Abstract
In innate immunity, pattern recognition molecules recognize cell wall components of microorganisms and activate subsequent immune responses, such as the induction of antimicrobial peptides and melanization in Drosophila. The diaminopimelic acid (DAP)-type peptidoglycan potently activates imd-dependent induction of antibacterial peptides. Peptidoglycan recognition protein (PGRP) family members act as pattern recognition molecules. PGRP-LC loss-of-function mutations affect the imd-dependent induction of antibacterial peptides and resistance to Gram-negative bacteria, whereas PGRP-LE binds to the DAP-type peptidoglycan, and a gain-of-function mutation induces constitutive activation of both the imd pathway and melanization. Here, we generated PGRP-LE null mutants and report that PGRP-LE functions synergistically with PGRP-LC in producing resistance to Escherichia coli and Bacillus megaterium infections, which have the DAP-type peptidoglycan. Consistent with this, PGRP-LE acts both upstream and in parallel with PGRP-LC in the imd pathway, and is required for infection-dependent activation of melanization in Drosophila. A role for PGRP-LE in the epithelial induction of antimicrobial peptides is also suggested.
Keywords: antibacterial defense, Drosophila, innate immunity, non-self recognition, PGRP
Introduction
Innate immunity is a self-defense mechanism that is evolutionarily conserved throughout all metazoans (reviewed in Hoffmann and Reichhart, 2002). In Drosophila, a model system to study the principles of innate immunity (reviewed in Hultmark, 2003), epithelial tissues such as the epidermis, gut, and trachea (the insect respiratory organ) serve as the first line of self-defense against invading microorganisms by functioning as structural barriers and by producing antimicrobial peptides that inhibit microbial growth (Tzou et al, 2000). Microorganisms that pass through the epithelial barrier are countered in hemocoels filled with the hemolymph (blood) by systemic reactions, including cellular and humoral reactions (reviewed in Tzou et al, 2002). The humoral reactions depend on the primary and secondary responses. The primary response is mediated by the activation of cascades of constitutive proteins present in the hemolymph, such as the prophenoloxidase (proPO) cascade leading to localized wound healing and melanization. Biochemical studies of the proPO cascade in insects and crustaceans indicate that the serine protease cascade is initiated by the recognition of microbial cell wall components (reviewed in Ashida and Brey, 1998; Söderhäll and Cerenius, 1998). The secondary response requires transcriptional activation of defense proteins such as antimicrobial peptides. In response to microbial infections, the antimicrobial peptides are synthesized in the fat body, the functional equivalent of the mammalian liver, and secreted into the hemolymph, which is mainly regulated by two distinct signaling pathways, the Toll pathway and the imd pathway (reviewed in Engström, 1999). An antifungal peptide gene, Drosomycin (Drs), is predominantly activated by the Toll pathway through Toll membrane receptor and transcriptional factors, Dorsal and Dorsal-related immunity factor (Dif), in response to fungal and some Gram-positive bacterial infections, such as Enterococcus faecalis. An antibacterial peptide gene, Diptericin (Dpt), however, is predominantly activated by the imd pathway through the Imd death domain adaptor protein and Relish transcriptional activator in response to Gram-negative and other Gram-positive bacterial infections, such as Bacillus subtilis. The induction of other antimicrobial peptide genes, such as Attacin (Att), is thought to be regulated by input from both the Toll pathway and the imd pathway. Thus, Drosophila possess specific mechanisms to discriminate between microbes and activate the appropriate immune reactions to these infections (Lemaitre et al, 1997; reviewed in Hoffmann and Reichhart, 2002).
In contrast to the relatively good understanding of the signaling cascades leading to the activation of the antimicrobial peptide genes, little is known about how the Drosophila immune system recognizes different groups of invading microorganisms and activates the proper immune responses. In innate immunity, the pattern recognition molecules recognize the pathogen-associated molecular patterns of repeating structural motifs, and activate immune reactions (reviewed in Medzhitov and Janeway, 2002). In fact, cell wall components containing repeated structures such as lipopolysaccharides (LPSs), peptidoglycans, and β-1,3-glucans strongly induce innate immune reactions. In mammals, Toll-like receptors (TLRs), mammalian homologs of Drosophila Toll, act as pattern recognition molecules. For example, TLR4 is a recognition receptor for LPSs, which are components of Gram-negative bacterial cell walls (reviewed in Akira et al, 2001). Drosophila Toll, however, does not act as a pattern recognition molecule; it is activated by an endogenous ligand, Spätzle, which is cleaved by proteolytic enzymes after microbial infection (Levashina et al, 1999; Ligoxygakis et al, 2002a; Weber et al, 2003). Therefore, pattern recognition molecules have remained the missing pieces of the puzzle in Drosophila immunity. Recent reports of Drosophila genetic screens of loss-of-function and gain-of-function mutations highlight the important roles of peptidoglycan recognition protein (PGRP) family members as the pattern recognition molecules in Drosophila.
PGRP was first purified from silkworm hemolymph based on its high affinity for peptidoglycan and its ability to mediate peptidoglycan-dependent activation of the proPO cascade as demonstrated by in vitro reconstitution experiments (Yoshida et al, 1996). Subsequent cloning of PGRP genes demonstrated that PGRPs are conserved from insects to mammals, and there are four PGRPs in humans (reviewed in Dziarski, 2004). Of the 13 PGRP family members encoded by the Drosophila genome (Werner et al, 2000; reviewed in Dziarski, 2004), PGRP-SA, PGRP-LC, and PGRP-LE participate in the recognition of invading bacteria and activation of immune responses. PGRP-SA is a circulating hemolymph protein that binds to the lysine-containing peptidoglycan of Micrococcus luteus (Werner et al, 2000). A mutation of PGRP-SA, called semmelweis (seml), abolishes the Toll-dependent expression of Drs in response to M. luteus infection, and fails to resist Gram-positive bacterial infections (Michel et al, 2001). On the other hand, seml does not affect Toll-dependent resistance to fungi and imd-mediated resistance to bacteria, suggesting that, like the role of the TLR family in mammals, diverse PGRP members are involved in bacterial discrimination. Supporting this hypothesis, mutations in a putative transmembrane protein, PGRP-LC, affect activation of the imd pathway and resistance to Gram-negative bacteria (Choe et al, 2002; Gottar et al, 2002; Rämet et al, 2002). The PGRP-LC null mutant phenotypes have much less activation of antibacterial peptide genes and survival against Gram-negative bacterial infections than other loss-of-function mutants in the imd pathway (Gottar et al, 2002; Rämet et al, 2002). These findings suggest that there is an activator of the imd pathway in addition to PGRP-LC. Consistent with this conclusion, we identified PGRP-LE in a gain-of-function screen, and found that PGRP-LE also acts upstream of the imd pathway to activate antimicrobial peptide gene expression in both the systemic and local epithelial responses (Takehana et al, 2002). In addition to activation of the imd pathway, the overexpression of PGRP-LE also activates the proPO cascade in Drosophila upstream of Imd. Peptidoglycans, components of cell walls of almost all bacteria, have a diverse amino-acid composition, and the linking of stem peptides depends on the bacterial species (Schleifer and Kandler, 1972). PGRP-LE binds to the directly crosslinked diaminopimelic acid (DAP)-containing peptidoglycan, which is an extremely potent inducer of the imd pathway (Leulier et al, 2003), but not to the lysine-containing peptidoglycans with an interpeptide bridge of Gram-positive bacteria (Takehana et al, 2002). PGRP-LC also mediates the DAP-type peptidoglycan-dependent induction of Dpt (Leulier et al, 2003). In addition, a different isoform of PGRP-LC, PGRP-LCa, participates in the LPS-dependent activation of antibacterial peptide genes, at least in cell culture systems (Werner et al, 2003).
In the present paper, we generated a null mutant of PGRP-LE, and found that PGRP-LE functioned synergistically with PGRP-LC to produce resistance to infection by Escherichia coli and Bacillus megaterium, which have DAP-type peptidoglycans. Consistent with this finding, epistatic analyses revealed that PGRP-LE acts both in parallel and upstream of PGRP-LC in the imd-mediated induction of the antimicrobial peptides. PGRP-LE is also required for infection-dependent activation of the proPO cascade. Therefore, PGRP-LE has important roles in the activation of systemic reactions in host defense. In the epithelial reactions, PGRP-LE is localized at the luminal surface of the trachea and has non-cell autonomous effects on the activation of the Drs promoter in tracheal cells, suggesting a role for PGRP-LE in the recognition and subsequent activation of the signaling at the first point of contact with invading bacteria.
Results
PGRP-LE and PGRP-LC synergistically induce resistance to bacterial infections
Gain-of-function screens utilizing P element insertions indicated that PGRP-LE activates antimicrobial responses in the absence of microbial infection. To investigate the requirement of PGRP-LE for the antimicrobial response, we generated a PGRP-LE-deficient mutant by mobilizing the P element (Supplementary Figure 1A). PGRP-LE expression was screened by reverse transcription–polymerase chain reaction (RT–PCR) with 149 excision lines, and in one line, PGRP-LE112, there was no expression of PGRP-LE (Supplementary Figure 1B). In PGRP-LE112, the expression of the neighboring genes of PGRP-LE, CG8974, CG32581, CG15602, and CG8509, was not affected (Supplementary Figure 1B). Sequencing analysis after genomic PCR of PGRP-LE112 revealed that a 2542-bp sequence, including the PGRP-LE start codon, was deleted in PGRP-LE112. Consistent with the molecular characterization, PGRP-LE protein was not expressed in PGRP-LE112 (Figure 6A). PGRP-LE112 flies were fertile and viable, suggesting that PGRP-LE is not critical for development. In wild type, the expression of PGRP-LE did not change after E. coli, B. subtilis, or E. faecalis infections, indicating that PGRP-LE is a constitutive protein (Supplementary Figure 1C).
Figure 6.
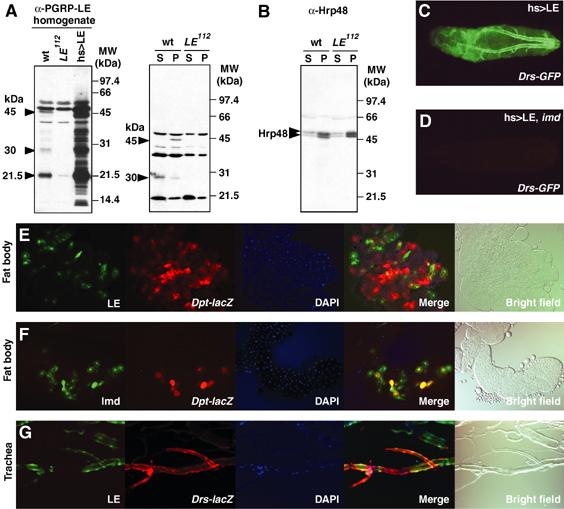
Non-cell autonomous effects of PGRP-LE on the activation of antimicrobial peptide genes in systemic and epithelial reactions. (A, B) Western blotting analyses using antibody against PGRP-LE (A) and Hrp48 (B). The homogenates (20 μg) prepared from wild-type (wt), PGRP-LE112, and UAS-PGRP-LE/+; hs-GAL4/+ larvae after heat shock (35°C, 20 min) were applied to the analysis. The hemolymph (plasma) fraction (35 μg, S) and the hemocyte fraction (P) prepared from wild-type (wt) and PGRP-LE112 larvae were analyzed. The arrowheads indicate 45-, 30-, and 21.5-kDa proteins (A). Molecular size markers are indicated on the right. (C, D) The Drs-GFP expression in GS1068; hs-GAL4/Drs-GFP larvae (C) and GS1068; imd1; hs-GAL4/Drs-GFP larvae (D) 12 h after heat shock (35°C, 20 min). (E) Non-cell autonomous effects of PGRP-LE on the expression of Dpt-lacZ in the fat body. (F) Cell autonomous effects of Imd on the expression of Dpt-lacZ in the fat body. (G) Non-cell autonomous effects of PGRP-LE on the expression of Drs-lacZ in the trachea. The overexpression of PGRP-LE and Imd was monitored by the expression of GFP (green). The expression of antimicrobial peptide genes was monitored by the expression of reporter genes using Cy3-labeled antibody (red). The GFP, Cy3, and DAPI (nuclear staining, blue) signals are merged.
We compared the survival rate of PGRP-LE112 with various mutant flies including PGRP-LC, which also acts upstream of the imd pathway, after Gram-negative bacterial infections, E. coli and Erwinia carotovora carotovora, and Gram-positive bacterial infections, B. megaterium, B. subtilis, E. faecalis, and M. luteus (Figure 1). The survival rate of PGRP-LE112 to bacterial infections was similar to that of wild type except for infection by E. carotovora carotovora and B. subtilis. In this experiment, E. carotovora carotovora, which naturally infects Drosophila (Basset et al, 2000), was injected into the flies. The susceptibility of PGRP-LE112 to E. carotovora was much weaker than that of PGRP-LC7454 and imd1; however, against B. subtilis, PGRP-LE112 had reduced resistance, similar to PGRP-LC7454 and imd1 (Figure 1B and D). These results suggest that PGRP-LE contributes to sensing these bacteria. Both PGRP-LE112 and PGRP-LC7454 had complete resistance to E. coli infections, but a double PGRP-LE112/PGRP-LC7454 mutant had reduced resistance, and the susceptibility was much greater than that of imd1 (Figure 1A). These results indicate that PGRP-LE and PGRP-LC function synergistically in the self-defense against E. coli infection. There is a similar synergy between PGRP-LE and PGRP-LC in the resistance to B. megaterium but not in the resistance to E. faecalis and M. luteus (Figure 1C, E, and F). B. megaterium and B. subtilis have the DAP-type peptidoglycan similar to Gram-negative bacteria such as E. coli, but E. faecalis and M. luteus have the lysine-containing peptidoglycan. These results are consistent with the previous conclusion that both PGRP-LE and PGRP-LC recognize the DAP-type peptidoglycan.
Figure 1.
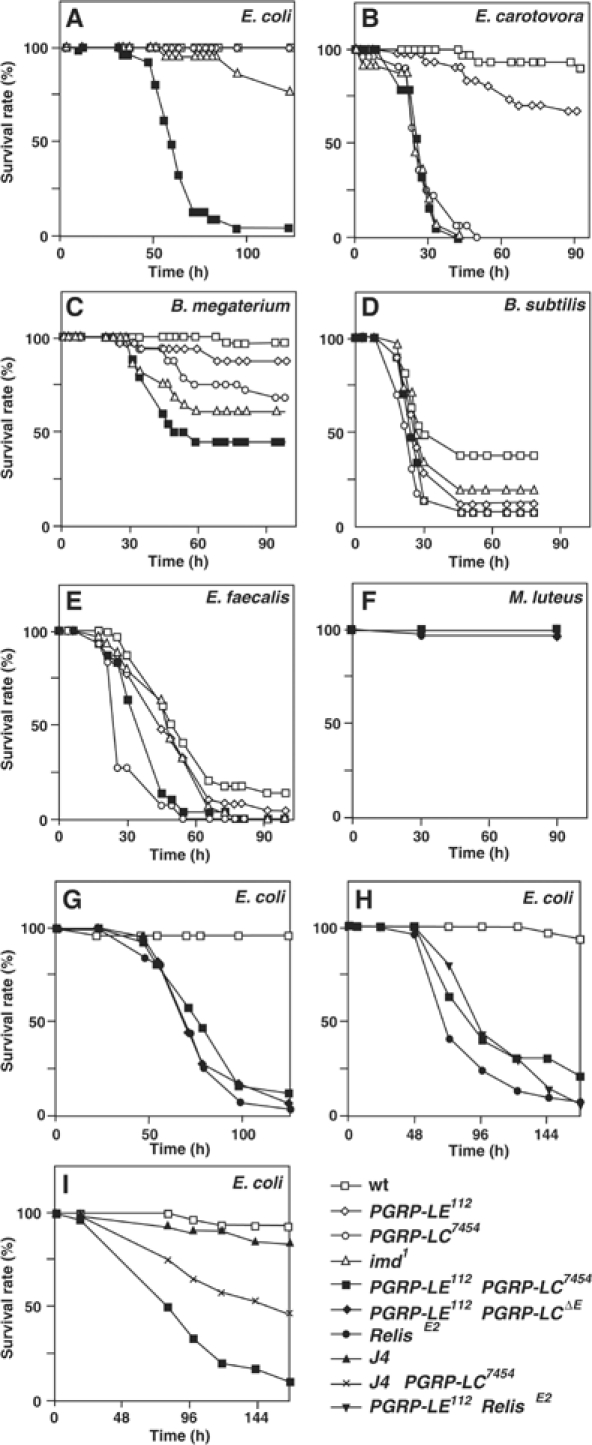
Survival rate of PGRP-LE mutant flies after different types of infections. (A–F) Survival rate of wild-type flies (wt), PGRP-LE112, PGRP-LC7454, imd1, and the double PGRP-LE112/PGRP-LC7454 mutant (PGRP-LE112;+; PGRP-LC7454) infected with the indicated bacteria. In (F), the survival rate of RelishE20 is also presented. (G) Survival rate of wild type, RelishE20, the double PGRP-LE112/PGRP-LC7454 mutant, and the double PGRP-LE112/PGRP-LCΔE mutant against E. coli infection. (H) Survival rate of wild type, RelishE20, the double PGRP-LE112/RelishE20 mutant, and the double PGRP-LE112/PGRP-LC7454 mutant against E. coli infection. (I) Survival rate of wild type, J4, the double J4/PGRP-LC7454 mutant, and the double PGRP-LE112/PGRP-LC7454 mutant against E. coli infection. The survival experiments were performed at 25°C.
The double PGRP-LE112/PGRP-LC7454 mutant was much more susceptible to E. coli than imd1. To determine whether PGRP-LE and PGRP-LC regulate other host defense reactions in addition to activation of the imd pathway, we compared the survival rate of the double mutant to that of RelishE20, which is a null mutation of the transcriptional activator of the imd pathway (Hedengren et al, 1999), because imd1 is a hypomorphic mutation (Georgel et al, 2001). The survival rate of the double PGRP-LE112/PGRP-LC7454 mutant was similar to that of RelishE20 (Figure 1G). The result was confirmed with a PGRP-LCΔE null mutation. These results suggest that the two pattern recognition molecules are major regulators of the imd pathway that sense E. coli. Corresponding to this, the double PGRP-LE112/RelishE20 mutant was not more susceptible to E. coli than RelishE20 (Figure 1H). Moreover, to determine whether PGRP-LE participates in the resistance to E. coli infection through regulating the Toll pathway, we compared the survival rate of the double PGRP-LE112/PGRP-LC7454 mutant to that of the double J4/PGRP-LC7454 mutant in which two transcriptional activators of the Toll pathway, dorsal and Dif, were deleted (Meng et al, 1999). The double PGRP-LE112/PGRP-LC7454 mutant was more susceptible to E. coli infection than the double J4/PGRP-LC7454 mutant (Figure 1I). These results suggest that PGRP-LE participates in host defense, mainly through regulating the imd pathway.
Rescue experiments using the artificial expression of PGRP-LE in the double PGRP-LE/PGRP-LC mutant could not be performed because the survival rate was decreased by the induction of PGRP-LE in the absence of bacterial infection, which is probably related to the PGRP-LE-mediated lethality described below.
Epistatic analyses of PGRP-LE and PGRP-LC in the induction of antimicrobial peptides
It was suggested that PGRP-LE and PGRP-LC were two major regulators of the imd pathway in the induction of antimicrobial peptides. To determine the relation between PGRP-LE and PGRP-LC, we analyzed the epistatic relation between PGRP-LE and PGRP-LC in the activation of antimicrobial peptide genes (Figure 2). Under the control of the c564-GAL4 driver, which expresses GAL4 in the fat body and the hemocytes (blood cells) (Harrison et al, 1995), PGRP-LE and PGRP-LCx, an isoform of PGRP-LC from the RA transcript (Choe et al, 2002), similarly induced strong expression of Dpt and weak expression of Att, whereas PGRP-LE more strongly induced Drs than PGRP-LCx. Green fluorescent protein (GFP) and PGRP-SA overexpression did not induce expression of these antimicrobial peptide genes. PGRP-LCx-mediated activation of these antimicrobial peptide genes was not affected in the PGRP-LE null mutant background, indicating that PGRP-LE is not downstream of PGRP-LC. In the reciprocal experiments, the PGRP-LE-mediated activation of antimicrobial peptide genes was reduced but still existed in the PGRP-LC null mutant. In the Relish null mutant, PGRP-LE-mediated activation of antimicrobial peptide genes was abolished or more reduced. These results indicate that PGRP-LE acts partially upstream and partially in parallel with PGRP-LC upstream of Relish, suggesting that there is an additional receptor on the cell surface in the imd pathway other than PGRP-LC.
Figure 2.
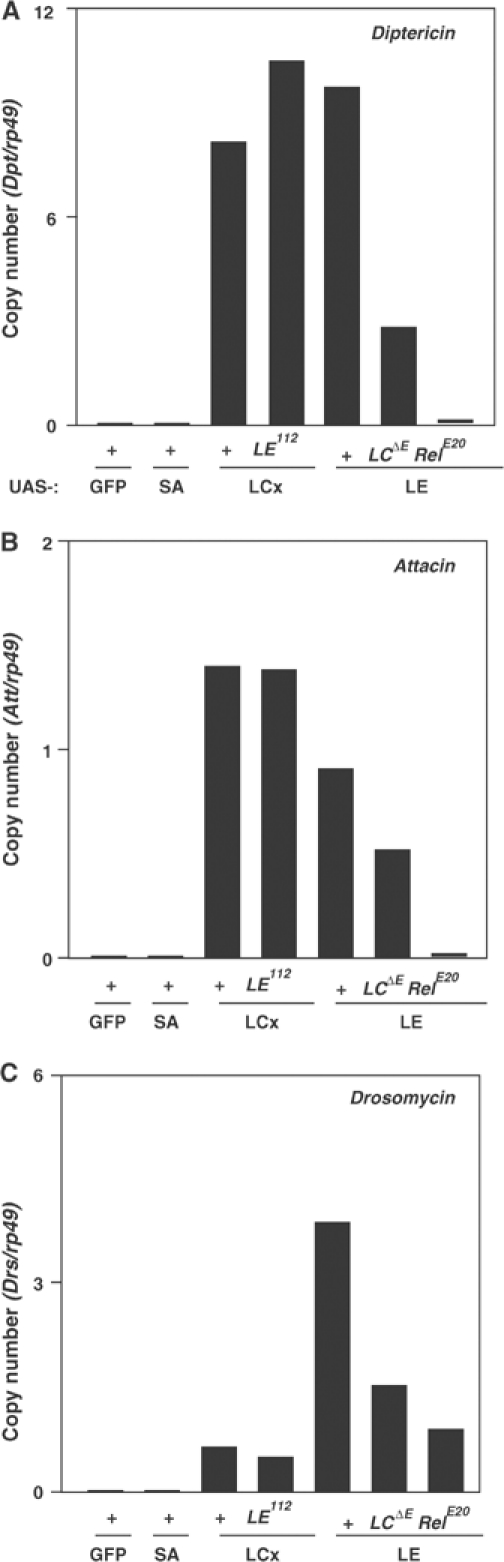
Epistatic analyses of PGRP-LE with PGRP-LC on antimicrobial peptide gene expression. Under the control of c564-GAL4, GFP, PGRP-SA, PGRP-LCx, and PGRP-LE were overexpressed in the wild-type (+), PGRP-LE112, PGRP-LCΔE, and RelishE20 larvae. The amount of mRNA of Diptericin (A), Attacin (B), and Drosomycin (C), and rp49 internal control was quantified by real-time RT–PCR in each sample.
Expression of antimicrobial peptide genes in the PGRP-LE mutant and in the double PGRP-LE/PGRP-LC mutant after bacterial challenge and natural infection
To determine the requirement for PGRP-LE and the synergy of PGRP-LE with PGRP-LC on the induction of antimicrobial peptides, we used kinetics after bacterial challenges of B. subtilis, E. faecalis, and E. coli to analyze the expression of the seven classes of inducible antimicrobial peptides so far identified (Figure 3). B. subtilis-dependent induction of antimicrobial peptides was reduced in PGRP-LE112 except for Drosocin, particularly in later stage such as 10–24 h after bacterial injection, indicating a requirement of PGRP-LE for the induction of antimicrobial peptides in response to B. subtilis (Figure 3A), whereas E. faecalis-dependent induction of antimicrobial peptides was not affected in PGRP-LE112 except for Metchnikowin (Figure 3B). After B. subtilis infection, Drosocin expression was greatly enhanced in spätzlerm7. These results are consistent with the results of survival experiments indicating that PGRP-LE112 is susceptible than wild type to B. subtilis but not to E. faecalis infections. Interestingly, there was no obvious synergy between PGRP-LE and PGRP-LC in the induction of antimicrobial peptides after E. coli infection (Figure 3C). Although the double PGRP-LE112/PGRP-LC7454 mutant and PGRP-LC7454 had similar antimicrobial peptide expression patterns after E. coli infection, only the PGRP-LE112/PGRP-LC7454 mutant was susceptible to E. coli infection. These results suggest that, in addition to antimicrobial peptide induction, other self-defense reactions were affected in the PGRP-LE and PGRP-LC double mutant.
Figure 3.
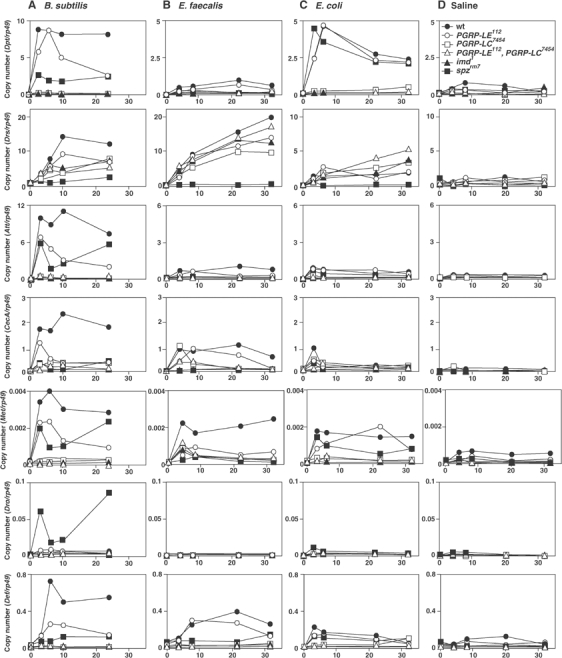
Expression of seven classes of antimicrobial peptide genes in different mutants after various bacterial challenges. After B. subtilis (A), E. faecalis (B), and E. coli (C) infection, the amount of mRNA of seven classes of inducible antimicrobial peptides, Diptericin (Dip), Drosomycin (Drs), Attacin (Att), Cecropin A (CecA), Metchnikowin (Mtk), Drosocin (Dro), and Defensin (Def), and the rp49 internal control in the indicated mutant flies, wild type (wt), PGRP-LE112, PGRP-LC7454, spätzlerm7, imd1, and the double PGRP-LE112/PGRP-LC7454 mutant, was quantified by real-time RT–PCR. As a negative control, pyrogen-free saline was used (D). Each experiment is representative of at least two independent experiments.
E. carotovora naturally infects Drosophila and induces local expression of antimicrobial peptides in surface epithelia such as the trachea via the imd pathway (Tzou et al, 2000). We investigated the effects of PGRP-LE mutation on the tracheal induction of Drs after natural infection. Compared to wild type, the number of Drs-GFP-expressing larvae was not changed in PGRP-LE112; however, Drs expression was reduced in PGRP-LC7454 and was abolished in the double PGRP-LE112/PGRP-LC7454 mutant (Figure 4). These results suggest that, in some contexts such as those occurring during natural infections, PGRP-LE and PGRP-LC synergistically induce antimicrobial peptides.
Figure 4.
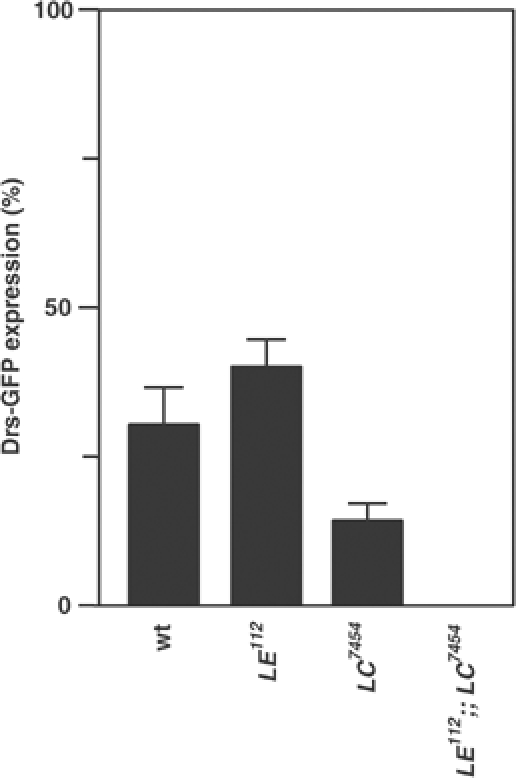
Activation of Drosomycin promoter in various mutants after natural infection of E. carotovora carotovora. The number of Drs-GFP-expressing larvae was counted in wild type (wt), PGRP-LE112, PGRP-LC7454, and the double PGRP-LE112/PGRP-LC7454 mutant. Bars indicate standard deviation of duplicate measurements.
Requirement of PGRP-LE for infection-dependent activation of the proPO cascade
In a previous paper, we reported that overexpression of PGRP-LE activates the proPO cascade. We investigated the PGRP-LE requirement for the activation of the proPO cascade in response to bacterial infection. Phenoloxidase (PO) is proteolytically activated from an inactive precursor, proPO, by the proPO-activating enzyme, a terminal serine protease of the proPO cascade, following bacterial infections. There was significant PO activity in the hemolymph of wild-type flies after E. coli infection, whereas the control challenge (saline) produced only background levels of proPO cascade activation (Figure 5A and B). The infection-dependent activation of the proPO cascade was abolished in PGRP-LE112. The artificial expression of PGRP-LE, based on GAL4/UAS targeted expression in PGRP-LE112, rescued infection-dependent activation of the proPO cascade. Consistent with these results, after E. coli infection, weak melanization was induced at the injury site of PGRP-LE112, whereas strong melanization was induced at the injury site of PGRP-LE112 after forced expression of PGRP-LE (Figure 5C–F). These results indicate that PGRP-LE is required for activation of the proPO cascade in response to bacterial infections. As described below, PGRP-LE is produced in both hemocytes (Figure 6A) and the fat body (Figure 8G). The tissue specificity requirement for PGRP-LE function in proPO cascade activation requires further analysis. In the rescue experiments, we used UAS-PGRP-LE flies that had a low expression of PGRP-LE and, in combination with hs-GAL4, the flies had the cuticle defect at the midline of the dorsal abdomen in the absence of heat shock, suggesting that the leaky expression of PGRP-LE causes the cuticle defect (Figure 5D). Consistent with these results, UAS-PGRP-LE flies that had strong expression of PGRP-LE had pupal lethality in combination with hs-GAL4 without any heat shock. The strong expression of PGRP-LE using UAS-PGRP-LE induces the proPO cascade activation in the larvae in the absence of bacterial challenge (Takehana et al, 2002), and the low level of PGRP-LE induction does not activate the proPO cascade in either larvae or adults.
Figure 5.
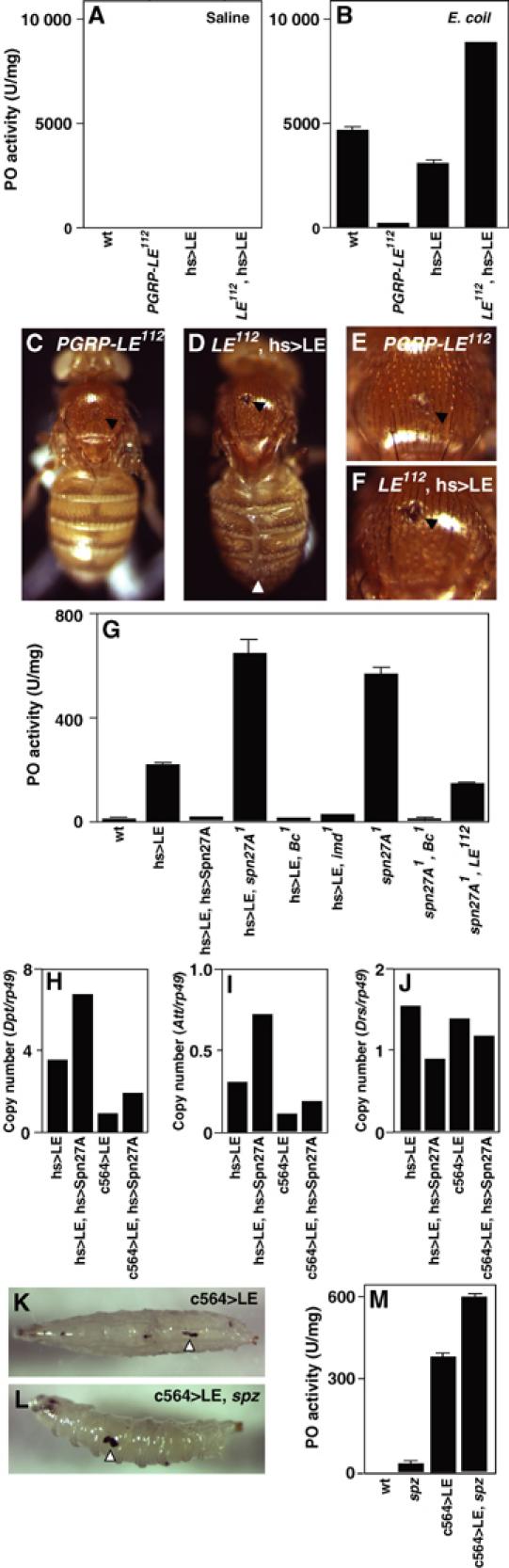
Requirement of PGRP-LE on the infection-dependent activation of the proPO cascade. (A, B) PO activity in the hemolymph after E. coli infection. The hemolymph was collected from wild-type (wt), PGRP-LE112, UAS-PGRP-LE/+; hs-GAL4/+, and PGRP-LE112; UAS-PGRP-LE/+; hs-GAL4/+ flies. (C–F) Melanization at the injury site (black arrowhead) of the indicated flies after E. coli challenge. White arrowhead indicates the cuticle defect (D). (E, F) Higher magnifications of (C, D) respectively. (G) PO activity in various mutant larvae. PO activity was assayed with homogenates of wild-type (wt), GS1068; +; hs-GAL4/+, GS1068; UAS-Serpin27A/+; hs-GAL4/+, GS1068; serpin27A1; hs-GAL4/+, GS1068; Bc1; hs-GAL4/+, GS1068; imd1; hs-GAL4/+, serpin27A1, serpin27A1Bc1, PGRP-LE112; serpin27A1 larvae. (H–J) The PGRP-LE-mediated induction of antimicrobial peptide genes. The amount of mRNA of Diptericin (H), Attacin (I), and Drosomycin (J), and rp49 internal control was quantified by real-time RT–PCR in GS1068; +; hs-GAL4/+, GS1068; +; hs-GAL4/UAS-Serpin27A, GS1068; c564-GAL4/+, GS1068; c564-GAL4/+; UAS-Serpin27A/+ larvae. mRNA was recovered from the larvae at 14 h after heat shock (31°C, 60 min). (K, L) The PGRP-LE-mediated melanization of GS1068; c564-GAL4/+ larvae (K) and GS1068; c564-GAL4/+; spätzlerm7 larvae (L). Arrowheads indicate melanization. (M) PGRP-LE-mediated activation of the proPO cascade. PO activity was assayed with homogenates of wild-type larvae (wt), spätzlerm7, GS1068; c564-GAL4/+ larvae, and GS1068; c564-GAL4/+; spätzlerm7 larvae. Bars indicate standard deviation of duplicate measurements.
Figure 8.
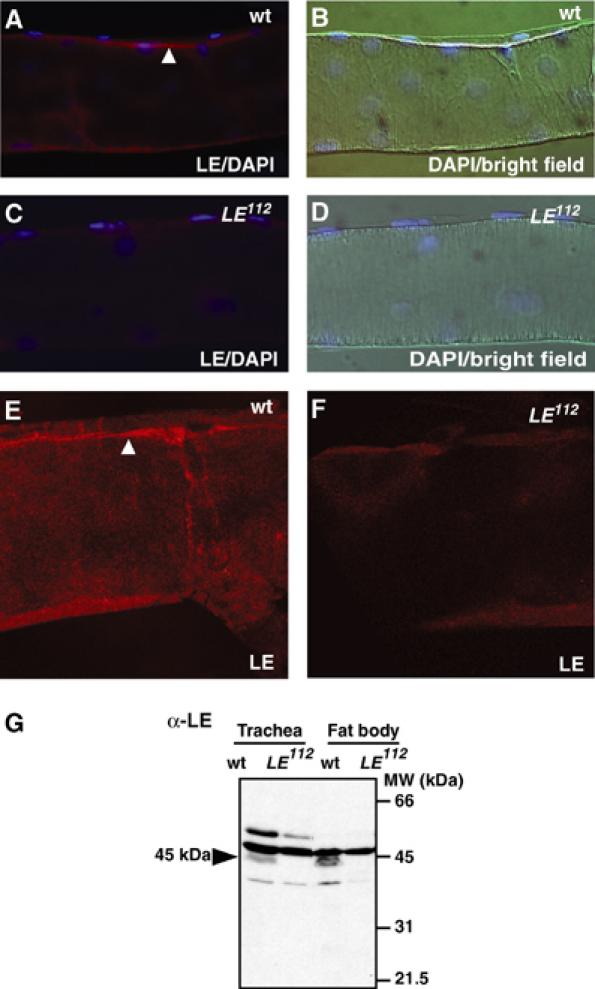
Localization of PGRP-LE in the trachea. (A–F) Immunostaining was performed in wild type (wt, A, B, E) and PGRP-LE112 (C, D, F) using anti-PGRP-LE antibody. The antibody staining is merged with DAPI staining (A, C). The bright-field micrograph is merged with DAPI staining (B, D). Confocal analysis in wild type (E) and PGRP-LE112 (F) using anti-PGRP-LE antibody. Arrowheads indicate PGRP-LE signal. (G) Western blotting analysis was performed with trachea and fat body homogenates prepared from wild type (wt) and PGRP-LE112 using anti-PGRP-LE antibody. Arrowhead indicates 45-kDa protein.
To determine the relation between PGRP-LE and other components of the proPO cascade, epistatic analysis was performed with serpin27A and Black cells (Bc). Serpin27A codes a serine protease inhibitor that inhibits the terminal protease proPO-activating enzyme, and the Bc mutant has no detectable PO activity (De Gregorio et al, 2002a). PGRP-LE-mediated proPO cascade activation was totally inhibited by the overexpression of Serpin27A, a negative regulator of the proPO cascade, and was enhanced by a mutation of serpin27A (Figure 5G). PGRP-LE-mediated proPO cascade activation was also abolished in Bc mutants (Figure 5G). These results indicate that PGRP-LE activates the proPO cascade upstream of Serpin27A and PO. The serpin27A mutation causes constitutive proPO cascade activation (De Gregorio et al, 2002a; Ligoxygakis et al, 2002b). The constitutive proPO cascade activation by serpin27A mutation was reduced in the PGRP-LE112 mutant background and was abolished in the Bc mutant background (Figure 5G). These results are consistent with the conclusion that PGRP-LE is one of the activators of the proPO cascade upstream of Serpin27A and PO.
As described above, overexpression of serpin27A inhibits PGRP-LE-mediated activation of the proPO cascade. Overexpression of serpin27A, however, did not inhibit the PGRP-LE-mediated induction of antimicrobial peptides, except Drs (Figure 5H–J, data not shown for Cecropin A, Metchnikowin, Drosocin, and Defensin). PGRP-LE-mediated induction of Drs is slightly inhibited by the overexpression of serpin27A. These results suggest that the PGRP-LE-dependent pathway of antimicrobial peptide induction branched out from the PGRP-LE-dependent proPO cascade upstream of Serpin27A.
Ligoxygakis et al (2002b) reported that proPO cascade activation is regulated by the Toll pathway through the induction of Serpin27A. We investigated the relation between PGRP-LE-mediated activation of the proPO cascade and the Toll pathway. PGRP-LE-mediated melanization was also induced in the spätzle mutant background (Figure 5K and L). Consistent with this finding, significant PO activity was detected in the homogenate of the larvae when PGRP-LE was overexpressed in the spätzle mutant background (Figure 5M). These results suggest that PGRP-LE-mediated proPO activation is independent of the Toll pathway. PGRP-LE-mediated proPO cascade activation was reduced by imd mutation (Figure 5G). We previously reported that there is proPO activation-induced melanization at the cuticle of imd background larvae when PGRP-LE is expressed under the control of a heat-shock promoter (Takehana et al, 2002). Therefore, further study is required to determine the involvement of the imd pathway in the PGRP-LE-dependent proPO cascade activation.
Non-cell autonomous effects of PGRP-LE in the induction of antimicrobial peptides in systemic and epithelial reactions
PGRP-LE was required for the infection-dependent proPO cascade activation upstream of Serpin27A hemolymph protein, and PGRP-LE induced antimicrobial peptide synthesis in the fat body upstream of Imd, an adaptor molecule, and partially upstream of PGRP-LC, a cell surface receptor. These results suggest that PGRP-LE recognizes invading bacteria and activates subsequent signaling in the hemolymph, although PGRP-LE lacks the predicted signal peptide for secretion (Werner et al, 2000). To determine whether endogenous PGRP-LE is present in the hemolymph, we generated an antibody against the PGRP domain of PGRP-LE. After affinity purification of the antiserum with the antigen, antibody specificity was analyzed by Western blotting using wild-type larvae and PGRP-LE-deficient larvae in which the transcript of PGRP-LE was not detected (Supplementary Figure 1B). Bands of 45, 30, and 21.5 kDa were detected in the wild-type homogenate but not in the PGRP-LE112 homogenate (Figure 6A). Forced expression of PGRP-LE increased band intensity. As described below, the 45-kDa protein was detected mainly in the hemocytes that express the PGRP-LE transcript. Although the molecular size of PGRP-LE was estimated to be 39.4 kDa, the 45-kDa band was probably the full-length PGRP-LE protein and the others were limited proteolysis products. After collecting the hemolymph from the larvae, the hemolymph (plasma) fraction and the floating hemocyte fraction were analyzed by Western blotting with antibody against PGRP-LE and antibody against Hrp48, an abundant heterogeneous nuclear RNA-associated protein (Hammond et al, 1997). The 45-kDa band and the 30-kDa band were detected in the wild-type hemocyte and hemolymph fractions, respectively (Figure 6A). These two bands were specific to the wild-type samples and were not detected in the PGRP-LE112 samples. In the control experiment, the nuclear protein Hrp48 was detected mainly in the hemocyte fraction, and was observed in both the wild-type and PGRP-LE112 samples (Figure 6B). Because the proPO cascade, a serine protease cascade, is activated spontaneously during the collection of hemolymph (Takehana et al, 2002), PGRP-LE was probably cleaved to the 30-kDa product in the hemolymph during preparation of the samples. These results indicate that endogenous PGRP-LE is a constitutive hemolymph protein.
We then confirmed the non-cell autonomous effects of PGRP-LE on the induction of antimicrobial peptide synthesis in the fat body. For this analysis, we applied a cell lineage tracer technique using a combination of the flp/FRT and GAL4/UAS recombinase systems (Ito et al, 1997). In this technique, the actin-promoter-GAL4 is nonfunctional due to the insertion of transcriptional termination signals with two FRT sequences in the initial state (Ay-GAL4). Flippase induces recombination at the two FRT sequences and removes the termination signal under the control of the hsp70 promoter, generating a functional Actin-GAL4 gene that drives the expression of PGRP-LE or imd under the control of UAS, which is monitored by UAS-GFP. Therefore, the PGRP-LE- or Imd-overexpressing cells are labeled with GFP in this system. Induction of the antimicrobial peptides was monitored by the expression of two reporter genes, Dpt-lacZ and Drs-lacZ, using an antibody against β-galactosidase. The double-labeling experiment revealed that Dpt-lacZ expression was not limited to the fat body cells overexpressing PGRP-LE (Figure 6E), whereas in the control experiments (Figure 6F), Dpt-lacZ expression was limited to the fat body cells overexpressing Imd, a cellular component of the imd pathway capable of inducing Dpt in the fat body (Georgel et al, 2001). These results indicate that PGRP-LE has non-cell autonomous effects on antibacterial peptide induction in systemic reactions.
A local epithelial reaction, activation of the Drs promoter in the trachea, is also induced by the overexpression of PGRP-LE (Takehana et al, 2002). The PGRP-LE-mediated expression of Drs-GFP in the trachea was abolished in an imd mutant background (Figure 6C and D). This result indicates that the PGRP-LE-mediated tracheal induction of Drs-GFP is imd dependent, consistent with the fact that induction of antimicrobial peptides in epithelial tissues depends on the imd pathway (Tzou et al, 2000). As PGRP-LE activates the epithelial reaction as well as the systemic reaction via the imd pathway, we examined whether PGRP-LE has a non-cell autonomous effect on the epithelial reaction using the same technique. In the trachea, Drs-lacZ expression was not limited to the cells overexpressing PGRP-LE (Figure 6G), whereas Drs-lacZ expression was limited to the cells overexpressing Imd (data not shown). These results indicate that PGRP-LE has a non-cell autonomous effect on the local epithelial reaction as well as on the systemic reaction.
Involvement of epithelial PGRP-LE in the epithelial immune reaction
PGRP-LE is present in the hemolymph and has non-cell autonomous effects on antimicrobial peptide synthesis in systemic and epithelial reactions. We investigated whether the epithelial Drs activation is mediated by the epithelial induction of PGRP-LE (Figure 7). As reported before, hs-GAL4-mediated ubiquitous expression of PGRP-LE activates Dpt-lacZ and Drs-GFP in the fat body and trachea, respectively (Takehana et al, 2002). The c564-GAL4-mediated induction of PGRP-LE activated Dpt-lacZ expression in the fat body, but did not activate Drs-GFP in the trachea. In the reciprocal experiments using NP2610-GAL4 as a driver that expresses GAL4 in the trachea, malpighian tubule, and gut but not in the fat body (Hayashi et al, 2002), Drs-GFP was activated in the trachea, whereas Dpt-lacZ was not activated in the fat body. These results suggest that the epithelial induction of PGRP-LE is sufficient to activate the epithelial response but not the systemic response.
Figure 7.
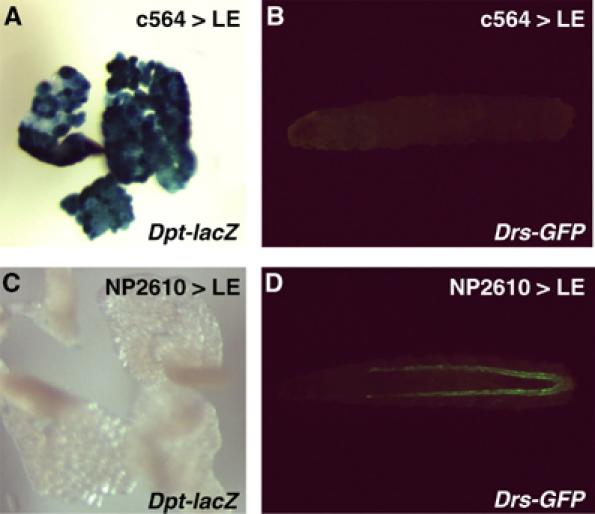
The epithelial induction of PGRP-LE is sufficient to activate the epithelial response. (A, B) Expression of Dpt-lacZ in the fat body (A) and Drs-GFP (B) in Dpt-lacZ, Drs-GFP; c564-GAL4/UAS-PGRP-LE larvae. (C, D) Expression of Dpt-lacZ in the fat body (C) and Drs-GFP (D) in Dpt-lacZ, Drs-GFP; NP2610-GAL4/UAS-PGRP-LE larvae.
Because it was suggested that there is a role for epithelial PGRP-LE in the epithelial immune reaction, we investigated whether PGRP-LE was present in the trachea by Western blotting (Figure 8G). The 45-kDa band was detected in the trachea of wild-type larvae but not in that of PGRP-LE112, indicating the presence of PGRP-LE in the trachea. The 45-kDa band was also detected in the wild-type fat body but not in the PGRP-LE112 fat body. Because the affinity-purified antibody against PGRP-LE crossreacted with three proteins that are present in the PGRP-LE112 trachea, the antibody was absorbed with PGRP-LE112 larval tissues. Immunostaining using absorbed antibody revealed the localization of PGRP-LE at the luminal surface of the trachea (Figure 8A–D). The trachea is a respiratory organ with a tube-like structure that can be visualized by DAPI nuclear staining. The anti-PGRP-LE antibody signal was detected at the surface of the wild-type trachea lumen. The signal was not detected at the surface of the tracheal lumen of PGRP-LE112, indicating the specificity of the PGRP-LE signal. Confocal microscopic analysis confirmed this observation (Figure 8E and F). These results imply that PGRP-LE recognizes invading bacteria and activates subsequent signaling at the luminal surface of the trachea, which is the first site of contact between the pathogens and the host.
Discussion
In the present paper, we report that two members of the PGRP-family, PGRP-LE and PGRP-LC, function synergistically to induce resistance to E. coli and B. megaterium infections. Consistent with these findings, PGRP-LE and PGRP-LC are two major upstream regulators of the imd pathway in response to E. coli infection, and PGRP-LE acts both in parallel and upstream of PGRP-LC in imd-mediated antibacterial peptide induction. Moreover, survival experiments suggest that PGRP-LE participates in host defense against E. coli, mainly through regulating the imd pathway of antimicrobial peptide induction. There is, however, no obvious synergy between PGRP-LE and PGRP-LC in the induction of the antibacterial peptides in response to E. coli infection, although, in some contexts such as natural infection, PGRP-LE and PGRP-LC synergistically induce antimicrobial peptides. A possible reason that the double PGRP-LE112/PGRP-LC7454 mutant has reduced resistance and PGRP-LC7454 has complete resistance to E. coli infection, although both mutants have similar antimicrobial peptide induction patterns, is that PGRP-LE112 lacks the infection-dependent proPO cascade activation. The lack of both antimicrobial peptide induction and proPO cascade activation probably leads to susceptibility to E. coli infection. Synergy between two pattern recognition molecules in Drosophila probably reflects the nature of innate immune systems in that, in innate immunity, pattern recognition molecules with a different pattern specificity, such as PGRP family members, are recruited to expand the immune repertoire and to establish an effective defense network with limited factors in response to new challenges during evolution.
Activation of the proPO cascade is the most immediate response following microbial infection or septic injury, and it produces cytotoxic reactive oxygen species and melanin by the catalytic conversion of dopamine into melanin. Biochemical in vitro reconstitution experiments indicate that the proPO cascade is triggered by the recognition of microbial cell wall components through pattern recognition molecules. We provide the first in vivo evidence that a pattern recognition molecule is required for bacterial infection-dependent activation of the proPO cascade. The lack of infection-dependent activation of the proPO cascade in PGRP-LE112 does not affect the survival of the host against E. coli infection. These results suggest that the proPO cascade is activated immediately after microbial infection and has an important role in the initial stage of host defense; however, for survival of the host extending to several days after infection, secondary responses that are mediated by transcriptional activation, such as antimicrobial peptide induction, are important.
The proPO cascade is regulated by Serpin27A, which is linked to the Toll pathway (De Gregorio et al, 2002a; Ligoxygakis et al, 2002b). PGRP-LE-mediated activation of the proPO cascade is suggested to be independent of Toll activation, because PGRP-LE overexpression induces proPO cascade activation and melanization in the spätzlerm7 mutant background. The spätzlerm7 is not a null allele, but a strong allele in which activation of the Toll pathway is severely affected (Lemaitre et al, 1996; De Gregorio et al, 2002b). Moreover, PGRP-LE activates the proPO cascade upstream of Serpin27A. Therefore, PGRP-LE participates in the primary activation of the proPO cascade upstream of Serpin27A and not in the secondary regulation, which depends on the regulation of Serpin27A by the Toll pathway.
Epistatic analyses suggest the existence of a cell surface receptor that acts downstream of PGRP-LE in the imd pathway in addition to PGRP-LC. This is consistent with the results that PGRP-LE is a constitutive hemolymph protein and has non-cell autonomous effects on activation of the antibacterial peptide gene. Because PGRP-LE-mediated activation of the antimicrobial peptide genes was reduced in the PGRP-LC mutant background, the putative receptor might make a functional complex with PGRP-LC, which is required for the full activation of PGRP-LE-mediated signaling. PGRP-LE overexpression had stronger activity to induce the Relish-dependent activation of Drs than that of PGRP-LCx. Thus, the putative receptor is also suggested to preferentially mediate the Relish-dependent activation of Drs. The identification of the putative receptor will provide further understanding of the details of signaling activation.
The epithelial induction of antimicrobial peptides is a highly conserved immune reaction observed in humans, insects, and plants, and is suggested to be the true ancestral antimicrobial defense (reviewed in Hoffmann and Reichhart, 2002). In contrast to the relatively good understanding of the systemic induction of antimicrobial peptides, the mechanisms of epithelial induction of the antimicrobial peptides are largely unknown in Drosophila. Thus, an understanding of the mechanisms of the epithelial responses will aid in the advancement of this field. In this paper, we demonstrated that the epithelial induction of PGRP-LE is sufficient to activate imd-mediated tracheal expression of Drs-GFP, but not to activate the systemic activation of antimicrobial peptide genes. Moreover, PGRP-LE is localized at the surface of the tracheal lumen and has non-cell autonomous effects on the induction of Drs-GFP in tracheal cells. Therefore, PGRP-LE is suggested to recognize invading bacteria and activate subsequent signaling at the front line of the epithelial barrier.
Materials and methods
Fly strains
Stocks were raised on a standard cornmeal-yeast agar medium at 25°C. Oregon R flies were used as a standard wild-type strain. UAS-LCx, PGRP-LC7454, PGRP-LCΔE, c564-GAL4, NP2610-GAL4, RelishE20, Ay-GAL4, imd1, spätzlerm7, J4, PGRP-SAseml, GS1068, UAS-PGRP-LE, UAS-Serpin27A, serpin27A1, and Bc1 are described elsewhere (Harrison et al, 1995; Lemaitre et al, 1995; 1996; Ito et al, 1997; Hedengren et al, 1999; Meng et al, 1999; Michel et al, 2001; Choe et al, 2002; Gottar et al, 2002; Hayashi et al, 2002; Takehana et al, 2002; De Gregorio et al, 2002a; Ligoxygakis et al, 2002b). The deletion of PGRP-LE was generated by P element mobilization of the GS1068 using standard protocols. To characterize the PGRP-LE112 mutation, genomic PCR was performed with two primers (LE loss F-1, 5′-CTGGCCCAACTCGCTTCAGT-3′, and LE loss R-2, 5′-GAGAGACGTCTGCGAGCTCT-3′, in Supplementary Figure 1A) after preparation of genomic DNA from adults, and the obtained PCR products were sequenced with a primer (PGRP-1, 5′-GTTCCTCCTCCTCGATATTG-3′; Supplementary Figure 1A).
Infection experiments
Bacterial infections were performed by challenging adult flies with a thin tungsten needle previously dipped into a concentrated culture of the bacterial strains. Survival experiments were performed with 30 flies for each genotype tested at 25°C. Surviving flies were transferred daily into fresh vials. E. coli K-12, E. carotovora carotovora 15, M. luteus (IFO13867), E. faecalis (IFO12964), B. subtilis (IFO3134), and B. megaterium (IFO13498) were used. For natural infection experiments, approximately 200 second instar larvae were placed into a 2-ml tube containing 200 μl of overnight cultured E. carotovora carotovora 15 pellet and 400 μl of crushed banana. The larvae, bacteria, and banana were mixed well, incubated at room temperature for 30 min, and transferred to standard fly medium. After incubation at 18°C for 5 days, observation of GFP expression was performed. Each experiment is representative of at least three independent experiments.
Measurements of PO activity
Hemolymph was collected 4 h after bacterial challenge from 50 flies for each genotype tested into 20 μl phosphate-buffered saline (PBS) containing protease inhibitors (Complete Cocktail, Roche, Germany) using glass capillaries with a handmade mouthpiece. Protein was determined by Protein Assay (Bio-Rad, Hercules, CA), using BSA as a standard. PO activity of 5-μl samples (ca 5 μg) was assayed as described previously (Takehana et al, 2002). For the rescue experiments, hemolymph was recovered from PGRP-LE112; UAS-PGRP-LE/+; hs-GAL4/+ flies at 12 h after heat shock (29°C, 2 h). Larval homogenate was prepared as described previously (Takehana et al, 2002). Each experiment is representative of at least three independent experiments.
Clonal analyses
For the clonal analyses, four kinds of trans-heterozygous larvae were used as follows: Dpt-lacZ/hs-flp; Ay-GAL4,UAS-GFP/UAS-PGRP-LE, Drs-lacZ/hs-flp; Ay-GAL4,UAS-GFP/UAS-PGRP-LE, Dpt-lacZ/hs-flp; Ay-GAL4,UAS-GFP/GS9049 and Drs-lacZ/hs-flp; Ay-GAL4,UAS-GFP/GS9049. In GS9049, the GAL4-dependent activation of imd is induced through a GS vector (unpublished data). Heat shock (37°C, 1 h) was applied twice at 24–48 h (the first instar larval stage) and at 48–62 h (the early second instar larval stage) after egg deposition.
Supplementary Material
Supplementary Material
Supplementary Figure 1
Acknowledgments
We are grateful to KV Anderson, D Ferrandon, Y Hiromi, JA Hoffmann, D Hultmark, K Ito, YT Ip, B Lemaitre, N Perrimon, J-M Reichhart, J Royet, the Bloomington Stock Center, Drosophila Genetic Resource Center in Kyoto Institute of Technology, and Genetic Strain Research Center of National Institute of Genetics for fly stocks and bacterial strains. We also thank M Ashida, S Iwanaga, and S Natori for helpful discussions and advice. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, the Japan Society for the Promotion of Science, the Program for promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN), and the Naito Foundation.
References
- Akira S, Takeda K, Kaisho T (2001) Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol 2: 675–680 [DOI] [PubMed] [Google Scholar]
- Ashida M, Brey PT (1998) Recent advances in research on the insect prophenoloxidase cascade. In Molecular Mechanisms of Immune Response in Insects, Brey PT, Hultmark D (eds) pp 135–172. London: Chapman and Hall [Google Scholar]
- Basset A, Khush RS, Braun A, Gardan L, Boccard F, Hoffmann JA, Lemaitre B (2000) The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc Natl Acad Sci USA 97: 3376–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV (2002) Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 296: 359–362 [DOI] [PubMed] [Google Scholar]
- De Gregorio E, Han SJ, Lee WJ, Baek MJ, Osaki T, Kawabata S, Lee BL, Iwanaga S, Lemaitre B, Brey PT (2002a) An immune-responsive Serpin regulates the melanization cascade in Drosophila. Dev Cell 3: 581–592 [DOI] [PubMed] [Google Scholar]
- De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B (2002b) The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J 21: 2568–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski R (2004) Peptidoglycan recognition proteins (PGRPs). Mol Immunol 40: 877–886 [DOI] [PubMed] [Google Scholar]
- Engström Y (1999) Induction and regulation of antimicrobial peptides in Drosophila. Dev Comp Immunol 23: 345–358 [DOI] [PubMed] [Google Scholar]
- Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, Kopczynski C, Duyk G, Reichhart JM, Hoffmann JA (2001) Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev Cell 4: 503–514 [DOI] [PubMed] [Google Scholar]
- Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J (2002) The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416: 640–644 [DOI] [PubMed] [Google Scholar]
- Hammond LE, Rudner DZ, Kanaar R, Rio DC (1997) Mutations in the hrp48 gene, which encodes a Drosophila heterogeneous nuclear ribonucleoprotein particle protein, cause lethality and developmental defects and affect P-element third-intron splicing in vivo. Mol Cell Biol 17: 7260–7267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DA, Binari R, Nahreini TS, Gilman M, Perrimon N (1995) Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J 14: 2857–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Ito K, Sado Y, Taniguchi M, Akimoto A, Takeuchi H, Aigaki T, Matsuzaki F, Nakagoshi H, Tanimura T, Ueda R, Uemura T, Yoshihara M, Goto S (2002) GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis 34: 58–61 [DOI] [PubMed] [Google Scholar]
- Hedengren M, Asling B, Dushay MS, Ando I, Ekengren S, Wihlborg M, Hultmark D (1999) Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol Cell 4: 827–837 [DOI] [PubMed] [Google Scholar]
- Hoffmann JA, Reichhart JM (2002) Drosophila innate immunity: an evolutionary perspective. Nat Immunol 3: 121–126 [DOI] [PubMed] [Google Scholar]
- Hultmark D (2003) Drosophila immunity: paths and patterns. Curr Opin Immunol 15: 12–19 [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D (1997) The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development 124: 761–771 [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart JM, Hoffmann JA (1995) A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc Natl Acad Sci USA 92: 9465–9469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA (1996) The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86: 973–983 [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Reichhart JM, Hoffmann JA (1997) Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci USA 94: 14614–14619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, Lee WJ, Mengin-Lecreulx D, Lemaitre B (2003) The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol 4: 478–484 [DOI] [PubMed] [Google Scholar]
- Levashina EA, Langley E, Green C, Gubb D, Ashburner M, Hoffmann JA, Reichhart JM (1999) Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science 285: 1917–1919 [DOI] [PubMed] [Google Scholar]
- Ligoxygakis P, Pelte N, Hoffmann JA, Reichhart JM (2002a) Activation of Drosophila Toll during fungal infection by a blood serine protease. Science 297: 114–116 [DOI] [PubMed] [Google Scholar]
- Ligoxygakis P, Pelte N, Ji C, Leclerc V, Duvic B, Belvin M, Jiang H, Hoffmann JA, Reichhart JM (2002b) A serpin mutant links Toll activation to melanization in the host defence of Drosophila. EMBO J 21: 6330–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA Jr (2002) Decoding the patterns of self and nonself by the innate immune system. Science 296: 298–300 [DOI] [PubMed] [Google Scholar]
- Meng X, Khanuja BS, Ip YT (1999) Toll receptor-mediated Drosophila immune response requires Dif, an NF-kappaB factor. Genes Dev 13: 792–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel T, Reichhart JM, Hoffmann JA, Royet J (2001) Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 414: 756–759 [DOI] [PubMed] [Google Scholar]
- Rämet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA (2002) Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416: 644–648 [DOI] [PubMed] [Google Scholar]
- Schleifer KH, Kandler O (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36: 407–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderhäll K, Cerenius L (1998) Role of the prophenoloxidase-activating system in invertebrate immunity. Curr Opin Immunol 10: 23–28 [DOI] [PubMed] [Google Scholar]
- Takehana A, Katsuyama T, Yano T, Oshima Y, Takada H, Aigaki T, Kurata S (2002) Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc Natl Acad Sci USA 99: 13705–13710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzou P, De Gregorio E, Lemaitre B (2002) How Drosophila combats microbial infection: a model to study innate immunity and host–pathogen interactions. Curr Opin Microbiol 5: 102–110 [DOI] [PubMed] [Google Scholar]
- Tzou P, Ohresser S, Ferrandon D, Capovilla M, Reichhart JM, Lemaitre B, Hoffmann JA, Imler JL (2000) Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity 13: 737–748 [DOI] [PubMed] [Google Scholar]
- Weber AN, Tauszig-Delamasure S, Hoffmann JA, Lelievre E, Gascan H, Ray KP, Morse MA, Imler JL, Gay NJ (2003) Binding of the Drosophila cytokine Spatzle to Toll is direct and establishes signaling. Nat Immunol 4: 794–800 [DOI] [PubMed] [Google Scholar]
- Werner T, Borge-Renberg K, Mellroth P, Steiner H, Hultmark D (2003) Functional diversity of the Drosophila PGRP-LC gene cluster in the response to lipopolysaccharide and peptidoglycan. J Biol Chem 278: 26319–26322 [DOI] [PubMed] [Google Scholar]
- Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D (2000) A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc Natl Acad Sci USA 97: 13772–13777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Kinoshita K, Ashida M (1996) Purification of a peptidoglycan recognition protein from hemolymph of the silkworm, Bombyx mori. J Biol Chem 271: 13854–13860 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplementary Figure 1


