U50 subdomains is accommodated by the flexible loops and linkers that maintain close proximity of the subdomains at their point of rotation (Figure 6 and Supplementary Movie 5). These loops and linkers (defined in Figure 6 and in Supplementary Table 8) interact with each other and undergo coupled distortions, which suggests that while their sequences are variable among myosin isoforms, they must be altered in a coordinated manner within a given myosin. Sequence variations in these loops and linkers likely allow kinetic tuning by altering rates of transitions between states of the actomyosin ATPase cycle. However, kinetic tuning obviously can be achieved by alterations in any region of the motor that either facilitates or retards transitions from one state of subdomain arrangements to another.
Figure 6.
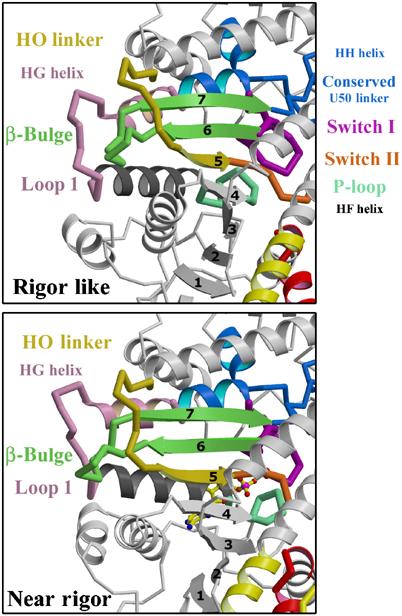
The transducer. The transducer is the central region of the motor domain near the nucleotide-binding site that includes the last three strands of the seven-stranded β-sheet that undergo distortion between the rigor-like and post-rigor states and the structural elements that accommodate this distortion. Among these elements are the previously studied loop, commonly referred to as loop 1 (residues 184–191), and the β-bulge (pale green) found at the end of the last two β-strands. Another of these elements is a linker that follows the HO helix, which provides a pathway of communication to the actin interface. We thus refer to this linker as the HO linker (residues 424–430); it leads to the fifth β-strand that is followed by switch II. Switch I and the P-loop are connected via three parallel α-helices that interact with the β-sheet. Loop 1 connects the ends of two of these helices (HF and HG). When the β-sheet undergoes distortion, these helices rotate and translate relative to each other.
We propose that these structural elements that accommodate the distortion of the β-sheet must interact in a coordinated manner to bring about kinetic tuning of product release. Thus, we refer to them collectively as the loop 1 tuning elements (Figure 6). Previous studies on myosin II (Sweeney et al, 1998) have shown that one of the elements, loop 1, alters the rate of transition between the actomyosin state that strongly binds ADP and the state that releases ADP rapidly. The fact that loop 1 also affects the rate of actin-activated phosphate release from smooth muscle myosin II (Sweeney et al, 1998) suggests that some β-sheet distortion must also occur at that transition. Together with the β-sheet we refer to this entire region as the ‘transducer', since it truly belongs to no subdomain and is at the heart of chemo-mechanical transduction in actomyosin.
New interactions among the switch elements of the rigor-like state
In our myosin V rigor-like structures, specific and novel interactions have formed between the elements (P-loop, switch I and switch II) of the nucleotide-binding pocket (Figure 7 and Supplementary Movie 6). One consequence of this is the loss of high-affinity nucleotide binding. Switch I has been repositioned by a movement of the U50 subdomain so that it is approximately 6.5 Å from the P-loop. While a similar movement of switch I was also seen in the DdNF structure (Reubold et al, 2003), the P-loop conformation, switch II position and resulting interactions are quite different than in the rigor-like structures.
Figure 7.
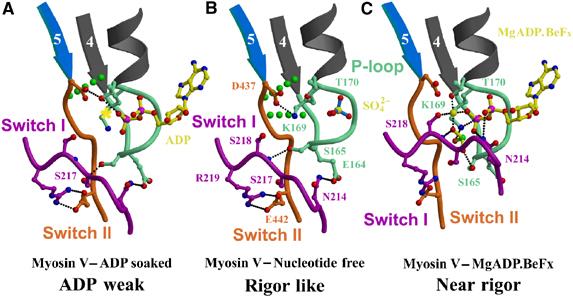
Active site. Interactions between the nucleotide-binding elements are shown for the M5 MDE in the rigor-like state (A), the ADP weak state (B) and the post-rigor state with MgADP.BeFx bound (C). The yellow star in the ADP weak state marks the position of the Mg2+ in the post-rigor state. In the rigor-like state, switch II interacts with the fourth β-strand and the P-loop of the N-terminal subdomain (broken green lines represent two hydrogen bonds found between the main-chain hydrogen of the amide of Ile438 and Gly163 and the carbonyls of Gly161 and Ile438, respectively). These bonds are progressively lost when P-loop and switch II rearrange upon nucleotide binding.
The new conformation of the P-loop, which is critical for decreasing nucleotide affinity, is stabilized by interactions with the unique switch II conformation. In the rigor-like structures, P-loop residue K169 (K185 in Dictyostelium myosin II) interacts with D437 (D454 equivalent) of switch II. (This lysine is normally involved in coordination of the β-phosphate of ADP or ATP.) A P-loop/switch II interaction also occurs in nucleotide-free structures of G proteins bound to exchange factors (Renault et al, 2003). Thus, actin is acting as an exchange factor, in that it favors a state in which the nucleotide-binding elements cannot strongly bind the nucleotide and Mg2+ ion. The P-loop also helps stabilize the myosin V conformation via interactions with switch I, via residues E164, S165 and N214 (E180, S181 and N233 in Dictyostelium myosin II) that are involved in γ- and β-phosphate coordination in the near-rigor and pre-powerstroke states (Figure 7).
Interestingly, in some of the rigor-like structures (PDB code: 1W8J; molecules A, B and D), the α-phosphate site of the nucleotide-binding pocket within the P-loop is occupied by a sulfate ion (see Figure 7). In previous post-rigor structures without nucleotide (Rayment et al, 1993), sulfate has been seen at the β-phosphate site. In the myosin V rigor-like structure, this site is unavailable due to the P-loop conformation, which suggests that nucleotide binding will trigger rearrangements of the active site.
Role of switch II in subdomain coupling
A major difference in the DdNF and rigor-like structures is the switch II conformation. In the rigor-like structure, it forms direct interactions with the N-terminal subdomain allowing the fifth β-strand of the sheet to be extended by two additional hydrogen bonds (green dots in Figure 7). This is significant since this unique switch II conformation facilitates cleft closure and subdomain coupling, as discussed below.
As shown in Figure 4 and Supplementary Movie 2, the unique switch II conformation of the rigor-like state involves a downward movement that allows interactions with the beginning of the SH1 helix. Concomitantly, the relay and the L50 subdomain are also moved down by the switch II movement. The relay and the SH1 helix accommodate this movement by a relative sliding on a hydrophobic interface (Figure 4b and Supplementary Movie 3). Compared to the post-rigor state in which switch II has no interactions and is extended, its conformation in the rigor-like state facilitates contacts between the subdomains (including cleft closure), allowing tight coupling of all four subdomains. This is likely necessary for a force-bearing state. Such coupling would prevent melting of the SH1 helix when myosin is bound to actin, consistent with the observed crosslinking studies (Wells and Yount, 1979; Nitao et al, 2003).
Starting from a post-rigor structure, the movement of switch II in the pre-powerstroke state is in the opposite direction as that seen in the rigor-like state. In the pre-powerstroke state, this movement allows partial cleft closure near the γ-phosphate and is essential for the positioning of a water molecule necessary for ATP hydrolysis. The conformation of switch II in this state forces a bending of the relay helix to avoid clashes with the SH1 helix (Figure 4). In contrast, the rigor-like position of switch II allows the relay helix to be straight, as in post-rigor, and dictates a ‘down' position for the converter and lever arm that is quite different from the ‘primed' position found in the pre-powerstroke state. The relay and the SH1 helix are two connectors, found at the interface between the L50 and N-terminal subdomains, that control the rotation of the converter. But switch II is also an important specifier of lever arm position since its conformation dictates the relative position of the subdomains and the conformation of these two connectors. Found at the bottom of the 50 kDa cleft and near the γ-phosphate-binding site, switch II is likely to be the sensor of phosphate release and the controller of lever arm position during or following that step.
Myosin V rigor-like crystals soaked with MgADP
To visualize possible rearrangements of the nucleotide-binding elements upon introduction of MgADP, rigor-like crystals of myosin V MDE were soaked in 10 mM MgADP. The crystals diffracted to 3 Å resolution (PDB code: 1W7I). As expected, a small movement at the end of the P-loop created space for the β-phosphate of ADP, but switch I remained far from the nucleotide-binding site as in the rigor-like state. Interestingly, the small change in P-loop is sufficient to modify the set of interactions between the switch elements (see Figure 7 and Supplementary Movie 6). The site for the Mg2+ ion is empty and binding of this cation is weak in this site because switch I is not in position to coordinate it (via S218). Surprisingly, no interaction is observed between lysine K169 of the P-loop and the β-phosphate of ADP. Thus, the ADP binding is weak, promoting rapid nucleotide exchange. This explains why ADP affinity is very weak for the actomyosin rigor state, in that both Mg2+ and specific P-loop interactions with the nucleotide are removed by the interactions among the nucleotide-binding elements that are unique to the rigor-like state. Furthermore, since weak ADP binding occurs with minimal alterations to the rigor-like structure, it gives support to previous suggestions that the weak ADP-binding state of myosin on actin is similar in its gross structure (lever arm position) to the rigor state (Rosenfeld et al, 2000). A clear and important implication of this structure is that Mg2+ is released prior to ADP in the actomyosin ATPase cycle. Thus, altered levels of free Mg2+ should affect the rate of ADP release from myosin V, as well as other myosins that have both strong and weak actomyosin ADP states.
To accommodate ATP in the active site, the nucleotide-binding site must undergo more rearrangements. The γ-phosphate cannot be accommodated by the P-loop without loss of some interactions between the P-loop and switch I (at the minimal, the bond between the hydroxyl groups of S165 and S217). Indeed rearrangements in the active site are supported by experiments in which soaking of rigor-like crystals with ATP leads to disorganization of the crystals.
Discussion
We have now recapitulated the rigor-like structure of myosin V with a different myosin V construct (MD). Comparing it to the Dictyostelium myosin II nucleotide-free structure of Reubold et al has reinforced our assertion that myosin V may be unique in its ability to populate a true rigor-like state in the absence of actin and nucleotide. It appears that many regions throughout the molecule contribute to this, which is not surprising since attaining the rigor-like structure requires concerted rearrangements of all of the subdomains of the motor. These rearrangements include the distortion of the β-sheet that is accommodated by the transducer region, the sliding of subdomains on hydrophobic patches and precise new interactions of the connectors.
One implication of the extensive communication that exists between the subdomains of the myosin motor is that the rates of transition between the structural states of the motor in the actomyosin cycle can potentially be altered by changes in many different regions. Thus, there is no unique strategy for kinetic tuning of myosin. For example, while alternative splice forms of loop 1 constitute the major tuning strategy exploited by vertebrate smooth and nonmuscle myosin isoforms, Drosophila striated muscle myosin II isoforms do not contain any alterations of the transducer region (Bernstein and Milligan, 1997).
An interesting new finding presented above is that ADP can bind to the myosin V rigor-like structure by inducing a small rearrangement of the P-loop. This occurs without disrupting the unique interactions between the P-loop, switch I and switch II that are characteristic of the rigor-like state. In this structure, we have for the first time visualized the myosin conformation that has weak ADP affinity and likely a high affinity for actin (AM.ADPw), and which facilitates nucleotide exchange by rapid ADP dissociation. The fact that myosin V, unlike myosin II, can populate the rigor-like state in the absence of actin may explain the otherwise puzzling observation that ADP inhibits the K-EDTA-ATPase activity of myosin V, but not that of myosin II (De La Cruz et al, 1999). This structure further suggests that loss of Mg2+ coordination is essential to achieving this weak ADP binding. Physiological relevance for the release of Mg2+ prior to ADP by actomyosin is supported by recent kinetic experiments (S Rosenfeld and HL Sweeney, unpublished data).
Furthermore, we demonstrate that the active site of the rigor-like structure is compatible with ADP but not ATP binding. This may imply that ATP binds first via the purine and ribose moiety, positioning the γ-phosphate to rapidly induce movement of switch I toward its post-rigor state position. This transition leads to the dissociation of myosin from actin when MgATP binds. When ADP is introduced, its phosphates are not in position to influence the movement of switch I, and thus Mg2+ would be critical to cause a switch I movement. Thus, this study allows us to understand why ATP but not ADP binding leads to the destabilization of the rigor complex.
What cannot be deduced from the myosin V rigor structure are details of the myosin state that immediately precedes the weak ADP-binding state (AM.ADPW) in the actin–myosin ATPase cycle (Figure 1). This is a state that binds strongly to actin and has MgADP bound strongly in the nucleotide pocket (AM.ADPs). In this state, the lever arm (and converter) assumes a position that is in between the pre-powerstroke state and rigor positions (Whittaker et al, 1995). While this state may not exist for all myosin isoforms (Rosenfeld et al, 2000), it does exist for myosin V (De La Cruz et al, 1999; Rosenfeld and Sweeney, 2004). Thus, another structural state must exist in which there are changes in the relative position of the U50 and L50 subdomains to achieve a high-affinity actin interface without significantly perturbing nucleotide binding. This could be achieved from a different type of distortion of the β-sheet and other regions within the U50 subdomain itself to accommodate a positioning of the P-loop, switch I and switch II that maintains strong MgADP binding and allows cleft closure.
A second missing structural state is that from which phosphate is released when myosin is bound to actin (Houdusse and Sweeney, 2001; Rosenfeld and Sweeney, 2004). Actin binding is the trigger for the structural rearrangements necessary to open an escape route for phosphate after rebinding in the pre-powerstroke state. While the structural details of this state cannot be guessed, it is likely that it also involves some type of β-sheet distortion, as supported by the fact that loop 1 changes can alter the rate of phosphate release (Sweeney et al, 1998). Sequential distortion of the β-sheet of the F1-ATPase has been proposed to accompany sequential product release by that enzyme (Menz et al, 2001). Interestingly, the β-sheet distortions observed in that structure are in the opposite direction as compared to what is seen in myosin V. The proposed sequential β-sheet rearrangements in myosin would facilitate both cleft closure and changes in the actin–myosin interface. This could underlie the observation that the myosin affinity for actin becomes tighter with each product release step.
As noted, switch II is seen in a different conformation in each of the myosin structural states that have been seen at high resolution. In the pre-powerstroke and rigor-like states, the switch II position is constrained by sets of interactions described above that ultimately determine the position of the lever arm. These positions represent the beginning and end of the myosin powerstroke on actin. The interesting question is how switch II alters its interactions to create the yet not seen myosin V structural states: (1) the state from which phosphate is released and (2) the state that maintains strong ADP and actin binding. In each of these states, it is likely that switch II will control the lever arm position.
While we are still missing structures of at least two states in the actomyosin ATPase cycle, the myosin V structures have provided the first detailed insights into the structural changes driven by actin binding. These insights allow clear predictions of rearrangements that must have preceded the rigor-like state, allowing the formulation of more precise and testable predictions of structural elements that are involved in the product release steps on actin. Actin crosslinking experiments suggest that concomitant actin structural changes are involved in triggering the myosin structural changes (Kim et al, 2002). Structural studies on actomyosin complexes will be necessary to elucidate the details of this mechanism. Thus, while the rigor-like structure of myosin V provides major insights into the actomyosin chemo-mechanical coupling mechanism, the final pieces of the puzzle are not yet in hand.
Materials and methods
Protein engineering and preparation
Myosin V was expressed using the baculovirus/SF9 cell expression system. The expression and purification were as previously described (De La Cruz et al, 1999). To create the recombinant virus used for expression, the cDNA coding for chicken myosin V was truncated after the codon corresponding to Arg792. This construct encompassed the motor domain and the first light chain/calmodulin-binding site of myosin V. A ‘Flag' tag DNA sequence (encoding GDYKDDDDK) (Hopp et al, 1988) was appended to the truncated myosin V coding sequence to facilitate purification. A truncated cDNA for the LC1-sa light chain was coexpressed with the truncated myosin V heavy chain. The construct of myosin V MD was the same as MDE but the sequence was truncated after residue K766.
Crystallization, X-ray data collection and processing
Myosin V MDE ADP-BeFx. Crystals were grown at 4°C in hanging drops by vapor diffusion using equal amounts of reservoir solution (containing 12% PEG 8000, 50 mM MES pH 6.3, 2 mM DTT and 2 mM NaN3) and stock solution of myosin V MDE complexed with BeFx at 6.9 mg/ml. Nice crystals grew after 4 weeks. Prior to freezing and data collection, the crystals were transferred stepwise into a final cryoprotectant solution containing 18% PEG 8000, 50 mM MES pH 6.3 and 27% ethylene glycol. The crystals belonged to the C2 space group (cell dimensions: a=169.7 Å, b=96.0 Å, c=84.3 Å and β=96.1°, one molecule per asymmetric unit). An X-ray data set was collected up to 2 Å resolution at 100 K at the European Synchrotron Radiation Facility (ESRF) Beamline ID-14-1 with an ADSC detector.
Myosin V MD NF. Crystals were grown at 4°C in hanging drops by vapor diffusion using equal amounts of reservoir solution (containing 10% PEG 8000, 50 mM MES pH 6.3, 100 mM ammonium sulfate, 2 mM DTT and 2 mM NaN3) and stock solution of myosin V MD at 7 mg/ml. The protein/precipitant drops were microseeded the next day by streak-seeding from previous crystallizations. Small crystals appeared within 5 days and grew to maximum size over a period of 4 weeks. Crystals were flash-cooled using ethylene glycol as cryoprotectant. The crystals belonged to the P212121 space group (cell dimensions: a=134.5 Å, b=162.3 Å and c=174.7 Å, four molecules per asymmetric unit). An X-ray data set was collected up to 2.7 Å resolution at 100 K at the ESRF Beamline ID-14-1 with an ADSC detector.
Myosin V MDE NF+soak MgADP. Crystals of myosin V MDE NF were obtained as previously described. Such crystals were then soaked for 8 h in a solution containing 8% PEG 8000, 50 mM MES pH 6.5 and 10 mM MgADP. Crystals were flash-cooled using ethylene glycol as cryoprotectant. The crystals belonged to the P21 space group (cell dimensions: a=54.9 Å, b=99.2 Å, c=112.5 Å and β=101.8°, one molecule per asymmetric unit). An X-ray data set was collected up to 3.0 Å resolution at 100 K at the ESRF Beamline ID-14-1 with an ADSC detector.
All the images of the three data sets were integrated and scaled with the programs DENZO and SCALEPACK (Otwinowski and Minor, 1997) (see Table I for statistics on the data collection).
Table 1.
Crystallographic data statistics
| Data collection | |||
| Crystal | MDE-BeFx | MD-NF | MDE NF+MgADP |
| PDB code | 1W7J | 1W8J | 1W7I |
| Space group | C2 | P212121 | P21 |
| Molecules/asymmetric unit | 1 | 4 | 1 |
| Resolution range | 84.50–2 (2.05–2) | 119.53–2.7 (2.84–2.7) | 111.8–3 (3.07–3) |
| Measured reflections | 536 171 | 1 500 948 | 362 803 |
| Unique reflections | 86 066 | 99 928 | 21 972 |
| Completeness (%) | 99.8 (100) | 99.9 (99.9) | 97.2 (88.3) |
| I/σ | 14.9 (3.9) | 15.7 (3.8) | 18.3 (3.1) |
| Rsym (%) | 6.5 (32.3) | 8.2 (35.3) | 8.1 (28.4) |
| Refinement | |||
| Resolution (Å) | 84.5–2 | 119.53–2.7 | 111.8–3 |
| Protein atoms | 6897 | 22 701 | 6872 |
| Ligand atoms | 32 | 40 | 27 |
| Water molecules | 525 | 40 | 11 |
| R.m.s.d. from ideal values | |||
| Bond length (Å) | 0.009 | 0.018 | 0.012 |
| Bond angle (deg) | 1.123 | 1.667 | 1.246 |
| Rcryst (%) | 19.5 | 25.4 | 24.8 |
| Rfree (%) | 22.5 | 30.7 | 31.8 |
Structure refinement
The structures were solved by molecular replacement using the myosin V MDE model (PDB code: 1OE9) with the program AmoRe (Navaza, 1994). In the case of myosin V MDE ADP-BeFx, several steps of rigid-body fitting were performed with AmoRe (each subdomain has been considered as a rigid group). We carried out model building and refinement using the program O (Jones et al, 1991) and Refmac5 (Collaborative Computational Project No. 4, 1994). The final statistics for all three models are presented in Table I. The quality of the models was assessed with PROCHECK (Laskowski et al, 1993). The statistics of the refinement are given in Table I.
Diagrams were computed using MOLSCRIPT (Kraulis, 1991) with the Dictyostelium myosin II structures (PDB codes: 1FMW, 1VOM, 1Q5G). The coordinates of the myosin V nucleotide-free MD, ADP-soaked MDE and ADP.BeFx-bound MDE have been deposited in the Protein Data Bank with accession codes 1W8J, 1W7I and 1W7J, respectively.
Supplementary Material
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Movie 4
Supplementary Movie 5
Supplementary Movie 6
Supplementary Table 7
Supplementary Table 8
Supplementary Legends
Acknowledgments
We are grateful to Jocelyn Kibbe for help in protein purification. We thank Dietmar Manstein for the coordinates of the nucleotide-free Dictyostelium myosin II structure. This work was supported by grants from the National Institutes of Health (HLS) and the CNRS (AH). We thank the staff of the European Synchrotron Radiation Facility for assistance during data collection. PDC gratefully thanks the Association pour la Recherche sur le Cancer for financial support.
References
- Bernstein SI, Milligan RA (1997) Fine tuning a molecular motor: the location of alternative domains in the Drosophila myosin head. J Mol Biol 271: 1–6 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Coureux P-D, Wells AL, Menetrey J, Yengo CM, Morris CA, Sweeney HL, Houdusse A (2003) A structural state of the myosin V motor without bound nucleotide. Nature 425: 419–423 [DOI] [PubMed] [Google Scholar]
- De La Cruz EM, Wells AL, Safer D, Ostap EM, Sweeney HL (1999) The kinetic mechanism of myosin V. Proc Natl Acad Sci USA 96: 13726–13731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AJ, Smith CA, Thoden JB, Smith R, Sutoh K, Holden HM, Rayment I (1995) X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4−. Biochemistry 34: 8960–8972 [DOI] [PubMed] [Google Scholar]
- Forkey JN, Quinlan ME, Shaw MA, Corrie JE, Goldman YE (2003) Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature 422: 399–404 [DOI] [PubMed] [Google Scholar]
- Geeves MA, Holmes KC (1999) Structural mechanism of muscle contraction. Annu Rev Biochem 68: 687–728 [DOI] [PubMed] [Google Scholar]
- Hopp TP, Prickett KS, Price V, Libby RT, March CJ, Cerretti P, Urdal DL, Conlon PJ (1988) A short polypeptide marker sequence useful for recombinant protein identification and purification. Biotechnology 6: 1205–1210 [Google Scholar]
- Houdusse A, Kalabokis VN, Himmel D, Szent-Gyorgyi AG, Cohen C (1999) Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. Cell 97: 459–470 [DOI] [PubMed] [Google Scholar]
- Houdusse A, Sweeney HL (2001) Myosin motors: missing structures and hidden springs. Curr Opin Struct Biol 11: 182–194 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kim E, Bobkova E, Hegyi G, Muhlrad A, Reisler E (2002) Actin cross-linking and inhibition of the actomyosin motor. Biochemistry 41: 86–93 [DOI] [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950 [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemistry of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Mehta AD, Rock RS, Rief M, Spudich JA, Mooseker MS, Cheney RE (1999) Myosin-V is a processive actin-based motor. Nature 400: 590–593 [DOI] [PubMed] [Google Scholar]
- Menz RI, Walker JE, Leslie AG (2001) Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell 106: 331–341 [DOI] [PubMed] [Google Scholar]
- Navaza J (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr A 50: 157–163 [Google Scholar]
- Nitao LK, Loo RR, O'Neall-Hennessey E, Loo JA, Szent-Gyorgyi AG, Reisler E (2003) Conformation and dynamics of the SH1–SH2 helix in scallop myosin. Biochemistry 42: 7663–7674 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–325 [DOI] [PubMed] [Google Scholar]
- Rayment I, Rypniewski WR, Schmidt-Bäde K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM (1993) Three-dimensional structure of myosin subfragment-1: a molecular motor. Science 261: 50–58 [DOI] [PubMed] [Google Scholar]
- Renault L, Guibert B, Cherfils J (2003) Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature 426: 525–530 [DOI] [PubMed] [Google Scholar]
- Reubold TF, Eschenburg S, Becker A, Kull FJ, Manstein DJ (2003) A structural model for actin-induced nucleotide release in myosin. Nat Struct Biol 10: 826–830 [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, Sweeney HL (2004) A model of myosin V processivity. J Biol Chem 279: 40100–40111 [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, Xing J, Whitaker M, Cheung HC, Brown F, Wells A, Milligan RA, Sweeney HL (2000) Kinetic and spectroscopic evidence for three actomyosin:ADP states in smooth muscle. J Biol Chem 275: 25418–25426 [DOI] [PubMed] [Google Scholar]
- Sasaki N, Ohkura R, Sutoh K (2000) Insertion or deletion of a single residue in the strut sequence of Dictyostelium myosin II abolishes strong binding to actin. J Biol Chem 275: 38705–38709 [DOI] [PubMed] [Google Scholar]
- Smith CA, Rayment I (1996) X-ray structure of the magnesium(II).ADP.vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 Å resolution. Biochemistry 35: 5404–5417 [DOI] [PubMed] [Google Scholar]
- Sweeney HL, Rosenfeld SS, Brown F, Faust L, Smith J, Xing J, Stein LA, Sellers JR (1998) Kinetic tuning of myosin via a flexible loop adjacent to the nucleotide binding pocket. J Biol Chem 273: 6262–6270 [DOI] [PubMed] [Google Scholar]
- Wells JA, Yount RG (1979) Active site trapping of nucleotides by crosslinking two sulfhydryls in myosin subfragment 1. Proc Natl Acad Sci USA 76: 4966–4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker M, Wilson-Kubalek EM, Smith JE, Faust L, Milligan RA, Sweeney HL (1995) A 35-Å movement of smooth muscle myosin on ADP release. Nature 378: 748–751 [DOI] [PubMed] [Google Scholar]
- Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR (2003) Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 300: 2061–2065 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Movie 4
Supplementary Movie 5
Supplementary Movie 6
Supplementary Table 7
Supplementary Table 8
Supplementary Legends


